Abstract
Probiotics are the most promising alternative to antibiotics for improving animal production and controlling pathogenic infections, while strains derived from natural hosts are considered highly desirable due to their good adaptation to the gastrointestinal tract. The aim of this study was to screen Lactobacillus with broad-spectrum antibacterial activity from broilers fed an antibiotic-free diet and evaluate their potential as poultry probiotics. A total of 44 lactic acid bacteria (LAB) strains were isolated from the intestines of healthy broilers, among which 3 strains exhibited outstanding antimicrobial activity and were subsequently identified through 16S rRNA sequencing as Enterococcus faecium L8, Lactiplantibacillus plantarum L10, and Limosilactobacillus reuteri H11. These three isolates demonstrated potent bacteriostatic activity against Staphylococcus aureus, Listeria monocytogenes, Escherichia coli, and Salmonella cholerae, with inhibition zones ranging from 15.67 ± 1.53 to 21.33 ± 0.58 mm. The selected LAB strains exhibited high tolerance to acid and bile salts, with L. reuteri H11 displaying the highest survival rate (ranging from 34.68% to 110.28%) after exposure to 0.3% (w/v) bile salts for 6 h or a low pH environment (pH 2, 2.5, and 3) for 3 h. Notably, L. reuteri H11 outperformed other strains in terms of hydrophobicity (84.31%), auto-aggregation (53.12%), and co−aggregation with E. coli ATCC 25922 (36.81%) and S. aureus ATCC 6538 (40.20%). In addition, the three LAB isolates were either fully or moderately susceptible to the tested antibiotics, except for strain L8, which resisted gentamycin and vancomycin. Consequently, these three LAB strains, especially L. reuteri H11, isolated from the intestines of broiler chickens, represent promising probiotic candidates that can be employed as feed additives to enhance production performance and control poultry pathogens.
1. Introduction
In recent decades, the breeding industry has witnessed the excessive and irrational use of antibiotics, significantly contributing to the growing threat of bacterial antimicrobial resistance. This issue poses risks to both animal and human health [1]. Recognizing this challenge, the European Union took a significant step by prohibiting antibiotics in animal feed in 2006 [2]. Similarly, China’s Ministry of Agriculture and Rural Affairs, through Notice No. 194, enforced a ban on commercial feed containing growth-promoting drug additives (except for Chinese herbal medicines), effective from 1 July 2020 [3]. However, the use of subtherapeutic antibiotics in the breeding industry is still permitted in some countries, like China, to combat bacterial diseases, which can provoke greater bacterial resistance [4,5]. As a result, researchers have been exploring safe, cost-effective, and efficient alternatives to ensure the long-term sustainability of the livestock and poultry industry [3]. Unquestionably, probiotics continue to be the most promising natural candidates [5]. In response, there has been a surge of interest in employing probiotics as an alternative to antibiotics in livestock and poultry production for disease prevention and enhanced growth performance [6].
Probiotics are non-pathogenic living microorganisms that confer health benefits when administered in appropriate quantities [7]. They are primarily sourced from dairy products, food, fermented products, animal and human gastrointestinal tracts, and so on [5,6,8]. Lactic acid bacteria (LAB), encompassing various species such as Enterococcus, Lactobacillus, Pediococcus, Streptococcus, Lactococcus, Vagococcus, Leuconostoc, Oenococcus, Weissella, Carnobacterium, and Tetragenococcus, have been extensively researched as probiotics over the years [9]. Extensive studies have demonstrated that LAB can adhere to and thrive in the gastrointestinal tract of animals by competitively inhibiting pathogen binding and producing antimicrobial substances like organic acids, bacteriocins, and hydrogen peroxide, thereby playing a stabilizing and protective role within this ecosystem [10,11]. Furthermore, LAB can influence digestive and metabolic processes, regulate immune responses and cytokine production, and enhance the integrity of the intestinal mucosal barrier through competitive exclusion [11,12,13]. These attributes contribute to improved animal health, enhancing production performance [14] and bolstering host immunity [15]. Nevertheless, the effectiveness of probiotics is contingent on the specific species and strains used [11]. Therefore, any potential probiotic candidate must meet the criteria of safety (non-pathogenic and susceptibility to antimicrobials), in vivo benefits (lactic acid production and antagonism against pathogens), and resilience within the host’s gastrointestinal environment (tolerance to acidic pH and high bile salt concentrations) [16]. Furthermore, an ideal candidate must display effective adhesion to the intestinal epithelium and establish a symbiotic colonization with the natural gut microbiota [17]. Consequently, rigorous selection criteria must be implemented to assess the probiotic properties of candidate strains [18].
Despite the development of a substantial number of commercially available probiotic strains in recent decades, there remains a pressing need for superior strains compared to the existing ones [6]. Moreover, the viability and inconsistent results of probiotic strains in many cases hinder their widespread use [5]. By comparison, isolating probiotic strains from their natural hosts is highly desirable, as such native strains are already adapted to the gastrointestinal tract. Consequently, when administered to animals, they exhibit better adherence and survivability, enabling them to exert the expected beneficial effects more effectively than strains from other sources [19]. Thus, developing host-specific probiotic strains is imperative to maximize health benefits and enhance animal production performance [20]. Accordingly, we isolated LAB strains from the intestinal tracts of healthy broilers fed an antibiotic-free diet rich in prebiotics. We evaluated their probiotic characteristics in vitro and screened potential probiotic isolates for application in poultry farming.
2. Materials and Methods
2.1. Isolation of LAB Strains
LAB strains were isolated from healthy broilers on antibiotic-free chicken farms. Briefly, the birds were humanely euthanized by CO2 inhalation followed by cervical dislocation. The intestinal tracts were aseptically excised and promptly transported to the laboratory for microbial isolation. After removing the intestinal contents, the mucosa of each intestinal segment was meticulously scraped into 40 mL of sterile normal saline and homogenized for 30 min at 37 °C with continuous shaking. The samples were appropriately diluted, and 0.1 mL of the diluted suspension was evenly spread onto de Man, Rogosa, Sharpe (MRS) (AOBOX, Beijing, China) screening plates containing 0.004% (w/v) bromocresol purple (Sigma-Aldrich, St. Louis, MO, USA) and 2% (w/v) CaCO3 (Sigma-Aldrich, St. Louis, MO, USA). Subsequently, the plates were incubated for 48 h at 37 °C under anaerobic conditions. Only colonies displaying a yellow zone and a calcium-dissolving zone were selected (presumed to be LAB) and subjected to three successive transfers on MRS agar plates using the dilution-streaking method to obtain individual colonies.
The presumed LAB isolates were then subjected to further characterization, including cell morphology assessment, Gram staining, and catalase reaction. Isolates that were catalase-negative and Gram-positive were chosen and preserved at −80 °C in MRS broth with 20% glycerol (Sigma-Aldrich, St. Louis, MO, USA). Before each use, these isolates were resuscitated through sub-culturing in MRS broth.
2.2. Antimicrobial Activity
To evaluate the antimicrobial activity of the cell-free supernatant (CFS) of the selected LAB against pathogens, the Oxford cup agar diffusion method [21] was employed. In brief, the LAB isolates were cultured in MRS broth at 37 °C for 24 h, and the CFS was obtained by centrifugation at 5867× g for 10 min, followed by filtration sterilization using a sterilized 0.22 µm MILLEX®GP filter unit (PES membrane) (Merck Millipore, St. Louis, MO, USA). Subsequently, 200 μL of the overnight cultures of indicator pathogens (107 CFU/mL) was evenly spread onto the surface of nutrient agar plates. Sterile Oxford cups (outer diameter of about 8.0 mm) (Shanghai Jinpan Biotech Co., Ltd., Shanghai, China) were placed on the inoculated plates and filled with 100 μL of LAB CFS. Following incubation at 37 °C for 18–24 h, the inhibition zones (including disc size) of the LAB were measured by using a pair of Vernier calipers. The indicator pathogens included Staphylococcus aureus ATCC 6538, Listeria monocytogenes ATCC 10403S, Escherichia coli ATCC 25922, and Salmonella cholerae ATCC 13312. LAB strains with noteworthy antimicrobial activity were selected based on comprehensive cluster analysis for further investigations.
2.3. Characterization of LAB Antimicrobial Compounds
The LAB isolates with higher antimicrobial activity were selected and tested to determine the nature of the produced antimicrobial substances—primarily organic acids, hydrogen peroxides, and bacteriocins. This characterization followed the method described by Reuben et al. [19] with some modifications. Specifically, the CFS of LAB, as prepared previously, was separated into five portions that underwent different treatments: one remained untreated as a control, the second was adjusted to a pH of 5.0 using NaOH, the third was treated with 1.5 mg/mL catalase (Sangon Biotech Co., Ltd., Shanghai, China) at 37 °C for 2 h, the fourth was treated with 1 mg/mL proteinase K (Sigma-Aldrich, St. Louis, MO, USA) at 37 °C for 2 h, and the fifth was heat-treated (boiled) for 20 min. Exactly 100 μL of CFS from each treatment was loaded into sterile Oxford cups placed on the agar plates previously inoculated with 200 μL of indicator pathogens (107 CFU/mL). These indicator pathogens included E. coli ATCC 25922 (representative of Gram-negative bacteria [G-]) and S. aureus ATCC 6538 (representative of Gram-positive bacteria [G+]). The plates were then incubated at 37 °C for 18–24 h, and the inhibition zones (including disc size) of the LAB were measured by using a pair of Vernier calipers.
2.4. Acid and Bile Tolerance Tests
The acid tolerance of the LAB isolates was assessed following the method described by Wang et al. [22], with some modifications. Overnight LAB cultures were resuspended in MRS broth at pH levels of 2.0, 2.5, and 3.0 after centrifugation and then adjusted to a concentration of 107 CFU/mL. The suspensions were subsequently incubated for 3 h at 37 °C, with samples collected at different intervals (0, 1, 2, and 3 h). These samples were serially diluted with PBS (Sangon Biotech Co., Ltd., Shanghai, China) and plated on MRS agar plates. After 48 h of incubation at 37 °C, the viable colonies were counted.
To evaluate bile salt tolerance, overnight LAB cultures were washed twice with sterile PBS at pH 7.2 after centrifugation. Subsequently, the cell pellets were resuspended in fresh MRS broth containing 0.3% and 0.5% bile salt (Solarbio, Beijing, China) and adjusted to a concentration of 107 CFU/mL. The cell suspensions were then incubated for 3 h at 37 °C, with samples taken at two different intervals. The gradient-diluted samples were spread on MRS agar plates and incubated at 37 °C for 48 h, after which the viable cells were counted.
2.5. Cell Surface Characteristics
The cell surface characteristics of LAB isolates, including hydrophobicity, auto-aggregation, and co-aggregation with pathogens, were determined as previously described [22] with some modifications. The cell pellets of LAB and pathogens (E. coli ATCC 25922 and S. aureus ATCC 6538) were washed twice and resuspended with sterile PBS. The cell suspensions were adjusted to an optical density at 600 nm (OD600) of 0.5 ± 0.05 (Ai).
2.5.1. Auto-Aggregation
Then, 5 mL of LAB cell suspension was vortexed for 10 s and then incubated for 4 h at 37 °C. The absorbance of the supernatant was read at 600 nm (At). The auto-aggregation rate was calculated as follows:
where Ai was the initial OD600 of LAB suspension, and At was OD600 after 4 h of incubation.
Auto-aggregation (%) = [1 − (At/Ai)] × 100
2.5.2. Co-Aggregation
To assess co-aggregation, 2 mL of each LAB cell suspension (As) was mixed with each pathogen suspension (Ap) in equal volume, vortexed for 30 s, and subsequently incubated for 4 h at 37 °C. The absorbance of the supernatant of each mixed suspension was measured at 600 nm (Am). The co-aggregation rate was calculated according to the formula below:
where As and Ap were initial OD600 of LAB and pathogenic bacteria and Am was OD600 of the mixed suspension after 4 h of incubation.
Co-aggregation (%) = [1 − Am/(As + Ap)/2] × 100
2.5.3. Cell Surface Hydrophobicity
Exactly 3 mL of LAB cell suspension was mixed with 1 mL of o-xylene (Sigma, USA) and then vortexed for 1 min. The mixture was stood at 37 °C for 20 min and separated into two phases. The aqueous phase was collected, and the OD600 was measured (At). The cell surface hydrophobicity was calculated as follows:
where Ai was the initial OD600 of LAB and At was OD600 of the aqueous phase after 20 min of incubation.
Hydrophobicity (%) = (1 − At/Ai) × 100
2.6. Safety Evaluation in Vitro of LAB Strains
2.6.1. Hemolytic Activity Assay
The LAB strains were inoculated on Columbia blood agar (Sangon Biotech Co., Ltd., Shanghai, China) and incubated for 24 h at 37 °C for hemolysis [23]. The hemolytic activity of the strain was shown by β-hemolysis (a clear, colorless/lightened yellow zone around the colony). In contrast, hemolytic inactivity was manifested as α-hemolysis (a zone of greenish to brownish discoloration surrounding the colony) and γ-hemolysis (no change).
2.6.2. Antibiotic Susceptibility Test
The antibiotic susceptibilities of the LAB isolates were evaluated through the disc diffusion method [24]. To elaborate, the overnight cultures of the LAB strains designated for testing were adjusted to achieve a concentration of 107 CFU/mL, and then they were evenly spread onto MRS agar plates. Subsequently, ten types of commercially available antibiotic discs (Hangzhou Microbial Reagent Co., Ltd., Hangzhou, China) were aseptically placed on the surface of the MRS agar. These antibiotics included chloramphenicol (30 μg), erythromycin (15 μg), tetracycline (30 μg), gentamicin (10 μg), ampicillin (10 μg), vancomycin (30 μg), streptomycin (10 μg), rifampicin (5 μg), kanamycin (30 μg), and novobiocin (30 μg). Following an incubation period of 24 h at 37 °C, the diameters (mm, including disc size) of the clear zones surrounding each antibiotic disc were measured by using a pair of Vernier calipers. The LAB isolates were classified as either resistant (≤15 mm), intermediately sensitive (16–20 mm), or sensitive (≥21 mm) in accordance with previously established criteria [25].
2.7. Molecular Identification of LAB Strains
The genomic DNA of the LAB isolates was extracted using a bacterial DNA extraction kit (Tiangen Biotech, Beijing, China) for molecular identification. The sequences of 16S rRNA were amplified using universal primers 27F and 1492R, as previously described [26]. The PCR products were sequenced by Sangon Biotech Co., Ltd. (Shanghai, China). The sequence data obtained were subjected to online homology analysis using NCBI BLASTn for the final identification of LAB.
2.8. Statistical Analysis
Statistical analyses were conducted using IBM SPSS Statistics version 27.0 software (IBM Corporation, Armonk, NY, USA). One-way ANOVA was employed to determine the significance of differences among the mean values. When necessary, Duncan’s multiple range test was used to identify differences between means. A significance level of p < 0.05 was considered statistically significant. All data were expressed as the mean values ± standard deviations (SDs) derived from triplicate samples. Cluster analysis was conducted using heatmap tools available at Hiplot Pro (https://hiplot.com.cn/, accessed on 30 May 2023), a comprehensive web service for analyzing and visualizing biomedical data, to classify the antimicrobial activities of the LAB isolates against pathogens.
3. Results
3.1. Isolation of LAB Strains
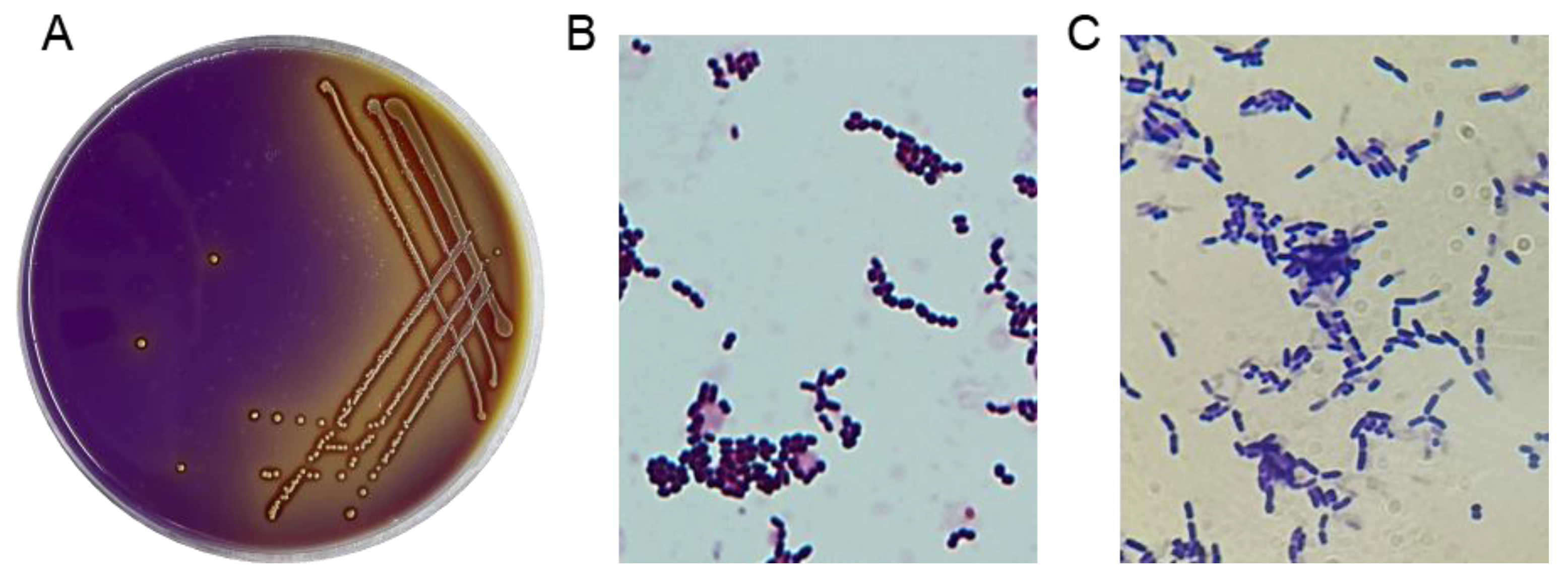
A total of 44 potential LAB strains were isolated from the intestinal tracts of healthy broilers based on the distinct calcium-dissolving zone and discoloration reaction (from purple to yellow) around the colonies on MRS screening plates (Figure 1A). All 44 isolates were catalase-negative and Gram-positive, displaying typical morphological features such as creamy-white and fruity colonies and coccus- or rod-shaped characteristics (Figure 1B,C), which enabled their morphological classification as LAB.
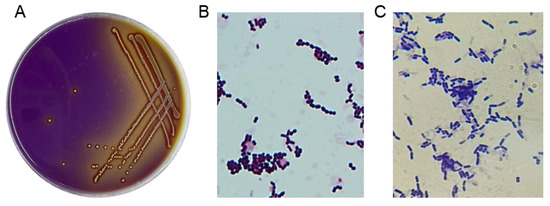
Figure 1.
Isolation of LAB strains. (A) LAB strains were visually screened using MRS agar plates with bromocresol blue and CaCO3. (B,C) The results of the gram staining test on the suspected LAB isolates (1000×).
3.2. Antimicrobial Activity In Vitro
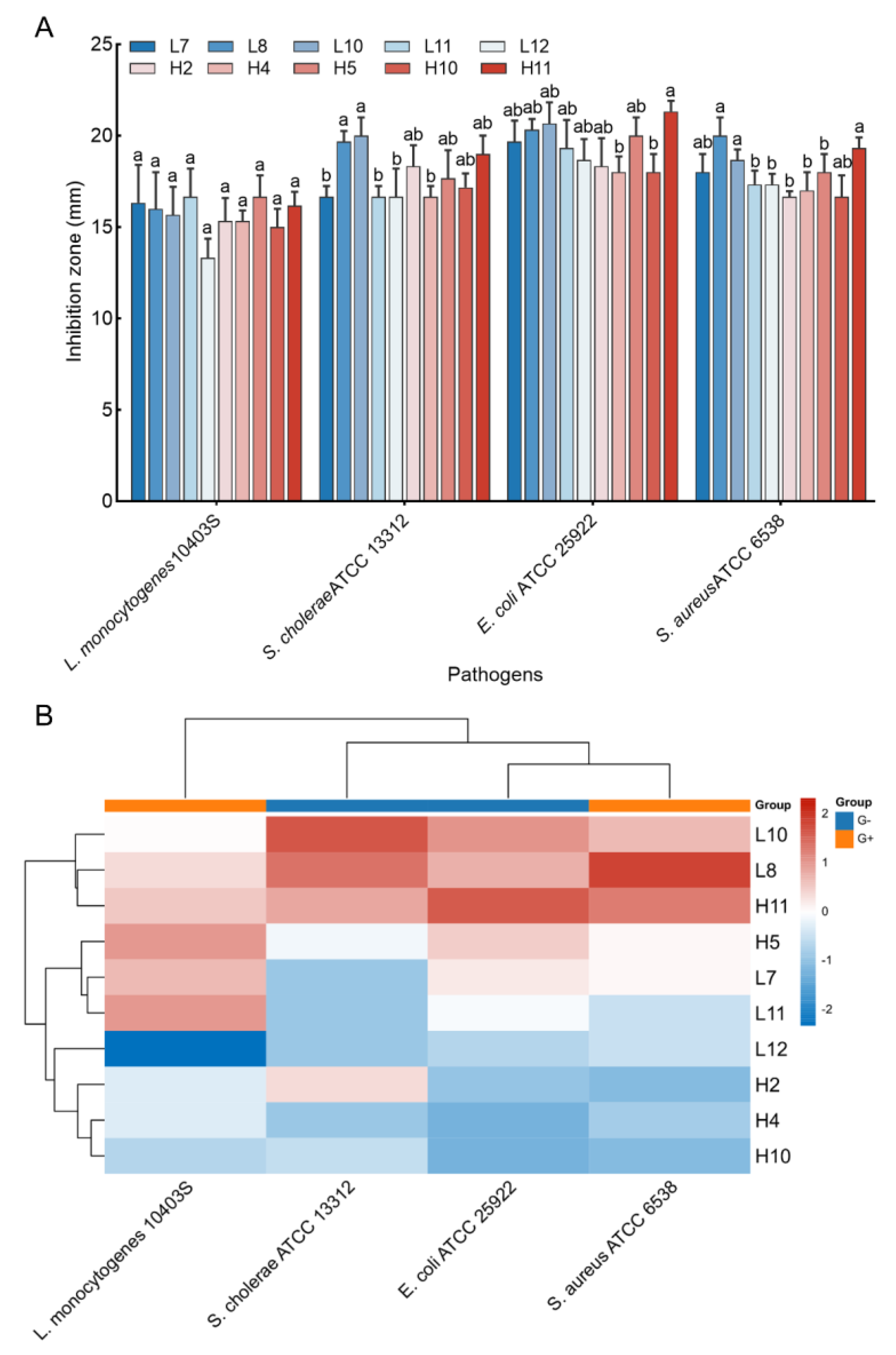
Antimicrobial activity against four pathogen indicators (S. aureus ATCC 6538, L. monocytogenes ATCC 10403S, E. coli ATCC 25922, and S. cholerae ATCC 13312) was assessed for the 44 LAB isolates. Results indicated that only 10 LAB isolates displayed significant inhibition activities against all tested pathogens, with inhibition zones ranging from 13.33 ± 1.04 to 21.33 ± 0.58 mm (Figure 2A). Notably, H5 and L11 exhibited the largest inhibition zone against L. monocytogenes ATCC 10403S (16.67 mm), while L8 and L10 yielded the most significant inhibition zone against S. cholerae ATCC 13312 (19.67 and 20.00 mm, respectively, p > 0.05). H11 exhibited the largest inhibition zone against E. coli ATCC 25922 (21.33 mm), and L8, H11, and L10 yielded larger inhibition zones against S. aureus ATCC 6538 (20.00, 19.33, and 18.67 mm, respectively, p > 0.05). Cluster analysis using the heatmap method demonstrated that three LAB strains (L10, L8, and H11) exhibited favorable antimicrobial activity against all tested pathogens (Figure 2B), leading to their selection for further analyses.
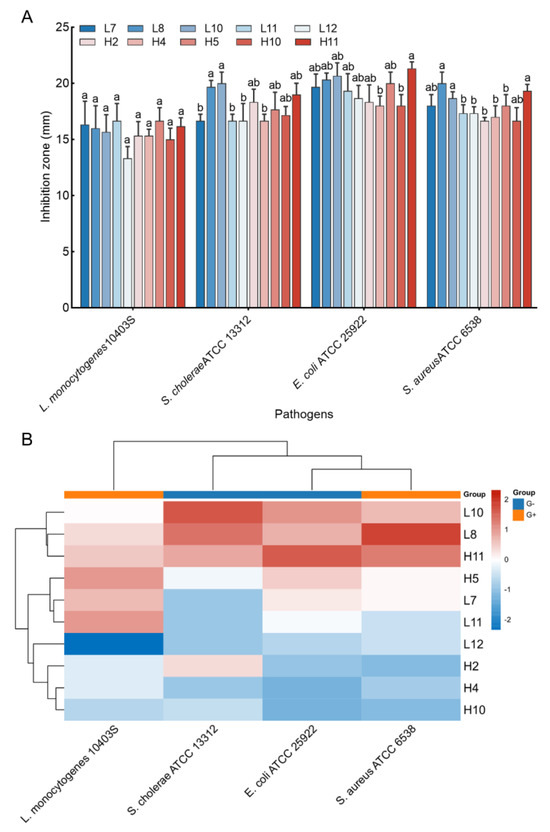
Figure 2.
Antimicrobial activity of the LAB strains against pathogens. (A) Inhibition zones of the CFS of the strains against four pathogen indicators. The results represent the mean ± SD of three replicates. For each pathogen indicator, different letters on each bar represent significant differences between values (p < 0.05). (B) Cluster analysis was undertaken to comprehensively assess the antimicrobial activities of LAB isolates against pathogens by using heat map tools in Hiplot Pro (https://hiplot.com.cn/, accessed on 30 May 2023).
3.3. Characterization of LAB Antimicrobial Compounds Produced by Strains
The selected LAB strains underwent characterization to produce antimicrobial compounds, including organic acids, hydrogen peroxide, and bacteriocins. The CFS of LAB strains, both before and after catalase treatment, exhibited nearly the same inhibitory activity against all tested pathogens, suggesting that the inhibition was not attributed to the production of hydrogen peroxides. However, the inhibitory effect of CFS after protease K treatment was significantly reduced, especially for strains H11 and L10, implying that the antimicrobial compounds are proteinaceous. Heat treatment did not affect the inhibitory activities of the CFS of LAB strains tested, indicating that the antimicrobial compounds produced by these strains were heat-stable. Furthermore, when the pH of the CFS of different LAB cultures was increased (pH 5.0), the antimicrobial activity dramatically decreased compared to the untreated CFS (Table 1). This effect disappeared when the CFS was neutralized at pH 5.5, suggesting that acid production significantly contributed to the inhibitory effect of these LAB strains, including the action of bacteriocins.

Table 1.
Characterization of antimicrobial compounds from three LAB isolates.
3.4. Acid and Bile Tolerance Ability of LAB Strains
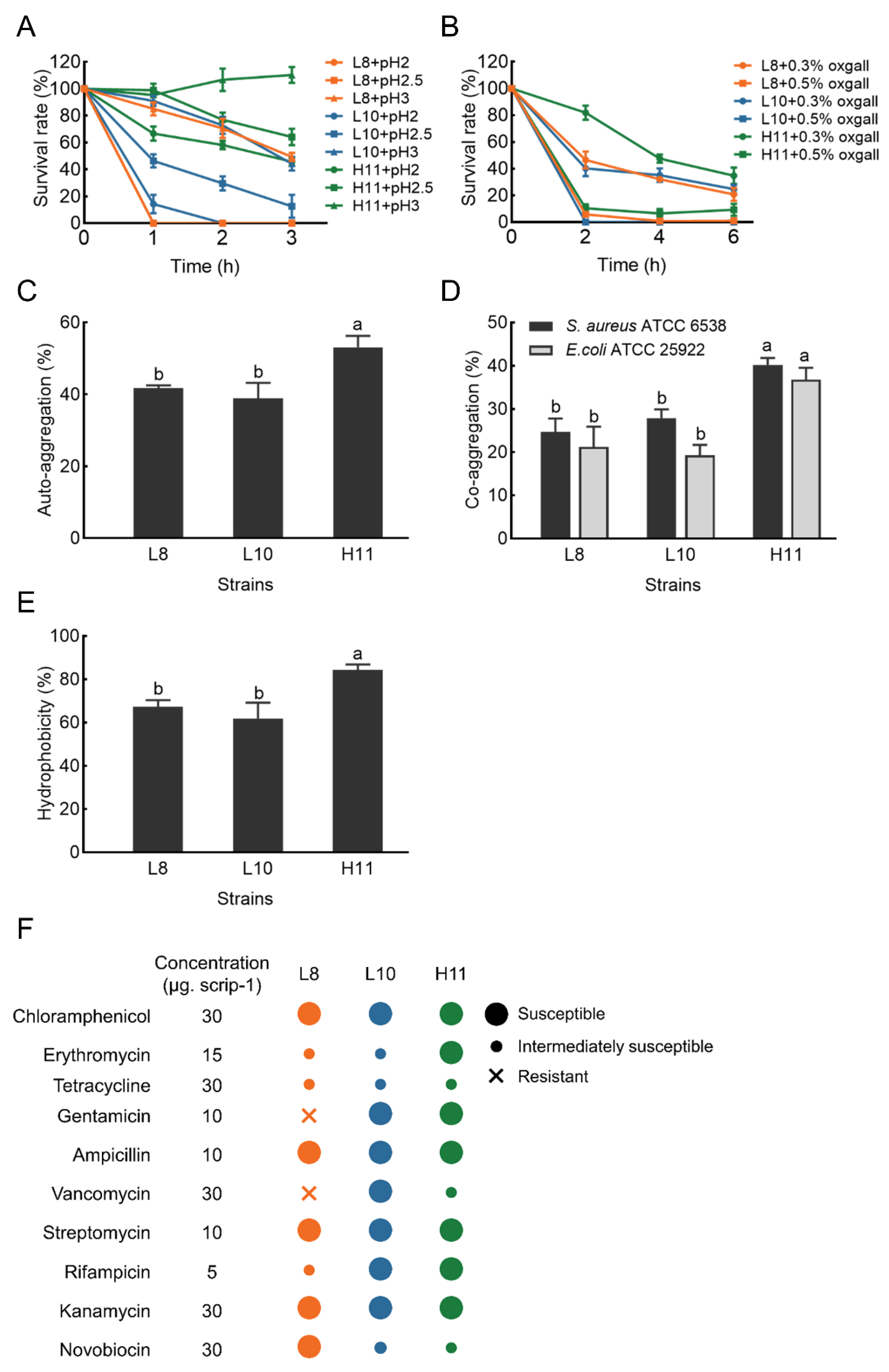
The acid tolerance ability of the three selected isolates is shown in Figure 3A. H11 yielded the highest survival rate after 3 h of incubation in an acidic environment (pH 2.0–3.0), with values ranging from 45% to 110%. L8 and L10 demonstrated moderate ability to survive at pH 3.0 after 3 h of incubation (49% and 44%, respectively). However, the survival rate of L8 and L10 was very low at pH < 3.0, particularly at pH 2.0.
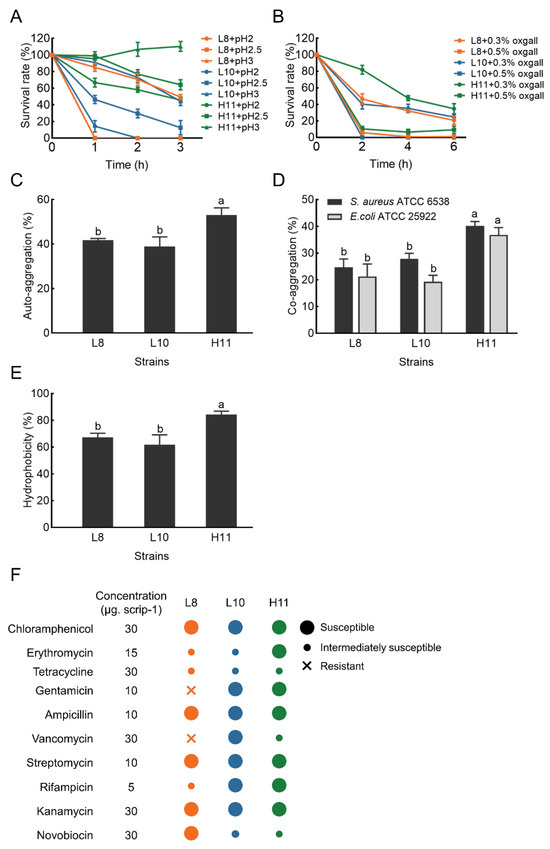
Figure 3.
The probiotic characteristics of strains L8, L10 and H11. (A) Acid tolerance; (B) bile tolerance; (C) Auto−aggregation; (D) Co−aggregation; (E) Hydrophobicity; (F) Antibiotic resistance dashboard. The results represent the mean ± SD of three replicates. Different letters on each bar represent significant differences between values (p < 0.05).
Simultaneously, H11 exhibited excellent tolerance in the presence of 0.3% bile salt and maintained a survival rate ranging from 35% to 81% after 2.0–6.0 h of incubation. Under the same conditions, L8 and L10 exhibited moderate bile salt tolerance, with a survival rate of 24−46%. However, the survival rate of the three LAB strains was less than 10% in the presence of 0.5% bile salt (Figure 3B).
3.5. Cell Surface Characteristics
3.5.1. Auto-Aggregation and Co-Aggregation Ability
Auto-aggregation results of the selected isolates are displayed in Figure 3C. The auto−aggregation of these isolates ranged from 38.90 ± 4.31 to 53.12 ± 3.14%. After 4 h of incubation, the auto-aggregation of strain H11 was significantly higher (p < 0.05) than strains L8 and L10.
Co−aggregation results of these LAB strains with two pathogens (E. coli ATCC 25922 and S. aureus ATCC 6538) are shown in Figure 3D. All three LAB strains exhibited stronger co−aggregation ability with S. aureus ATCC 6538 compared to E. coli ATCC 25922. The co−aggregation of strain H11 with pathogens was 40.20% (S. aureus ATCC 6538) and 36.81% (E. coli ATCC 25922), respectively. H11 demonstrated significantly higher co−aggregation ability with the two pathogens than L8 and L10 (p < 0.05).
3.5.2. Cell Surface Hydrophobicity
The cell surface hydrophobicity results of the three LAB strains are presented in Figure 3E. The hydrophobicity value of strain H11 was 84.31% in xylene, significantly higher than that of L8 (67.20%) and L10 (61.81%) (p < 0.05).
3.6. Safety Evaluation In Vitro of LAB Strains
The hemolytic activity of the three strains was examined based on the presence of hemolytic halos around the colonies on blood agar plates. None of the three strains exhibited a zone of hemolysis or a zone of greenish to brownish discoloration, indicative of γ−hemolysis (non−hemolytic activity). Figure 3F demonstrates the susceptibility profile of the three LAB isolates to several commonly used antibiotics, confirming their susceptibility to most antibiotics. Specifically, all three LAB isolates were susceptible to chloramphenicol, penicillin, ampicillin, streptomycin, and kanamycin. Two (L10 and H11) were susceptible to gentamicin and rifampicin, while one was susceptible to erythromycin (H11), vancomycin (L10), and novobiocin (L8). Nevertheless, strain L8 was resistant to gentamicin and vancomycin.
3.7. Molecular Identification of LAB Strains
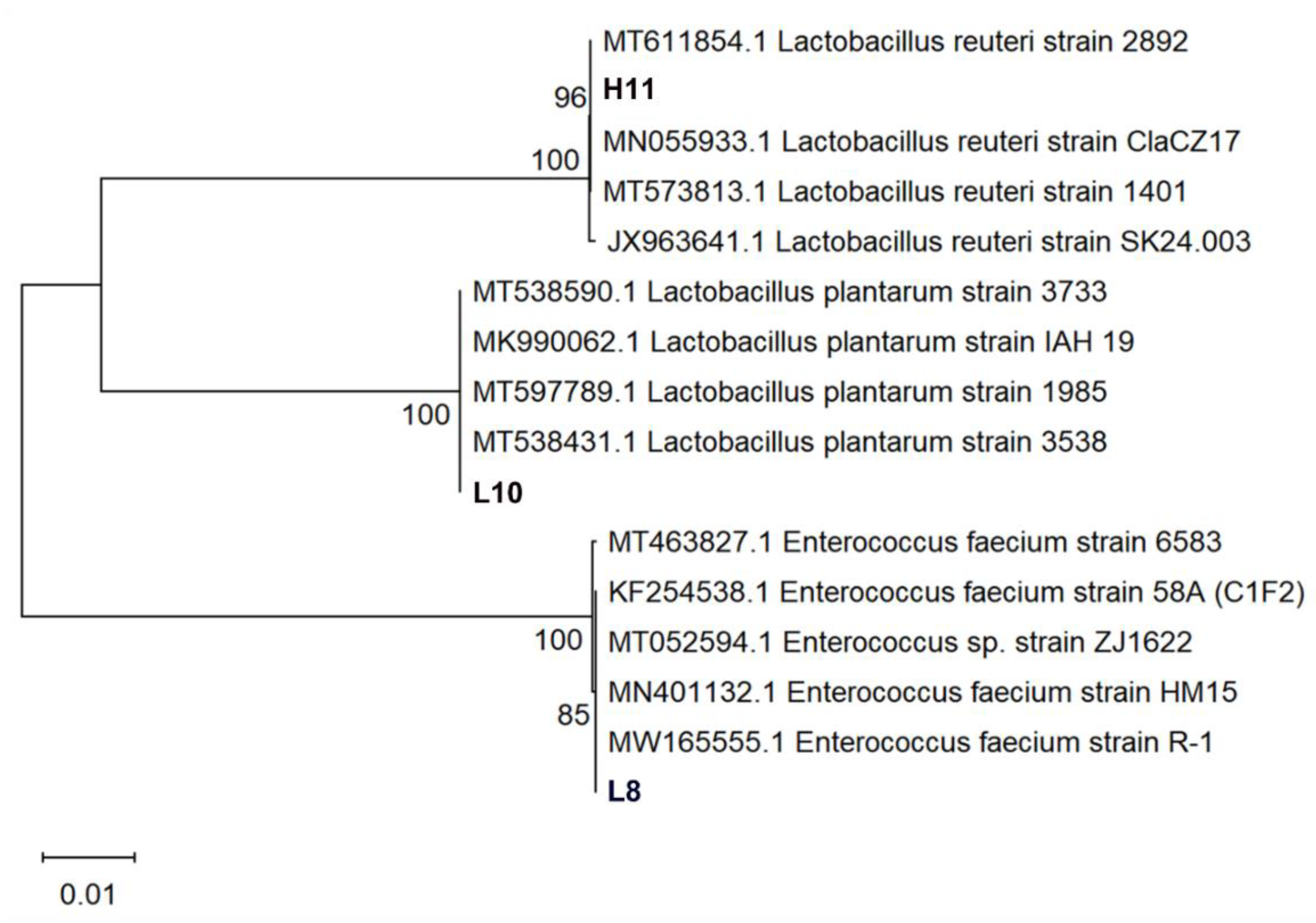
Three LAB strains were identified through 16S rRNA sequencing. Phylogenetic analyses of 16S rDNA sequences of the isolates revealed that strain L8 clustered into the clade of Enterococcus faecium (E. faecium), indicating a genetic homology of 99.72% with E. faecium strain HM15 (Accession No.:MN401132). Isolate L10 belonged to the cluster of Lactiplantibacillus plantarum (L. plantarum) with 99.66% genetic homology to L. plantarum IAH 19 (Accession No.: MK990062). Strain H11 was classified in the cluster of Limosilactobacillus reuteri (L. reuteri) with 99.59% genetic homology to L. reuteri strain ClaCZ17 (Accession No.: MN055933) isolated from chickens (Figure 4). The three LAB isolates were identified as E. faecium L8, L. plantarum L10, and L. reuteri H11. These lactobacilli are registered on the GenBank website (https://www.ncbi.nlm.nih.gov/genbank/, accessed on 30 November 2023.) with the accession numbers: OR879323 (L8), OR879324 (H11), and OR879325 (L10).
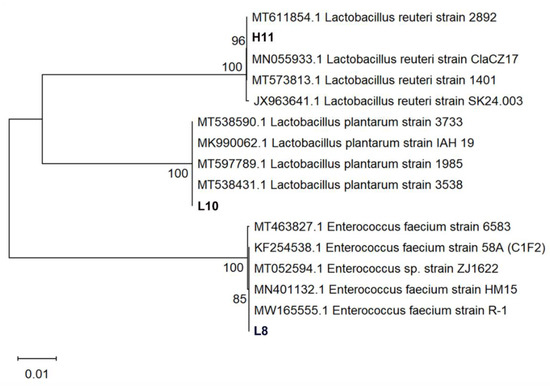
Figure 4.
Phylogenetic analysis of strains L8, L10, and H11 based on 16S rRNA gene sequences. The isolates sequenced in the study are depicted in bold font. The tree was constructed with the neighbor-joining method by using MEGA 11. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. The evolutionary distances were computed using the Kimura two-parameter method and are in the units of the number of base substitutions per site.
4. Discussion
In recent years, probiotics have attracted significant interest and are now considered a promising alternative to the widespread use of antibiotics in the poultry industry. Most probiotics are sourced from the microbial communities found in the intestines of animals and humans and in dairy products [8]. Typically, probiotic strains derived from their natural hosts are preferred over strains from other sources because of their increased host adaptation, higher viability, greater safety, and more effective delivery of probiotic effects [19]. Moreover, since the antagonistic effect of probiotics against pathogens is the main influential factor that hinders the colonization of heterochthonous bacteria in the GI tract, the use of indigenous microorganisms as probiotics is expected to make an important contribution to the control of pathogens [27]. Similarly, Jose et al. [28] stated that LAB strains from animal rumen yielded a better inhibitory effect on pathogen growth than those from dairy products. Therefore, developing host-specific probiotics is essential to facilitate animal health and optimal performance [20]. To this end, LAB strains were selectively isolated from healthy broilers, and their potential for development as poultry probiotics was evaluated in this work. Bromocresol purple and CaCO3 (as pH indicators) were supplemented into the MRS screening plates for visually selecting LAB strains. The emergence of a yellow area and a calcium-dissolving zone around a bacterial colony on the MRS screening plates indicated that the bacterium could produce acid or make the environment acidic [29]. After isolating 44 strains of LAB from the intestinal tracts of broilers, they were subjected to in vitro evaluation of probiotic properties and final molecular characterization.
It is widely acknowledged that infections caused by zoonotic and food−borne enteric pathogens can lead to high morbidity and mortality, ultimately resulting in significant economic losses in the poultry industry [30]. Thus, antimicrobial activity against these pathogens is a major requirement for selecting potential probiotics [5]. In this study, 10 of 44 LAB strains showed broad-spectrum antimicrobial activity against the four pathogens tested (Figure 2A). Moreover, these LAB strains had comparable antimicrobial activity against Gram−negative and Gram-positive bacteria, but the least-inhibitory zones were observed against Listeria monocytogenes. Our findings are similar to many previous studies that reported antimicrobial activity by LAB strains sourced from poultry against broad-spectrum pathogens [5,11,19,31]. Conversely, some researchers revealed that LABs isolated from the chicken gastrointestinal tract have high inhibitory activity against Gram-positive pathogens (Clostridium perfringens) but low inhibitory activity against Gram-negative pathogens (including Escherichia coli and Salmonella enteritidis) [32]. However, when the inhibitory activity of LAB is due to lactic acid, acetic acid, or hydrogen peroxide, it was independent of the Gram type of pathogens tested [33]. Admittedly, the antagonistic activity of probiotics against a broad spectrum of pathogens is a significant property, as it can provide valuable prospects for their use as feed additives, or in veterinary medicine [34]. Therefore, out of the LAB strains examined, three were selected as candidate probiotics for subsequent evaluation, given their favorable antimicrobial activities against the tested pathogens.
The antimicrobial activity of LAB is maintained by the secretion of different antibacterial components, such as hydrogen peroxide, organic acids (lactic and acetic acids), bacteriocins, alcohols, diacetyl, peptides, etc., which prevent or reduce the growth of pathogenic bacteria in the gut [30,35,36,37]. Our findings revealed that the antimicrobial activity of the LAB strains tested was not a result of hydrogen peroxide production, as it was almost the same in the untreated and catalase−treated supernatants of LAB strains. Many LAB strains can produce bacteriocins or bacteriocin-like components—a class of small molecules, cationic, hydrophobic, and heat−stable peptides [38]. LAB-derived bacteriocins have potent antibacterial properties and strong bactericidal or bacteriostatic effects against various pathogens through specific mechanisms, such as targeting bacterial membrane integrity and septum formation during mitosis [39]. Bacteriocin production has always been considered an important parameter in selecting probiotics [40]. The secretion of bacteriocins can promote the probiotic activity of intestinal LAB and, in some cases, may be directly responsible for beneficially regulating the intestinal microflora or inhibiting some pathogens [41]. Therefore, bacteriocins and bacteriocinogenic LAB are considered excellent alternatives to antibiotics [42] and have been widely used in the food industry, medicine, veterinary medicine, animal feed, and other fields [43,44]. So far, increasing numbers of bacteriocinogenic LAB strains have been isolated and characterized. For example, Miao et al. [45] characterized a novel bacteriocin F1 secreted by L. paracasei strain FX-6 isolated from Tibetan kefir. Our findings showed that the antimicrobial components of the selected LAB, especially strains H11 and L10, contained heat-stable substances sensitive to proteases, such as bacteriocins, suggesting that the selected LAB strains have huge prospects as potential probiotics. LAB can also produce a variety of organic acids in different concentrations, and lactic acid is usually one of its major metabolites [46]. Indeed, many LAB strains exert antimicrobial activity largely due to the secretion of lactic acid [47]. In addition, the antibacterial effects of other acids, such as citric acid and tartaric acid, have been reported previously [48]. The antimicrobial mechanisms of organic acids vary widely, affecting cell function and development by acidifying the cytoplasm and inhibiting the activity of acid-sensitive enzymes [49]. In the present study, the antimicrobial properties of the selected LAB strains were significantly inhibited in an acidic environment at pH 5.0, showing only weak to moderate inhibitory activity, which disappeared completely at pH 7.0. The above results indicate that the selected LAB strains contained antimicrobial substances other than organic acids and that these substances were acid-dependent, exerting antimicrobial activity only in an acidic environment. Our findings are consistent with those of Niku-Paavola et al. [50], who indicated that inhibitory substances produced by L. plantarum E76 were active only at low pH and in the presence of lactic acid. In addition, Makras et al. [51] reported that the antagonistic effect of the antimicrobial compounds produced by L. johnsonii La1 and L. plantarum ACA-DC 287 was significant only in the acidic environment of lactic acid, which may be due to lactic acid acting as a permeabilizer of the bacterial outer membrane, which may trigger the antimicrobial activity of other inhibitory compounds when a certain concentration is reached [52]. In future work, it is necessary to further characterize the antimicrobial components of the selected LAB and explore the molecular mechanisms underlying their antimicrobial properties.
The ability of potential probiotics to withstand various adverse factors in the gastrointestinal tract (digestive enzymes, bile salts, low pH gastric acid, etc.) is crucial to their colonization and proliferation in the host gut. Therefore, this is the major requirement for screening and evaluating probiotics [53,54]. According to previous studies, pH 2.0–3.0 [55,56] and 0.3% bile salts [11,57] were used as thresholds of acid and bile salt tolerance tests for potential probiotics, respectively. L. plantarum strain OF101, isolated from a fermented cereal beverage, showed 98.4% and 96.9% survival rates at pH 2.5 and 0.3% bile salts, respectively [58]. Jang et al. [23] reported that the survival rate of L. brevis KU15153, isolated from kimchi, was 52.48% and 101.91% under artificial gastric conditions (pH 2.5, 0.3% pepsin for 3 h) and bile salt conditions (0.3% bile salts for 24 h), respectively. In addition, MA2 showed a survival of 70% at pH 2.5 and 0.3% bile salt for 3 h [59]. In another study, six LAB strains isolated from chickens showed good tolerance to 0.3% bile salt after 6 h of exposure and moderate-to-good survival in simulated gastric juice with a pH of 2.0 [19]. In comparison, our isolate, H11, was found to possess moderate-to-good survivability under pH 2.0–3.0 and 0.3% bile salt conditions, indicating that it could be considered a promising candidate for use as a probiotic.
The hydrophobicity and aggregation abilities of probiotics correlate strongly with their adhesion to gastrointestinal epithelial cells. Therefore, these are two important characteristics for evaluating and selecting potential LAB probiotics [31]. The hydrophobicity of probiotics’ cell surface gauges their capacity to adhere to enterocyte cell lines [60], a highly desirable characteristic in probiotics. Some LAB strains have shown a correlation between hydrophobicity and adhesion [53]. In general, probiotics with high hydrophobicity values exhibited a greater ability to adhere to epithelial cells [61]. García-Hernández et al. [11] reported a hydrophobicity of 71.10% for L. pentosus LB-31 isolated from broiler excreta. In this work, all three LAB strains tested showed high hydrophobicity, ranging from 61.81% to 84.31%, indicating their strong adhesion to epithelial cells and mucosal surfaces. In two other studies, the hydrophobicity of LAB strains isolated from poultry ranged between 40.5–71.0% [19] and 21.18–95.27% [22]. Such discrepancy among the LAB strains tested may be attributed to differences in the hydrophobic moieties of surface proteins in the cell wall [62]. Significantly, a moderate level of hydrophobicity does not necessarily mean a lower adhesion ability of the bacteria, as hydrophilic domains may also engage in bacterial adhesion [63]. The aggregation ability of bacteria has usually been connected with adhesion properties [64]. Auto-aggregation facilitates probiotics to adhere to and colonize host intestinal cells, which is critical in several ecological niches [6,30]. According to Roghmann and McGrail [65], auto-aggregation above 40% is necessary for a strain to be a potential probiotic. In the current study, the auto−aggregation of the selected LAB strains was between 38.90 ± 4.31% and 53.12 ± 3.14%, indicating their acceptable adhesion ability. Our findings are consistent with a study by Reuben et al., who reported that the auto−aggregation of LAB strains isolated from poultry ranged from 32 ± 5.66% to 56.5 ± 3.54% [19]. Co-aggregation measures the adherence of tested strains to the enteric pathogens [6]. Therefore, the co-aggregation capacity of probiotic strains may make it possible to form a barrier that effectively hinders the adhesion and colonization of enteric pathogens on intestinal cells [30]. Chandran and Keerthi reported 63.66% co-aggregation for L. plantarum MBTU−HK1 with S. typhi [66]. In this work, the co-aggregation abilities between the three LAB strains and the pathogens were significantly different, and the co−aggregation value ranged between 21.21 and 36.81% for E. coli and 24.71 and 40.20% for S. aureus. In another investigation, Sophatha et al. [67] also reported that the co-aggregation value ranged between 21 and 32% for different LAB strains with E. coli. In addition, the co-aggregation abilities between the three LAB strains tested and pathogens were strain- and pathogen-specific, and the co-aggregation value of LAB strains and S. aureus was higher than that of E. coli. Findings from the work of Reuben et al. [19] also reported strain- and pathogen-specific co-aggregation abilities between the potential LAB probiotics and the six pathogens.
Probiotic candidates should not serve as reservoirs for antibiotic resistance genes [68], as such genes may further be transferred to enteric pathogens. Therefore, assessing the antimicrobial susceptibility profile is considered essential for selecting and safely using potential probiotic strains [69]. All three LAB isolates were susceptible to chloramphenicol, penicillin, ampicillin, streptomycin, and kanamycin; two were susceptible to gentamicin and rifampicin, while one was susceptible to erythromycin, vancomycin, and novobiocin. It has been reported that lactobacilli are generally susceptible to ampicillin [70]. In a recent study, Reuben et al. reported high susceptibility to chloramphenicol, penicillin, ampicillin, and novobiocin in LAB strains isolated from poultry [19]. Likewise, Dowarah et al. [20] mentioned that LAB strains isolated from pigs and poultry were highly susceptible to penicillin, ampicillin, and chloramphenicol. Interestingly, only one strain of LAB in this study was resistant to gentamycin and vancomycin. It has been documented that LAB is intrinsically resistant to gentamicin and vancomycin due to their impermeable membranes [28]. Meanwhile, the absence of hemolytic activity has been recommended as a safety characteristic for selecting probiotics [54]. Our results revealed that the three LAB strains showed non-hemolytic activities, depicting their non-virulent nature and safety in vivo. Similar findings showed that most LAB strains were non-hemolytic, as previously reported [6]. Building upon their antibiotic sensitivity profile and hemolytic activity in vitro, the three LAB strains could meet the basic requirements of probiotics, which makes them safe candidates for further verification in future probiotic studies in vivo.
5. Conclusions
Among the three LAB isolates from poultry, Limosilactobacillus reuteri H11 showed favorable in vitro probiotic characteristics, including broad-spectrum antimicrobial activity against pathogens, high cell surface properties, good acid (pH 2.0–3.0) and bile salt tolerance, and no antibiotic resistance or hemolytic activity. Therefore, L. reuteri H11 may be a great candidate for probiotics, which can be used as a poultry feed supplement to promote animal health and production performance. Of course, in vivo animal tests and whole−genome analysis are recommended to confirm the application potential of this strain.
Author Contributions
Conceptualization, X.L. and P.C.; methodology, P.C., W.L. and X.L.; data curation, P.C. and X.L.; writing—original draft preparation, X.L.; writing—review and editing, X.L., K.D. and P.C.; supervision, W.H., L.Z. and Y.L.; project administration, X.L. and P.C.; funding acquisition, P.C. and K.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research is sponsored by the Natural Science Foundation of Henan Province in China (Project No. 162300410080) and the National Natural Science Foundation of China (Grant No. 32072771).
Institutional Review Board Statement
The animal study protocol was approved by the Animal Care and Use Committee of Henan University of Science and Technology (DWFL36894-2023, 4 April 2023).
Data Availability Statement
The data underlying this article will be shared upon reasonable request to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
Correction Statement
This article has been republished with a minor correction of the information included in the Institutional Review Board Statement. This change does not affect the scientific content of the article.
References
- Lekshmi, M.; Ammini, P.; Kumar, S.; Varela, M.F. The food production environment and the development of antimicrobial resistance in human pathogens of animal origin. Microorganisms 2017, 5, 11. [Google Scholar] [CrossRef]
- Castanon, J.I. History of the use of antibiotic as growth promoters in European poultry feeds. Poult. Sci. 2007, 86, 2466–2471. [Google Scholar] [CrossRef]
- Gao, J.; Yang, Z.; Zhao, C.; Tang, X.; Jiang, Q.; Yin, Y. A comprehensive review on natural phenolic compounds as alternatives to in-feed antibiotics. Sci. China Life Sci. 2023, 66, 1518–1534. [Google Scholar] [CrossRef]
- Vieco-Saiz, N.; Belguesmia, Y.; Raspoet, R.; Auclair, E.; Gancel, F.; Kempf, I.; Drider, D. Benefits and inputs from lactic acid bacteria and their bacteriocins as alternatives to antibiotic growth promoters during food-animal production. Front. Microbiol. 2019, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Melara, E.G.; Avellaneda, M.C.; Rondón, A.J.; Rodríguez, M.; Valdivié, M.; Martínez, Y. Characterization of autochthonous strains from the cecal content of creole roosters for a potential use as probiotics. Animals 2023, 13, 455. [Google Scholar] [CrossRef] [PubMed]
- Makzum, S.; Ghadam, P.; Ramezani, M. Isolation, functional evaluation of probiotic properties and molecular identification of strains isolated from Iranian poultry’s gut. Iran J. Microbiol. 2023, 15, 267–277. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastro. Hepat. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Linares, D.M.; Gómez, C.; Renes, E.; Fresno, J.M.; Tornadijo, M.E.; Ross, R.P.; Stanton, C. Lactic acid bacteria and bifidobacteria with potential to design natural biofunctional health-promoting dairy foods. Front. Microbiol. 2017, 8, 846. [Google Scholar] [CrossRef]
- Pasolli, E.; De Filippis, F.; Mauriello, I.E.; Cumbo, F.; Walsh, A.M.; Leech, J.; Cotter, P.D.; Segata, N.; Ercolini, D. Large-scale genome-wide analysis links lactic acid bacteria from food with the gut microbiome. Nat. Commun. 2020, 11, 2610. [Google Scholar] [CrossRef]
- Negi, Y.K.; Pandey, C.; Saxena, N.; Sharma, S.; Garg, F.C.; Garg, S.K. Isolation of antibacterial protein from Lactobacillus spp. and preparation of probiotic curd. J. Food Sci. Technol. 2018, 55, 2011–2020. [Google Scholar] [CrossRef]
- García−Hernández, Y.; Pérez-Sánchez, T.; Boucourt, R.; Balcázar, J.L.; Nicoli, J.R.; Moreira-Silva, J.; Rodríguez, Z.; Fuertes, H.; Nuñez, O.; Albelo, N.; et al. Isolation, characterization and evaluation of probiotic lactic acid bacteria for potential use in animal production. Res. Vet. Sci. 2016, 108, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Guillermo, T.-I.; Christine, N.V.; Brittany, D.G.; Callie, M.S.; Lucas, E.G.; Roberto, S.-C.; Thaina, L.B.; Lesleigh, B.; Makenly, E.C.; Aaron, J.F.; et al. Developing probiotics, prebiotics, and organic acids to control Salmonella spp. in commercial turkeys at the University of Arkansas USA. Ger. J. Vet. Res. 2021, 1, 7–13. [Google Scholar]
- Riaz Rajoka, M.S.; Shi, J.; Zhu, J.; Shao, D.; Huang, Q.; Yang, H.; Jin, M. Capacity of lactic acid bacteria in immunity enhancement and cancer prevention. Appl. Microbiol. Biotechnol. 2017, 101, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Isolauri, E.; Salminen, S.; Ouwehand, A.C. Probiotics. Best Pract. Res. Clin. Gastroenterol. 2004, 18, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Shukla, R.; Goyal, A. Probiotics in valorization of innate immunity across various animal models. J. Funct. Foods 2015, 14, 549–561. [Google Scholar] [CrossRef]
- Gupta, M.; Raut, R.; Manandhar, S.; Chaudhary, A.; Shrestha, U.; Dangol, S.; Sudarshan, G.C.; Budha, K.R.; Karki, G.; Díaz-Sánchez, S.; et al. Identification and characterization of probiotics isolated from indigenous chicken (Gallus domesticus) of Nepal. PLoS ONE 2023, 18, e0280412. [Google Scholar] [CrossRef] [PubMed]
- Collins, J.K.; Thornton, G.; Sullivan, G.O. Selection of probiotic strains for human applications. Int. Dairy J. 1998, 8, 487–490. [Google Scholar] [CrossRef]
- Son, S.-H.; Jeon, H.-L.; Yang, S.-J.; Sim, M.-H.; Kim, Y.-J.; Lee, N.-K.; Paik, H.-D. Probiotic lactic acid bacteria isolated from traditional Korean fermented foods based on β-glucosidase activity. Food Sci. Biotechnol. 2018, 27, 123–129. [Google Scholar] [CrossRef]
- Reuben, R.C.; Roy, P.C.; Sarkar, S.L.; Alam, R.-U.; Jahid, I.K. Isolation, characterization, and assessment of lactic acid bacteria toward their selection as poultry probiotics. BMC Microbiol. 2019, 19, 253. [Google Scholar] [CrossRef]
- Dowarah, R.; Verma, A.K.; Agarwal, N.; Singh, P.; Singh, B.R. Selection and characterization of probiotic lactic acid bacteria and its impact on growth, nutrient digestibility, health and antioxidant status in weaned piglets. PLoS ONE 2018, 13, e0192978. [Google Scholar] [CrossRef]
- Ayeni, F.A.; Sánchez, B.; Adeniyi, B.A.; de los Reyes-Gavilán, C.G.; Margolles, A.; Ruas-Madiedo, P. Evaluation of the functional potential of Weissella and Lactobacillus isolates obtained from Nigerian traditional fermented foods and cow’s intestine. Int. J. Food Microbiol. 2011, 147, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Chen, S.; Liao, C.; Jia, Y.; Li, J.; Shang, K.; Chen, J.; Cao, P.; Li, W.; Li, Y.; et al. Probiotic properties of chicken-derived highly adherent lactic acid bacteria and inhibition of enteropathogenic bacteria in Caco-2 cells. Microorganisms 2022, 10, 2515. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.J.; Lee, N.-K.; Paik, H.-D. Probiotic characterization of Lactobacillus brevis KU15153 showing antimicrobial and antioxidant effect isolated from kimchi. Food Sci. Biotechnol. 2019, 28, 1521–1528. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.W.; Kirby, W.M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Abbasiliasi, S.; Tan, J.S.; Bashokouh, F.; Ibrahim, T.A.T.; Mustafa, S.; Vakhshiteh, F.; Sivasamboo, S.; Ariff, A.B. In vitro assessment of Pediococcus acidilactici Kp10 for its potential use in the food industry. BMC Microbiol. 2017, 17, 121. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, S.; Chen, S.; Wang, X.; Deng, X.; Liu, G.; Chang, W.; Beckers, Y.; Cai, H. Screening and characterization of Pediococcus acidilactici LC-9-1 toward selection as a potential probiotic for poultry with antibacterial and antioxidative properties. Antioxidants 2023, 12, 215. [Google Scholar] [CrossRef] [PubMed]
- Spanggaard, B.; Huber, I.; Nielsen, J.; Sick, E.B.; Pipper, C.B.; Martinussen, T.; Slierendrecht, W.J.; Gram, L. The probiotic potential against vibriosis of the indigenous microflora of rainbow trout. Environ. Microbiol. 2001, 3, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Jose, N.M.; Bunt, C.R.; Hussain, M.A. Comparison of microbiological and probiotic characteristics of lactobacilli isolates from dairy food products and animal rumen contents. Microorganisms 2015, 3, 198–212. [Google Scholar] [CrossRef] [PubMed]
- Sobrun, Y.; Bhaw-Luximon, A.; Jhurry, D.; Puchooa, D. Isolation of lactic acid bacteria from sugar cane juice and production of lactic acid from selected improved strains. Adv. Biosci. Biotechnol. 2012, 3, 398–407. [Google Scholar] [CrossRef]
- Venkatasatyanarayana, N.; Sadishkumar, V.; Jeevaratnam, K. Molecular characterization of antimicrobial Lactobacillus isolates and evaluation of their probiotic characteristics in vitro for use in poultry. Food Biotechnol. 2017, 31, 20–41. [Google Scholar]
- Taheri, H.R.; Moravej, H.; Tabandeh, F.; Zaghari, M.; Shivazad, M. Screening of lactic acid bacteria toward their selection as a source of chicken probiotic. Poult. Sci. 2009, 88, 1586–1593. [Google Scholar] [CrossRef]
- Kizerwetter-Swida, M.; Binek, M. Selection of potentially probiotic Lactobacillus strains towards their inhibitory activity against poultry enteropathogenic bacteria. Polish J. Microbiol. 2005, 54, 287–294. [Google Scholar]
- de Almeida Júnior, W.L.G.; Ferrari, Í.d.S.; de Souza, J.V.; da Silva, C.D.A.; da Costa, M.M.; Dias, F.S. Characterization and evaluation of lactic acid bacteria isolated from goat milk. Food Control 2015, 53, 96–103. [Google Scholar] [CrossRef]
- Cortés-Zavaleta, O.; López-Malo, A.; Hernández-Mendoza, A.; García, H.S. Antifungal activity of lactobacilli and its relationship with 3-phenyllactic acid production. Int. J. Food Microbiol. 2014, 173, 30–35. [Google Scholar] [CrossRef]
- Siroli, L.; Braschi, G.; Rossi, S.; Gottardi, D.; Patrignani, F.; Lanciotti, R. Lactobacillus paracasei A13 and high-pressure homogenization stress response. Microorganisms 2020, 8, 439. [Google Scholar] [CrossRef]
- Simons, A.; Alhanout, K.; Duval, R.E. Bacteriocins, antimicrobial peptides from bacterial origin: Overview of their biology and their impact against multidrug-resistant bacteria. Microorganisms 2020, 8, 639. [Google Scholar] [CrossRef]
- Betancur, C.; Martínez, Y.; Tellez-Isaias, G.; Avellaneda, M.C.; Velázquez-Martí, B. In vitro characterization of indigenous probiotic strains isolated from colombian creole pigs. Animals 2020, 10, 1204. [Google Scholar] [CrossRef]
- Wang, L.; Zhang, H.; Rehman, M.U.; Mehmood, K.; Jiang, X.; Iqbal, M.; Tong, X.; Gao, X.; Li, J. Antibacterial activity of Lactobacillus plantarum isolated from Tibetan yaks. Microb. Pathog. 2018, 115, 293–298. [Google Scholar] [CrossRef]
- Cavera, V.L.; Arthur, T.D.; Kashtanov, D.; Chikindas, M.L. Bacteriocins and their position in the next wave of conventional antibiotics. Int. J. Antimicrob. Agents 2015, 46, 494–501. [Google Scholar] [CrossRef]
- Dobson, A.; Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocin production: A probiotic trait? Appl. Environ. Microbiol. 2012, 78, 1–6. [Google Scholar] [CrossRef]
- Cotter, P.D.; Ross, R.P.; Hill, C. Bacteriocins-a viable alternative to antibiotics? Nat. Rev. Microbiol. 2013, 11, 95–105. [Google Scholar] [CrossRef]
- Lone, A.; Mottawea, W.; Mehdi, Y.; Hammami, R. Bacteriocinogenic probiotics as an integrated alternative to antibiotics in chicken production—Why and how? Crit. Rev. Food Sci. Nutr. 2022, 62, 8744–8760. [Google Scholar] [CrossRef]
- Musikasang, H.; Sohsomboon, N.; Tani, A.; Maneerat, S. Bacteriocin-producing lactic acid bacteria as a probiotic potential from Thai indigenous chickens. Czech J. Anim. Sci. 2012, 57, 137–149. [Google Scholar] [CrossRef]
- Ali, M.S.; Lee, E.B.; Hsu, W.H.; Suk, K.; Sayem, S.A.J.; Ullah, H.M.A.; Lee, S.J.; Park, S.C. Probiotics and postbiotics as an alternative to antibiotics: An emphasis on pigs. Pathogens 2023, 12, 874. [Google Scholar] [CrossRef] [PubMed]
- Miao, J.; Guo, H.; Ou, Y.; Liu, G.; Fang, X.; Liao, Z.; Ke, C.; Chen, Y.; Zhao, L.; Cao, Y. Purification and characterization of bacteriocin F1, a novel bacteriocin produced by Lactobacillus paracasei subsp. tolerans FX-6 from Tibetan kefir, a traditional fermented milk from Tibet, China. Food Control 2014, 42, 48–53. [Google Scholar] [CrossRef]
- Muthusamy, K.; Soundharrajan, I.; Srisesharam, S.; Kim, D.; Kuppusamy, P.; Lee, K.D.; Choi, K.C. Probiotic characteristics and antifungal activity of Lactobacillus plantarum and its impact on fermentation of Italian Ryegrass at low moisture. Appl. Sci. 2020, 10, 417. [Google Scholar] [CrossRef]
- Zhang, X.; Ali Esmail, G.; Fahad Alzeer, A.; Valan Arasu, M.; Vijayaraghavan, P.; Choon Choi, K.; Abdullah Al-Dhabi, N. Probiotic characteristics of Lactobacillus strains isolated from cheese and their antibacterial properties against gastrointestinal tract pathogens. Saudi J. Biol. Sci. 2020, 27, 3505–3513. [Google Scholar] [CrossRef]
- Zalán, Z.; Hudáček, J.; Štětina, J.; Chumchalová, J.; Halász, A. Production of organic acids by Lactobacillus strains in three different media. Eur. Food Res. Technol. 2010, 230, 395–404. [Google Scholar] [CrossRef]
- Hirshfield, I.N.; Terzulli, S.; O’Byrne, C. Weak organic acids: A panoply of effects on bacteria. Sci. Prog. 2003, 86 Pt 4, 245–269. [Google Scholar] [CrossRef]
- Niku-Paavola, M.L.; Laitila, A.; Mattila-Sandholm, T.; Haikara, A. New types of antimicrobial compounds produced by Lactobacillus plantarum. J. Appl. Microbiol. 1999, 86, 29–35. [Google Scholar] [CrossRef]
- Makras, L.; Triantafyllou, V.; Fayol-Messaoudi, D.; Adriany, T.; Zoumpopoulou, G.; Tsakalidou, E.; Servin, A.; De Vuyst, L. Kinetic analysis of the antibacterial activity of probiotic lactobacilli towards Salmonella enterica serovar Typhimurium reveals a role for lactic acid and other inhibitory compounds. Res. Microbiol. 2006, 157, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Alakomi, H.L.; Skyttä, E.; Saarela, M.; Mattila-Sandholm, T.; Latva-Kala, K.; Helander, I.M. Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Appl. Environ. Microbiol. 2000, 66, 2001–2005. [Google Scholar] [CrossRef]
- Ehrmann, M.A.; Kurzak, P.; Bauer, J.; Vogel, R.F. Characterization of lactobacilli towards their use as probiotic adjuncts in poultry. J. Appl. Microbiol. 2002, 92, 966–975. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Gong, W.; Xu, C.; Zhu, Z.; Peng, Y.; Xie, C. Probiotic assessment and antioxidant characterization of Lactobacillus plantarum GXL94 isolated from fermented chili. Front. Microbiol. 2022, 13, 997940. [Google Scholar] [CrossRef] [PubMed]
- Sahadeva, R.P.K.; Leong, S.F.; Chua, K.H.; Tan, C.H.; Chan, H.Y.; Tong, E.V.; Wong, S.; Chan, H.K. Survival of commercial probiotic strains to pH and bile. Int. Food Res. J. 2011, 18, 1515–1522. [Google Scholar]
- Prasad, J.; Gill, H.; Smart, J.; Gopal, P.K. Selection and characterisation of Lactobacillus and Bifidobacterium strains for use as probiotics. Int. Dairy J. 1998, 8, 993–1002. [Google Scholar] [CrossRef]
- Shokryazdan, P.; Kalavathy, R.; Sieo, C.C.; Alitheen, N.B.; Liang, J.B.; Jahromi, M.F.; Ho, Y.W. Isolation and characterization of Lactobacillus strains as potential probiotics for chickens. Pertanika J. Trop. Agric. Sci. 2014, 37, 141–157. [Google Scholar]
- Adesulu-Dahunsi, A.T.; Jeyaram, K.; Sanni, A.I.; Banwo, K. Production of exopolysaccharide by strains of Lactobacillus plantarum YO175 and OF101 isolated from traditional fermented cereal beverage. PeerJ 2018, 6, e5326. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Xing, Z.; Li, C.; Wang, J.; Wang, Y. Molecular mechanisms and in vitro antioxidant effects of Lactobacillus plantarum MA2. Food Chem. 2017, 221, 1642–1649. [Google Scholar] [CrossRef]
- Sadeghi, M.; Panahi, B.; Mazlumi, A.; Hejazi, M.A.; Komi, D.E.A.; Nami, Y. Screening of potential probiotic lactic acid bacteria with antimicrobial properties and selection of superior bacteria for application as biocontrol using machine learning models. LWT 2022, 162, 113471. [Google Scholar] [CrossRef]
- Blajman, J.; Gaziano, C.; Zbrun, M.V.; Soto, L.; Astesana, D.; Berisvil, A.; Scharpen, A.R.; Signorini, M.; Frizzo, L. In vitro and in vivo screening of native lactic acid bacteria toward their selection as a probiotic in broiler chickens. Res. Vet. Sci. 2015, 101, 50–56. [Google Scholar] [CrossRef]
- Pang, R.; Wu, S.; Zhang, F.; Huang, J.; Wu, H.; Zhang, J.; Li, Y.; Ding, Y.; Zhang, J.; Chen, M.; et al. The genomic context for the evolution and transmission of community-associated Staphylococcus aureus ST59 through the food chain. Front. Microbiol. 2020, 11, 422. [Google Scholar] [CrossRef]
- Savage, D.C. Growth phase, cellular hydrophobicity, and adhesion in vitro of lactobacilli colonizing the keratinizing gastric epithelium in the mouse. Appl. Environ. Microbiol. 1992, 58, 1992–1995. [Google Scholar] [CrossRef]
- Waśko, A.; Polak-Berecka, M.; Paduch, R.; Jóźwiak, K. The effect of moonlighting proteins on the adhesion and aggregation ability of Lactobacillus helveticus. Anaerobe 2014, 30, 161–168. [Google Scholar] [CrossRef]
- Roghmann, M.-C.; McGrail, L. Novel ways of preventing antibiotic-resistant infections: What might the future hold? Am. J. Infect. Control 2006, 34, 469–475. [Google Scholar] [CrossRef]
- Chandran, C.H.; Keerthi, T.R. Probiotic potency of Lactobacillus plantarum KX519413 and KX519414 isolated from honey bee gut. FEMS Microbiol. Lett. 2018, 365, fnx285. [Google Scholar]
- Sophatha, B.; Piwat, S.; Teanpaisan, R. Adhesion, anti-adhesion and aggregation properties relating to surface charges of selected Lactobacillus strains: Study in Caco-2 and H357 cells. Arch. Microbiol. 2020, 202, 1349–1357. [Google Scholar] [CrossRef]
- Danielsen, M.; Wind, A. Susceptibility of Lactobacillus spp. to antimicrobial agents. Int. J. Food Microbiol. 2003, 82, 1–11. [Google Scholar] [CrossRef] [PubMed]
- D’Aimmo, M.R.; Modesto, M.; Biavati, B. Antibiotic resistance of lactic acid bacteria and Bifidobacterium spp. isolated from dairy and pharmaceutical products. Int. J. Food Microbiol. 2007, 115, 35–42. [Google Scholar] [CrossRef]
- Gueimonde, M.; Sánchez, B.; G de Los Reyes-Gavilán, C.; Margolles, A. Antibiotic resistance in probiotic bacteria. Front. Microbiol. 2013, 4, 202. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).