Prevalence and Preferred Niche of Small Eukaryotes with Mixotrophic Potentials in the Global Ocean
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Regions
2.2. Contextual Environmental Variables
2.3. 18S rRNA Gene Abundances and Corrected Cell Abundances
2.4. Assignment of Trophic Modes
2.5. Trophic Index and Trophic Composition
2.6. Statistical Analysis
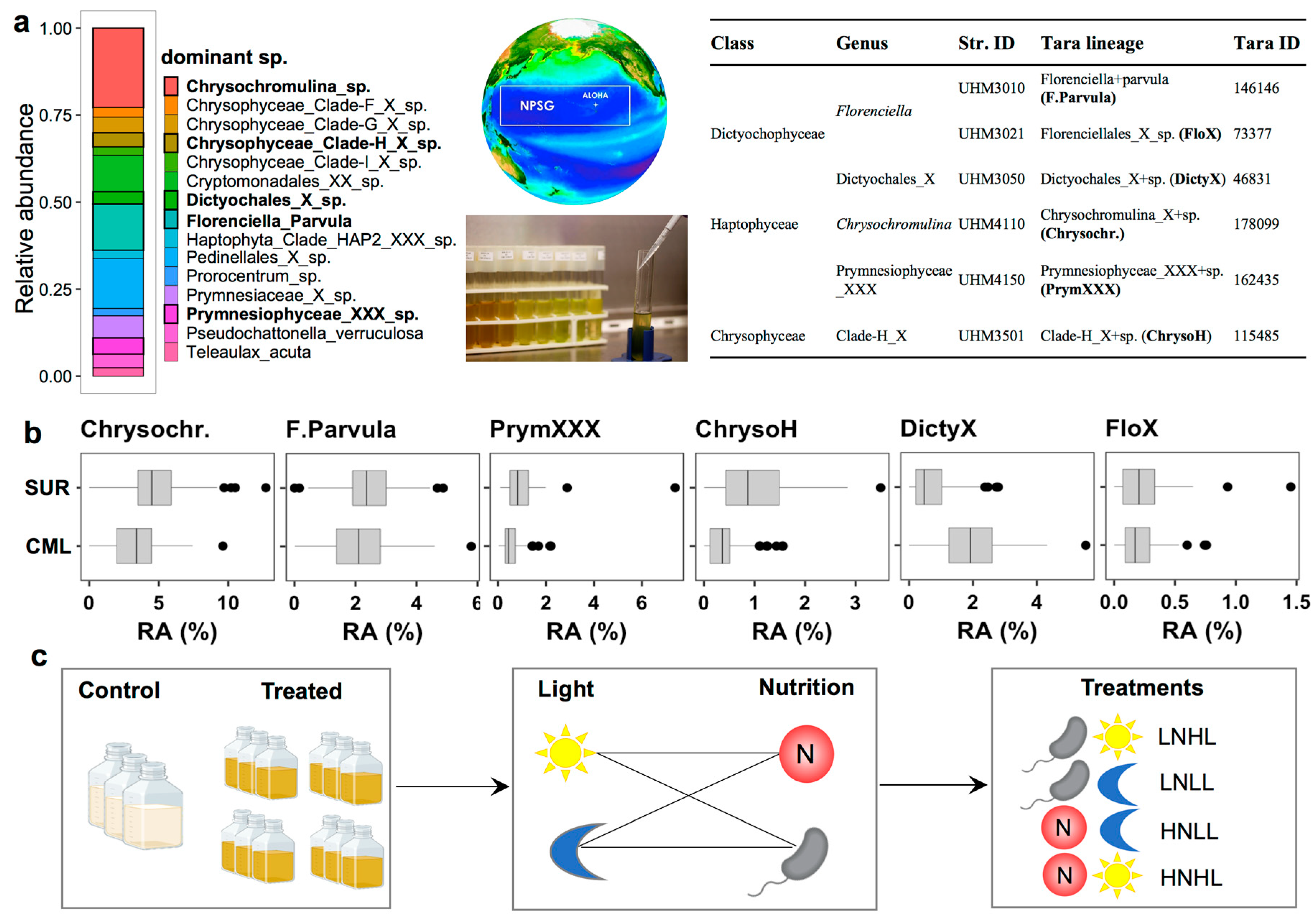
2.7. Culture Experiments
3. Results
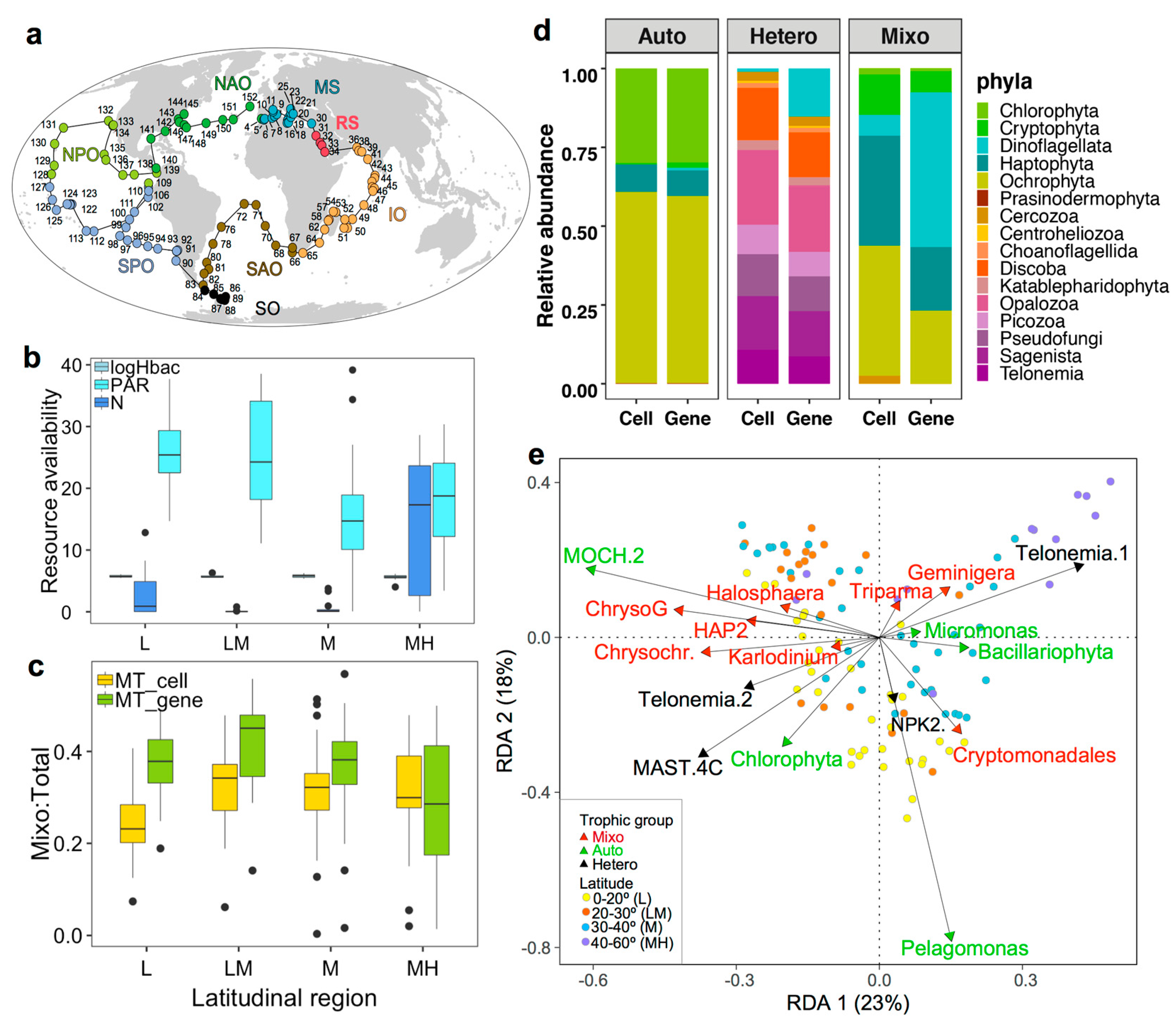
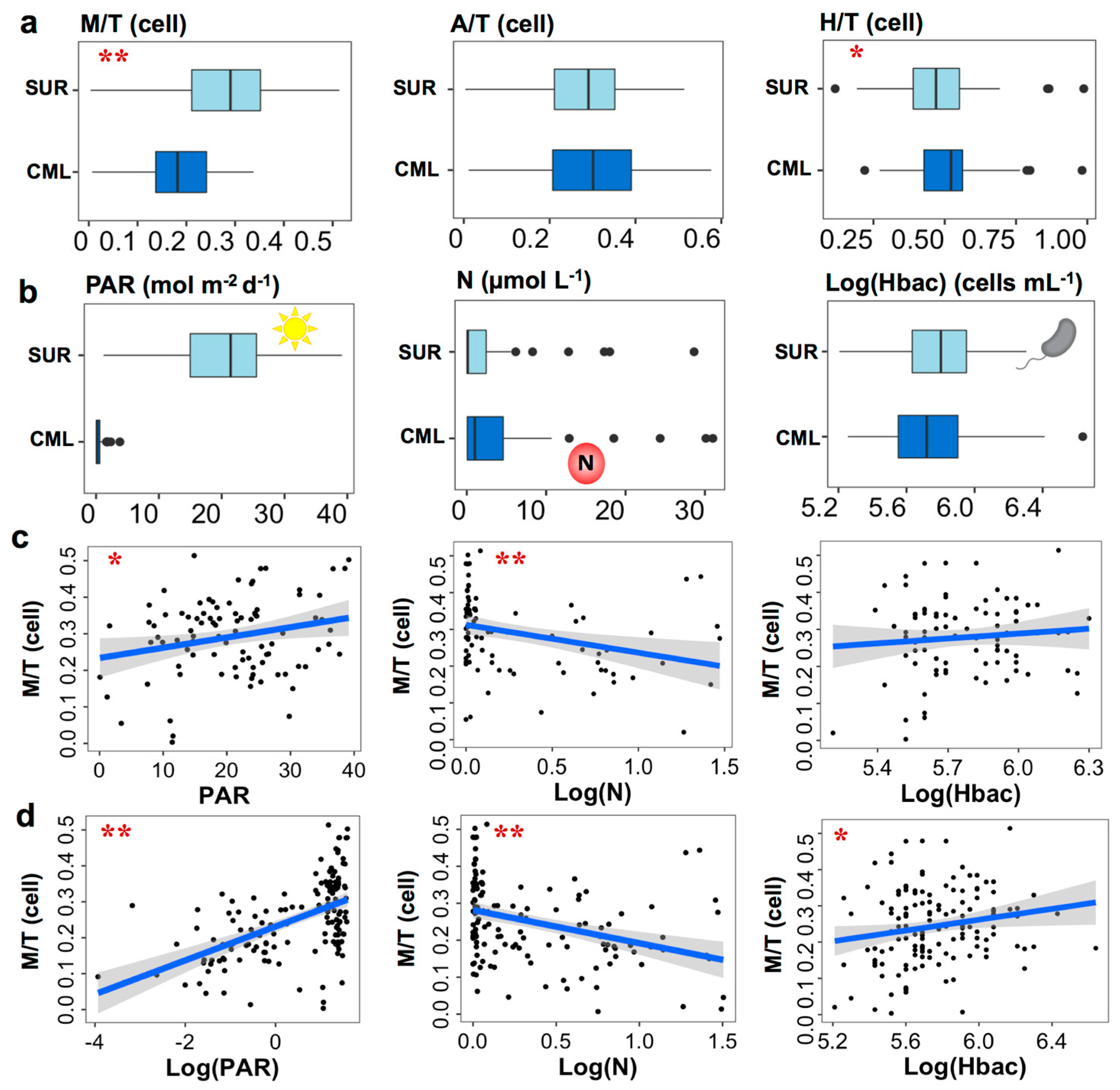
3.1. Oceanographic Characteristics across the Tara Oceans Sampling Regions
3.2. Relative Abundances and Taxonomic Compositions of Three Trophic Modes
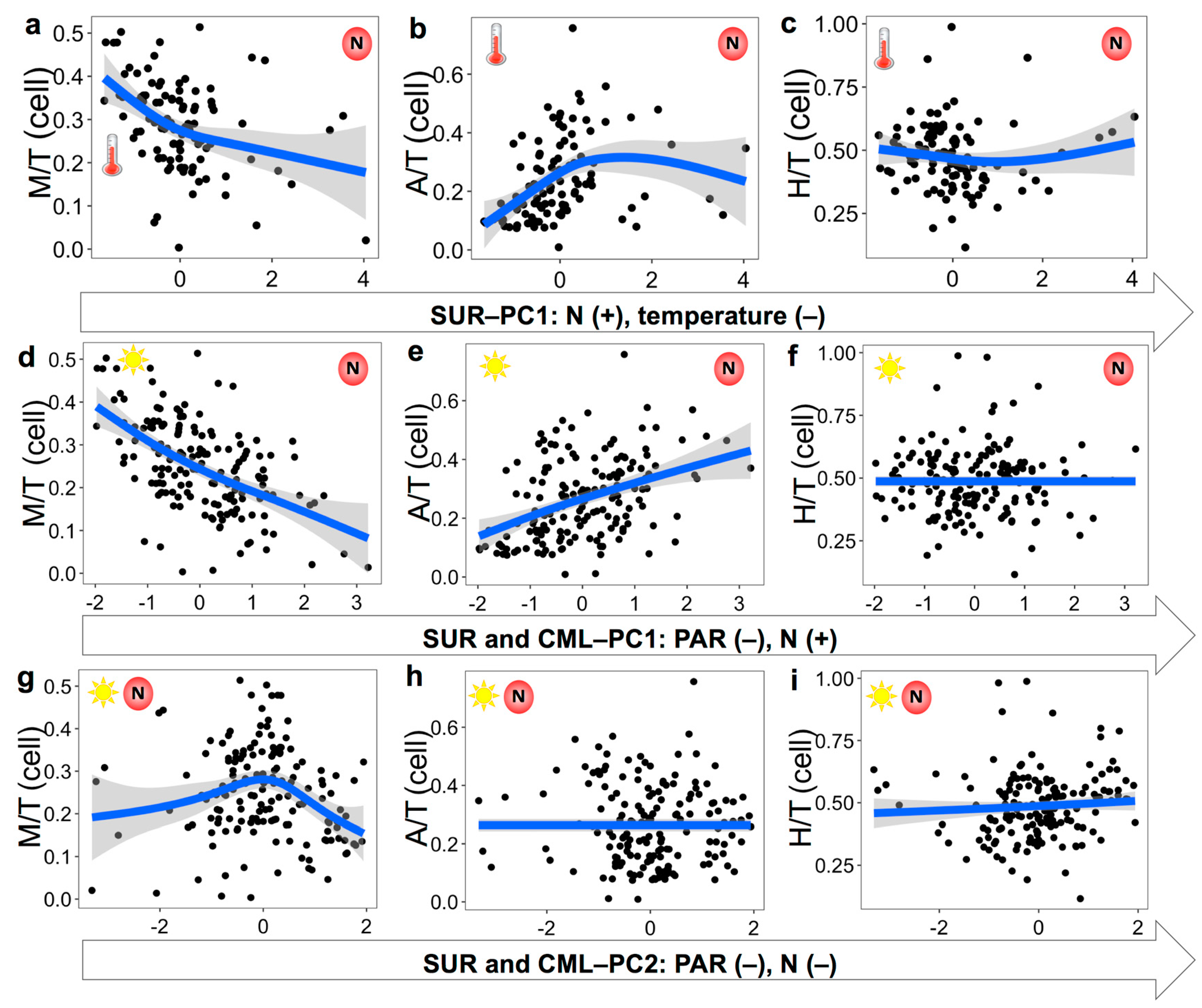
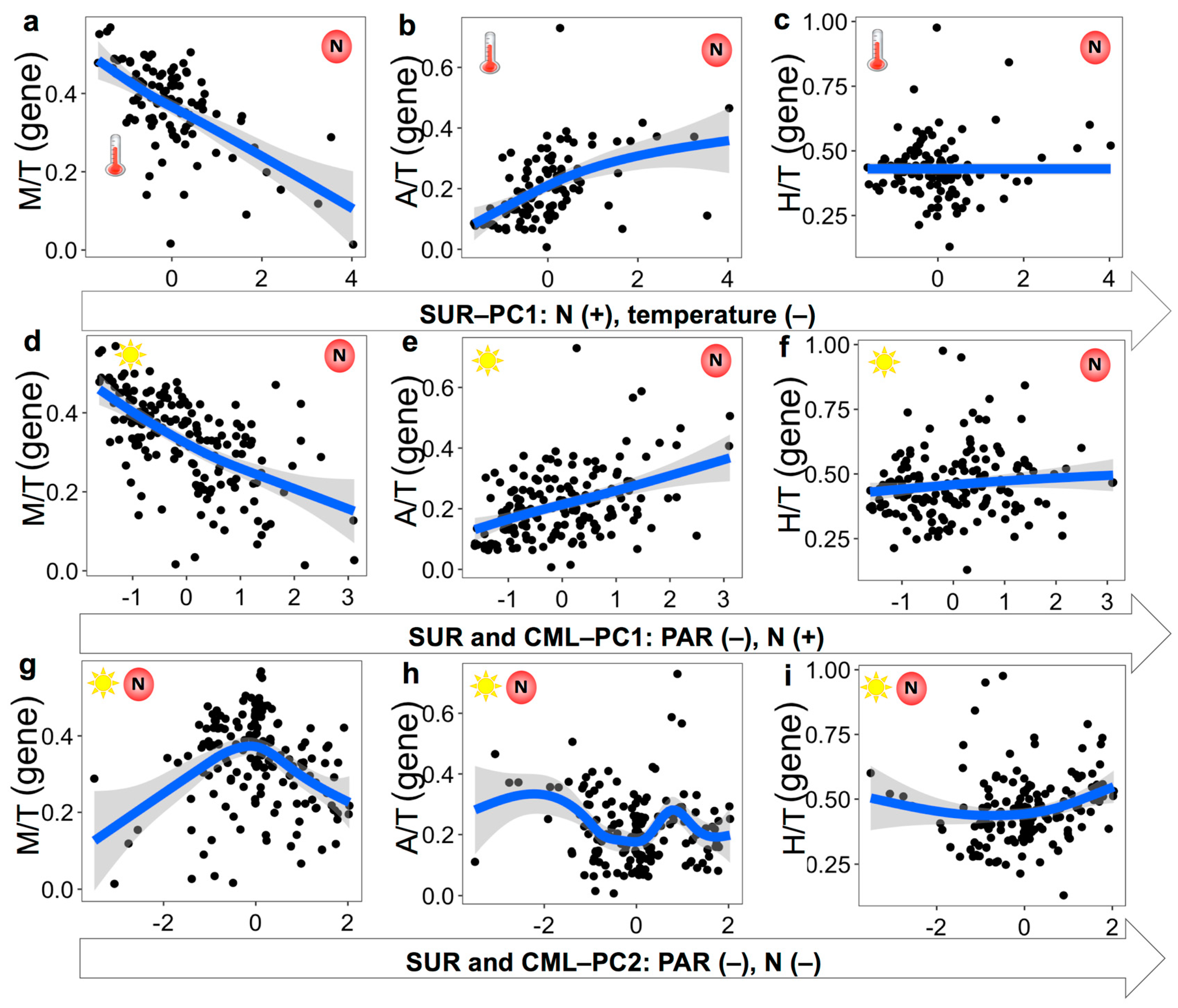
3.3. Nutritional Variables and the Prevalence of Three Trophic Modes
3.4. Mixotrophic Growth with Varying Nutritional Resource
4. Discussion
4.1. Mixotrophic Growth Responding to Changing Nutritional Resources
4.2. Relationship between Mixotrophy and Nutritional Resources on Large Spatial Scale
4.3. Environmental Variables Affecting the Prevalence of Trophic Strategies
4.4. Methodology Caveats
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sherr, E.B.; Sherr, B.F. Significance of predation by protists in aquatic microbial food webs. Antonie Leeuwenhoek 2002, 81, 293–308. [Google Scholar] [CrossRef] [PubMed]
- Stoecker, D.K.; Hansen, P.J.; Caron, D.A.; Mitra, A. Mixotrophy in the Marine Plankton. Annu. Rev. Mar. Sci. 2017, 9, 311–335. [Google Scholar] [CrossRef] [PubMed]
- Millette, N.C.; Gast, R.J.; Luo, J.Y.; Moeller, H.V.; Stamieszkin, K.; Andersen, K.H.; Brownlee, E.F.; Cohen, N.R.; Duhamel, S.; Dutkiewicz, S.; et al. Mixoplankton and mixotrophy: Future research priorities. J. Plankton Res. 2023, 45, 576–596. [Google Scholar] [CrossRef] [PubMed]
- Bock, N.A.; Charvet, S.; Burns, J.; Gyaltshen, Y.; Rozenberg, A.; Duhamel, S.; Kim, E. Experimental identification and in silico prediction of bacterivory in green algae. ISME J. 2021, 15, 1987–2000. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Edwards, K.F.; Schvarcz, C.R.; Steward, G.F. Broad phylogenetic and functional diversity among mixotrophic consumers of Prochlorococcus. ISME J. 2022, 16, 1557–1569. [Google Scholar] [CrossRef] [PubMed]
- Flynn, K.J.; Mitra, A.; Anestis, K.; Anschütz, A.A.; Calbet, A.; Ferreira, G.D.; Gypens, N.; Hansen, P.J.; John, U.; Martin, J.L.; et al. Mixotrophic protists and a new paradigm for marine ecology: Where does plankton research go now? J. Plankton Res. 2019, 41, 375–391. [Google Scholar] [CrossRef]
- Mitra, A.; Flynn, K.J.; Tillmann, U.; Raven, J.A.; Caron, D.; Stoecker, D.K.; Not, F.; Hansen, P.J.; Hallegraeff, G.; Sanders, R.; et al. Defining Planktonic Protist Functional Groups on Mechanisms for Energy and Nutrient Acquisition: Incorporation of Diverse Mixotrophic Strategies. Protist 2016, 167, 106–120. [Google Scholar] [CrossRef] [PubMed]
- Rothhaupt, K.O. Laboratorary Experiments with a Mixotrophic Chrysophyte and Obligately Phagotrophic and Photographic Competitors. Ecology 1996, 77, 716–724. [Google Scholar] [CrossRef]
- Thingstad, T.F.; Havskum, H.; Garde, K.; Riemann, B. On the Strategy of ‘Eating Your Competitor’: A Mathematical Analysis of Algal Mixotrophy. Ecology 1996, 77, 2108–2118. [Google Scholar] [CrossRef]
- Anderson, R.; Charvet, S.; Hansen, P.J. Mixotrophy in Chlorophytes and Haptophytes—Effect of Irradiance, Macronutrient, Micronutrient and Vitamin Limitation. Front. Microbiol. 2018, 9, 1704. [Google Scholar] [CrossRef]
- Edwards, K.F. Mixotrophy in nanoflagellates across environmental gradients in the ocean. Proc. Natl. Acad. Sci. USA 2019, 116, 6211–6220. [Google Scholar] [CrossRef] [PubMed]
- Ward, B.A. Mixotroph ecology: More than the sum of its parts. Proc. Natl. Acad. Sci. USA 2019, 116, 5846–5848. [Google Scholar] [CrossRef] [PubMed]
- Raven, J.A. Phagotrophy in phototrophs. Limnol. Oceanogr. 1997, 42, 198–205. [Google Scholar] [CrossRef]
- Mitra, A.; Flynn, K.J.; Burkholder, J.M.; Berge, T.; Calbet, A.; Raven, J.A.; Granéli, E.; Glibert, P.M.; Hansen, P.J.; Stoecker, D.K.; et al. The role of mixotrophic protists in the biological carbon pump. Biogeosciences 2014, 11, 995–1005. [Google Scholar] [CrossRef]
- Hansen, P.; Anderson, R.; Stoecker, D.; Decelle, J.; Altenburger, A.; Blossom, H.; Drumm, K.; Mitra, A.; Flynn, K. Mixotrophy among Freshwater and Marine Protists. In Encyclopedia of Microbiology; Elsevier: Amsterdam, The Netherlands, 2019. [Google Scholar]
- Leles, S.G.; Bruggeman, J.; Polimene, L.; Blackford, J.; Flynn, K.J.; Mitra, A. Differences in physiology explain succession of mixoplankton functional types and affect carbon fluxes in temperate seas. Prog. Oceanogr. 2021, 190, 102481. [Google Scholar] [CrossRef]
- Zubkov, M.V.; Tarran, G.A. High bacterivory by the smallest phytoplankton in the North Atlantic Ocean. Nature 2008, 455, 224–226. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Grob, C.; Tarran, G.A.; Martin, A.P.; Burkill, P.H.; Scanlan, D.J.; Zubkov, M.V. Mixotrophic basis of Atlantic oligotrophic ecosystems. Proc. Natl. Acad. Sci. USA 2012, 109, 5756–5760. [Google Scholar] [CrossRef] [PubMed]
- Tittel, J.; Bissinger, V.; Zippel, B.; Gaedke, U.; Bell, E.; Lorke, A.; Kamjunke, N. Mixotrophs combine resource use to outcompete specialists: Implications for aquatic food webs. Proc. Natl. Acad. Sci. USA 2003, 100, 12776–12781. [Google Scholar] [CrossRef]
- Hansson, T.H.; Grossart, H.-P.; del Giorgio, P.A.; St-Gelais, N.F.; Beisner, B.E. Environmental drivers of mixotrophs in boreal lakes. Limnol. Oceanogr. 2019, 64, 1688–1705. [Google Scholar] [CrossRef]
- Le Noac’h, P.; Cremella, B.; Kim, J.; Soria-Píriz, S.; del Giorgio, P.A.; Pollard, A.I.; Huot, Y.; Beisner, B.E. Nutrient availability is the main driver of nanophytoplankton phago-mixotrophy in North American lake surface waters. J. Plankton Res. 2024, 46, 9–24. [Google Scholar] [CrossRef]
- Wilken, S.; Huisman, J.; Naus-Wiezer, S.; Van Donk, E. Mixotrophic organisms become more heterotrophic with rising temperature. Ecol. Lett. 2013, 16, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Berge, T.; Chakraborty, S.; Hansen, P.J.; Andersen, K.H. Modeling succession of key resource-harvesting traits of mixotrophic plankton. ISME J. 2017, 11, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Lepori-Bui, M.; Paight, C.; Eberhard, E.; Mertz, C.M.; Moeller, H.V. Evidence for evolutionary adaptation of mixotrophic nanoflagellates to warmer temperatures. Glob. Change Biol. 2022, 28, 7094–7107. [Google Scholar] [CrossRef] [PubMed]
- Chu, T.; Moeller, H.V.; Archibald, K.M. Competition between phytoplankton and mixotrophs leads to metabolic character displacement. Ecol. Model. 2023, 481, 110331. [Google Scholar] [CrossRef]
- Pesant, S.; Not, F.; Picheral, M.; Kandels-Lewis, S.; Le Bescot, N.; Gorsky, G.; Iudicone, D.; Karsenti, E.; Speich, S.; Troublé, R.; et al. Open science resources for the discovery and analysis of Tara Oceans data. Sci. Data 2015, 2, 150023. [Google Scholar] [CrossRef] [PubMed]
- Picheral, M.; Searson, S.; Taillandier, V.; Bricaud, A.; Boss, E.; Stemmann, L.; Gorsky, G.; Tara Oceans Consortium, C.; Tara Oceans Expedition, P. Vertical profiles of environmental parameters measured from physical, optical and imaging sensors during station TARA_181 of the Tara Oceans expedition 2009–2013. Sci. Data 2014, 10, 347. [Google Scholar] [CrossRef]
- Ardyna, M.; Tara Oceans Consortium, C.; Tara Oceans Expedition, P. Environmental Context of All Stations from the Tara Oceans Expedition (2009–2013), about the Annual Cycle of Key Parameters Estimated Daily from Remote Sensing Products at a Spatial Resolution of 100 km. In Tara Oceans Consortium, Coordinators; Tara Oceans Expedition, Participants (2017): Registry of all Samples from the Tara Oceans Expedition (2009–2013). PANGAEA. 2017. Available online: https://b2find9.cloud.dkrz.de/dataset/ac50a5b1-afdf-5ce5-bb99-78bb1b446c82 (accessed on 23 March 2024).
- Ibarbalz, F.M.; Henry, N.; Brandão, M.C.; Martini, S.; Busseni, G.; Byrne, H.; Coelho, L.P.; Endo, H.; Gasol, J.M.; Gregory, A.C.; et al. Global Trends in Marine Plankton Diversity across Kingdoms of Life. Cell 2019, 179, 1084–1097.e1021. [Google Scholar] [CrossRef] [PubMed]
- Faure, E.; Not, F.; Benoiston, A.-S.; Labadie, K.; Bittner, L.; Ayata, S.-D. Mixotrophic protists display contrasted biogeographies in the global ocean. ISME J. 2019, 13, 1072–1083. [Google Scholar] [CrossRef] [PubMed]
- Pierella Karlusich, J.J.; Pelletier, E.; Zinger, L.; Lombard, F.; Zingone, A.; Colin, S.; Gasol, J.M.; Dorrell, R.G.; Henry, N.; Scalco, E.; et al. A robust approach to estimate relative phytoplankton cell abundances from metagenomes. Mol. Ecol. Resour. 2023, 23, 16–40. [Google Scholar] [CrossRef]
- de Vargas, C.; Audic, S.; Henry, N.; Decelle, J.; Mahé, F.; Logares, R.; Lara, E.; Berney, C.; Le Bescot, N.; Probert, I.; et al. Eukaryotic plankton diversity in the sunlit ocean. Science 2015, 348, 1261605. [Google Scholar] [CrossRef]
- Guillou, L.; Bachar, D.; Audic, S.; Bass, D.; Berney, C.; Bittner, L.; Boutte, C.; Burgaud, G.; de Vargas, C.; Decelle, J.; et al. The Protist Ribosomal Reference database (PR2): A catalog of unicellular eukaryote Small Sub-Unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2012, 41, D597–D604. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Massana, R.; Not, F.; Marie, D.; Vaulot, D. Mapping of picoeucaryotes in marine ecosystems with quantitative PCR of the 18S rRNA gene. FEMS Microbiol. Ecol. 2005, 52, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Gong, W.; Marchetti, A. Estimation of 18S Gene Copy Number in Marine Eukaryotic Plankton Using a Next-Generation Sequencing Approach. Front. Mar. Sci. 2019, 6, 219. [Google Scholar] [CrossRef]
- Martin, J.L.; Santi, I.; Pitta, P.; John, U.; Gypens, N. Towards quantitative metabarcoding of eukaryotic plankton: An approach to improve 18S rRNA gene copy number bias. Metabarcoding Metagenom. 2022, 6, e85794. [Google Scholar] [CrossRef]
- Li, Q.; Dong, K.; Wang, Y.; Edwards, K.F. Relative importance of bacterivorous mixotrophs in an estuary-coast environment. Limnol. Oceanogr. Lett. 2023, 9, 81–91. [Google Scholar] [CrossRef]
- Mitra, A.; Caron, D.A.; Faure, E.; Flynn, K.J.; Leles, S.G.; Hansen, P.J.; McManus, G.B.; Not, F.; do Rosario Gomes, H.; Santoferrara, L.F.; et al. The Mixoplankton Database (MDB): Diversity of photo-phago-trophic plankton in form, function, and distribution across the global ocean. J. Eukaryot. Microbiol. 2023, 70, e12972. [Google Scholar] [CrossRef] [PubMed]
- Stacklies, W.; Redestig, H.; Scholz, M.; Walther, D.; Selbig, J. pcaMethods—A bioconductor package providing PCA methods for incomplete data. Bioinformatics 2007, 23, 1164–1167. [Google Scholar] [CrossRef]
- Li, Q.; Edwards, K.F.; Schvarcz, C.R.; Selph, K.E.; Steward, G.F. Plasticity in the grazing ecophysiology of Florenciella (Dichtyochophyceae), a mixotrophic nanoflagellate that consumes Prochlorococcus and other bacteria. Limnol. Oceanogr. 2021, 66, 47–60. [Google Scholar] [CrossRef]
- Montagnes, D.J.S.; Barbosa, A.B.; Boenigk, J.; Davidson, K.; Jürgens, K.; Macek, M.; Parry, J.D.; Roberts, E.C.; Imek, K. Selective feeding behaviour of key free-living protists: Avenues for continued study. Aquat. Microb. Ecol. 2008, 53, 83–98. [Google Scholar] [CrossRef]
- Fenchel, T. The Ecology of Heterotrophic Microflagellates. In Advances in Microbial Ecology; Marshall, K.C., Ed.; Springer: Boston, MA, USA, 1986; pp. 57–97. [Google Scholar]
- Edwards, K.F.; Li, Q.; McBeain, K.A.; Schvarcz, C.R.; Steward, G.F. Trophic strategies explain the ocean niches of small eukaryotic phytoplankton. Proc. R. Soc. B Biol. Sci. USA 2023, 290, 20222021. [Google Scholar] [CrossRef]
- Stoecker, D.K. Conceptual models of mixotrophy in planktonic protists and some ecological and evolutionary implications. Eur. J. Protistol. 1998, 34, 281–290. [Google Scholar] [CrossRef]
- Jones, H. A classification of mixotrophic protists based on their behaviour. Freshw. Biol. 1997, 37, 35–43. [Google Scholar] [CrossRef]
- Fuhrman, J.A.; Noble, R.T. Viruses and protists cause similar bacterial mortality in coastal seawater. Limnol. Oceanogr. 1995, 40, 1236–1242. [Google Scholar] [CrossRef]
- Pernthaler, J. Predation on prokaryotes in the water column and its ecological implications. Nat. Rev. Microbiol. 2005, 3, 537–546. [Google Scholar] [CrossRef] [PubMed]
- Edwards, K.F.; Li, Q.; Steward, G.F. Ingestion kinetics of mixotrophic and heterotrophic flagellates. Limnol. Oceanogr. 2023, 68, 917–927. [Google Scholar] [CrossRef]
- Stukel, M.R.; Landry, M.R.; Selph, K.E. Nanoplankton mixotrophy in the eastern equatorial Pacific. Deep. Sea Res. Part II Top. Stud. Oceanogr. 2011, 58, 378–386. [Google Scholar] [CrossRef]
- Ptacnik, R.; Gomes, A.; Royer, S.-J.; Berger, S.A.; Calbet, A.; Nejstgaard, J.C.; Gasol, J.M.; Isari, S.; Moorthi, S.D.; Ptacnikova, R.; et al. A light-induced shortcut in the planktonic microbial loop. Sci. Rep. 2016, 6, 29286. [Google Scholar] [CrossRef]
- Millette, N.C.; Pierson, J.J.; Aceves, A.; Stoecker, D.K. Mixotrophy in Heterocapsa rotundata: A mechanism for dominating the winter phytoplankton. Limnol. Oceanogr. 2017, 62, 836–845. [Google Scholar] [CrossRef]
- Anderson, R.; Hansen, P.J. Meteorological conditions induce strong shifts in mixotrophic and heterotrophic flagellate bacterivory over small spatio-temporal scales. Limnol. Oceanogr. 2020, 65, 1189–1199. [Google Scholar] [CrossRef]
- Lambert, B.S.; Groussman, R.D.; Schatz, M.J.; Coesel, S.N.; Durham, B.P.; Alverson, A.J.; White, A.E.; Armbrust, E.V. The dynamic trophic architecture of open-ocean protist communities revealed through machine-guided metatranscriptomics. Proc. Natl. Acad. Sci. USA 2022, 119, e2100916119. [Google Scholar] [CrossRef]
- Jimenez, V.; Burns, J.A.; Le Gall, F.; Not, F.; Vaulot, D. No evidence of Phago-mixotropy in Micromonas polaris (Mamiellophyceae), the Dominant Picophytoplankton Species in the Arctic. J. Phycol. 2021, 57, 435–446. [Google Scholar] [CrossRef] [PubMed]
- McKie-Krisberg, Z.M.; Sanders, R.W. Phagotrophy by the picoeukaryotic green alga Micromonas: Implications for Arctic Oceans. ISME J. 2014, 8, 1953–1961. [Google Scholar] [CrossRef] [PubMed]
- Beisner, B.E.; Grossart, H.-P.; Gasol, J.M. A guide to methods for estimating phago-mixotrophy in nanophytoplankton. J. Plankton Res. 2019, 41, 77–89. [Google Scholar] [CrossRef]
- Godhe, A.; Asplund, M.E.; Härnström, K.; Saravanan, V.; Tyagi, A.; Karunasagar, I. Quantification of diatom and dinoflagellate biomasses in coastal marine seawater samples by real-time PCR. Appl. Environ. Microbiol. 2008, 74, 7174–7182. [Google Scholar] [CrossRef] [PubMed]

| Latitude | Oceanic Region † | Station | Temp (°C) | Salinity | PAR (mol m−2 d−1) | NO3−&NO2− (µmol L−1) | Hbac (Cells mL−1) | M/T (Cell/Gene) | A/T (Cell/Gene) | H/T (Cell/Gene) |
|---|---|---|---|---|---|---|---|---|---|---|
| 0–20° | SPO, IO, NPO, SAO, NAO | 36 | 26.6 | 35.1 | 25.4 | 0.89 | 4.9 × 105 | 0.23/ 0.38 | 0.32/ 0.24 | 0.43/ 0.38 |
| 20–30° | IO, NPO, SPO, SAO, RS | 17 | 24.7 | 36.2 | 24.3 | 0.03 | 3.9 × 105 | 0.34/ 0.45 | 0.10/ 0.08 | 0.51/ 0.44 |
| 30–40° | MS, SAO, NAO, SO | 37 | 19.5 | 36.4 | 14.7 | 0.13 | 5.3 × 105 | 0.32/ 0.38 | 0.19/ 0.16 | 0.48/ 0.43 |
| 40–60° | SO, SAO, MS | 14 | 7.1 | 34.3 | 18.8 | 17.3 | 4.5 × 105 | 0.30/ 0.29 | 0.18/ 0.22 | 0.47/ 0.46 |
| Global | SPO, IO, NPO, SAO, NAO, SO, RS, MS | 104 | 23.8 | 35.4 | 21.3 | 0.14 | 4.5 × 105 | 0.29/ 0.38 | 0.21/ 0.19 | 0.47/ 0.42 |
| SUR | SUR&CML | ||||
|---|---|---|---|---|---|
| PC1 | PC2 | PC1 | PC2 | Sum † | |
| PAR | −0.08 | −0.14 | −0.42 | −0.35 | 0.99 |
| NO3−&NO2− | 0.31 | −0.14 | 0.23 | −0.25 | 0.93 |
| Silicate | 0.23 | −0.10 | 0.17 | −0.17 | 0.67 |
| Temperature | −0.24 | 0.08 | −0.14 | 0.13 | 0.59 |
| Nitracline depth | −0.20 | −0.11 | −0.15 | 0.12 | 0.58 |
| CML | 0.06 | −0.17 | −0.10 | 0.07 | 0.40 |
| Mixed layer depth | 0.06 | −0.22 | −0.04 | −0.06 | 0.38 |
| R2 | 0.35 | 0.24 | 0.30 | 0.25 | |
| Growth Rates | Clearance Rates | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Single Factor | Multi-Factors | Single Factor | Multi-Factors | |||||||
| PAR | N | Sp. | PA and N | PAR and N and Sp. | PAR | N | Sp. | PAR and N | PAR and N and Sp. | |
| p † | * | >0.05 | >0.05 | * | * | >0.05 | >0.05 | >0.05 | * | 0.13 |
| F ‡ | 6.5 | 1.7 | 2.1 | 5.1 | 3.6 | 0.00 | 2.98 | 1.35 | 3.96 | 2.02 |
| Dfn § | 1 | 1 | 5 | 3 | 5 | 1 | 1 | 5 | 3 | 5 |
| Dfd ‖ | 20 | 20 | 17 | 18 | 18 | 20 | 20 | 17 | 18 | 18 |
| Effect ¶ | + | + | / | / | / | + | – | / | / | / |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, K.; Wang, Y.; Zhang, W.; Li, Q. Prevalence and Preferred Niche of Small Eukaryotes with Mixotrophic Potentials in the Global Ocean. Microorganisms 2024, 12, 750. https://doi.org/10.3390/microorganisms12040750
Dong K, Wang Y, Zhang W, Li Q. Prevalence and Preferred Niche of Small Eukaryotes with Mixotrophic Potentials in the Global Ocean. Microorganisms. 2024; 12(4):750. https://doi.org/10.3390/microorganisms12040750
Chicago/Turabian StyleDong, Kaiyi, Ying Wang, Wenjing Zhang, and Qian Li. 2024. "Prevalence and Preferred Niche of Small Eukaryotes with Mixotrophic Potentials in the Global Ocean" Microorganisms 12, no. 4: 750. https://doi.org/10.3390/microorganisms12040750




