Impact of MSMEG5257 Deletion on Mycolicibacterium smegmatis Growth
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Primers, and Media
2.2. Construction of msmeg5257 Mutant Strains
2.3. Western Blotting
2.4. RNA-Seq Experiments and Real-Time PCR
2.5. Drug Susceptibility Testing
2.6. Growth of msmeg5257 Mutant Strains in Iron-Depleted Agar
2.7. Subcellular Fractionation of msmeg5257
2.8. Label-Free Mass Spectrometry Protein Quantification
2.9. Growth of msmeg5257 Mutant Strains in Middlebrook 7H9 Media
2.10. Statistical Analysis
3. Results
3.1. Construction of msmeg5257 Deletion Strains
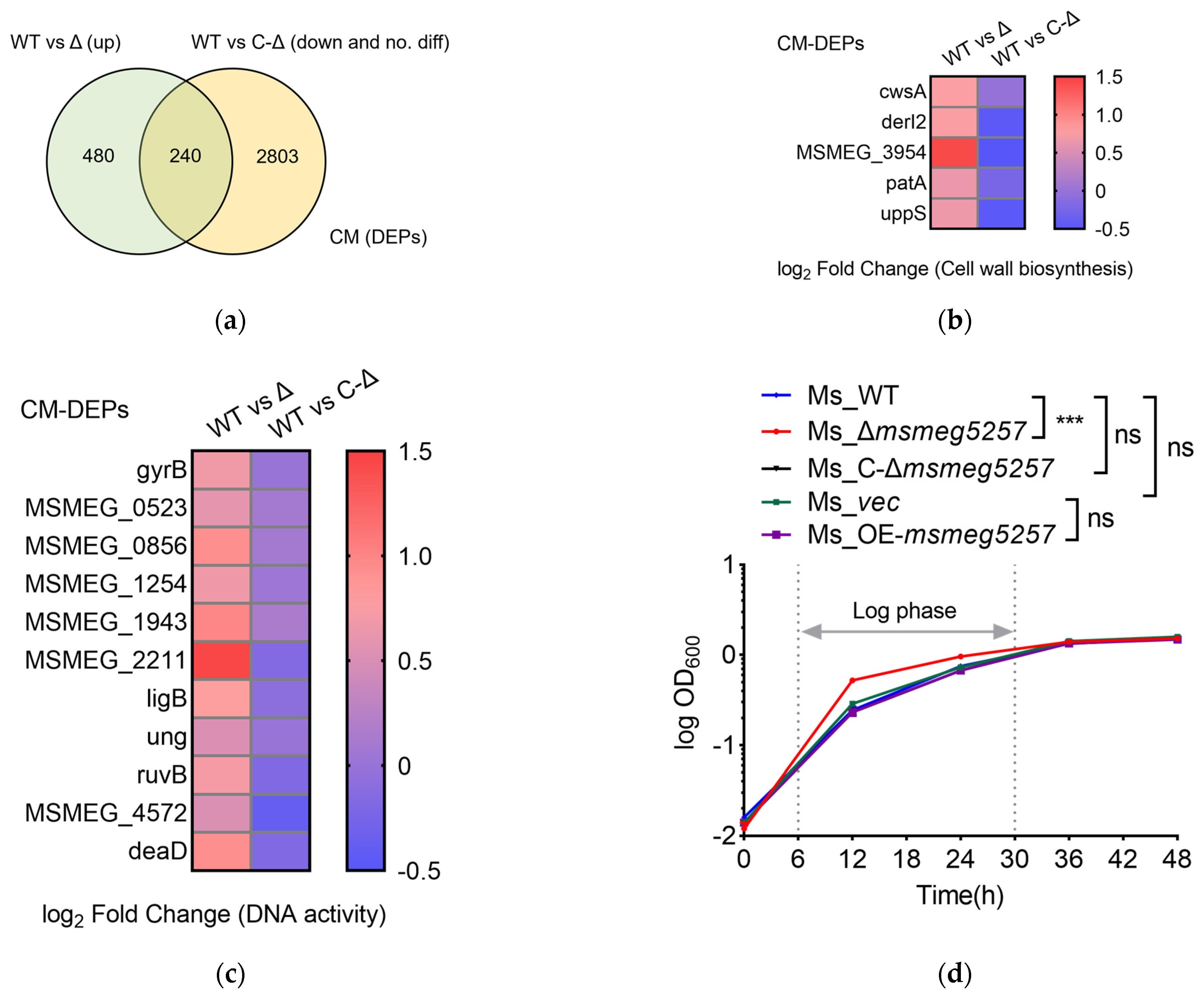
3.2. KEGG Enrichment Analysis of RNA Data in msmeg5257 Deletion Strains
3.3. The Correlative Analysis of LC-MS/MS and RNA Data in msmeg5257 Deletion Strains
3.4. Deletion of msmeg5257 Inhibits Expression in ABC Transporters of Cytomembrane
3.5. MSMEG5257 Localizes in the Cytomembrane of Ms
3.6. Deletion of msmeg5257 Inhibits the Growth of Ms in Iron-Depleted Media
3.7. Deletion of msmeg5257 Increases the Growth Rate of Ms In Vitro
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Global Tuberculosis Report 2023; Tedros, A.G., Tereza, K., Eds.; World Health Organizaiton: Geneva, Switzerland, 2023. [Google Scholar]
- Forrellad, M.A.; Klepp, L.I.; Gioffre, A.; Sabio y Garcia, J.; Morbidoni, H.R.; de la Paz Santangelo, M.; Cataldi, A.A.; Bigi, F. Virulence factors of the Mycobacterium tuberculosis complex. Virulence 2013, 4, 3–66. [Google Scholar] [CrossRef] [PubMed]
- Loiseau, C.; Windels, E.M.; Gygli, S.M.; Jugheli, L.; Maghradze, N.; Brites, D.; Ross, A.; Goig, G.; Reinhard, M.; Borrell, S.; et al. The relative transmission fitness of multidrug-resistant Mycobacterium tuberculosis in a drug resistance hotspot. Nat. Commun. 2023, 14, 1988. [Google Scholar] [CrossRef]
- Dufour, C.; Richard, C.; Pardons, M.; Massanella, M.; Ackaoui, A.; Murrell, B.; Routy, B.; Thomas, R.; Routy, J.-P.; Fromentin, R.; et al. Phenotypic characterization of single CD4+ T cells harboring genetically intact and inducible HIV genomes. Nat. Commun. 2023, 14, 3716. [Google Scholar] [CrossRef] [PubMed]
- Broset, E.; Saubi, N.; Guitart, N.; Aguilo, N.; Uranga, S.; Kilpelainen, A.; Eto, Y.; Hanke, T.; Gonzalo-Asensio, J.; Martin, C.; et al. MTBVAC-Based TB-HIV Vaccine Is Safe, Elicits HIV-T Cell Responses, and Protects against Mycobacterium tuberculosis in Mice. Mol. Ther. Methods Clin. Dev. 2019, 13, 253–264. [Google Scholar] [CrossRef]
- Faller, M.; Niederweis., M.; Schulz., G.E. The Structure of a Mycobacterial Outer-Membrane Channel. Science 2004, 303, 1189–1192. [Google Scholar] [CrossRef]
- Niederweis, M.; Danilchanka, O.; Huff, J.; Hoffmann, C.; Engelhardt, H. Mycobacterial outer membranes: In search of proteins. Trends Microb. 2010, 18, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Kwofie, S.K.; Hanson, G.; Sasu, H.; Enninful, K.S.; Mensah, F.A.; Nortey, R.T.; Yeboah, O.P.; Agoni, C.; Wilson, M.D. Molecular Modelling and Atomistic Insights into the Binding Mechanism of MmpL3 Mtb. Chem. Biodivers. 2022, 19, e202200160. [Google Scholar] [CrossRef]
- Pasca, M.R.; Guglierame, P.; Arcesi, F.; Bellinzoni, M.; De Rossi, E.; Riccardi, G. Rv2686c-Rv2687c-Rv2688c, an ABC fluoroquinolone efflux pump in Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2004, 48, 3175–3178. [Google Scholar] [CrossRef]
- Miao, J.; Liu, H.; Qu, Y.; Fu, W.; Qi, K.; Zang, S.; He, J.; Zhao, S.; Chen, S.; Jiang, T. Effect of peptidoglycan amidase MSMEG_6281 on fatty acid metabolism in Mycobacterium smegmatis. Microb. Pathog. 2020, 140. [Google Scholar] [CrossRef]
- Pecsi, I.; Hards, K.; Ekanayaka, N.; Berney, M.; Hartman, T.; Jacobs, W.R.; Cook, G.M.; Rubin, E.J.; Rhee, K. Essentiality of Succinate Dehydrogenase in Mycobacterium smegmatis and Its Role in the Generation of the Membrane Potential Under Hypoxia. mBio 2014, 5, e01014–e01093. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Yabaji, S.M.; Rukhlenko, O.S.; Bhattacharya, B.; Waligurski, E.; Vallavoju, N.; Ray, S.; Kholodenko, B.N.; Brown, L.E.; Beeler, A.B.; et al. Channeling macrophage polarization by rocaglates increases macrophage resistance to Mycobacterium tuberculosis. iScience 2021, 24, 102845. [Google Scholar] [CrossRef]
- Magazzù, A.; Marcuello, C. Investigation of Soft Matter Nanomechanics by Atomic Force Microscopy and Optical Tweezers: A Comprehensive Review. Nanomaterials 2023, 13, 963. [Google Scholar] [CrossRef] [PubMed]
- Kochanowski, J.A.; Carroll, B.; Asp, M.E.; Kaputa, E.C.; Patteson, A.E. Bacteria Colonies Modify Their Shear and Compressive Mechanical Properties in Response to Different Growth Substrates. ACS Appl. Bio. Mater. 2024. [Google Scholar] [CrossRef] [PubMed]
- Brewer, W.J.; Xet-Mull, A.M.; Yu, A.; Sweeney, M.I.; Walton, E.M.; Tobin, D.M. Macrophage NFATC2 mediates angiogenic signaling during mycobacterial infection. Cell Rep. 2022, 41, 11. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Mackenzie, J.; Tezera, L.; Krause, R.; Truebody, B.; Garay-Baquero, D.; Vallejo, A.; Govender, K.; Adamson, J.; Fisher, H.; et al. Mycobacterium tuberculosis senses host Interferon-gamma via the membrane protein MmpL10. Commun. Biol. 2022, 5, 1317. [Google Scholar] [CrossRef]
- Wang, M.; Nie, Y.; Wu, X.L. Extracellular heme recycling and sharing across species by novel mycomembrane vesicles of a Gram-positive bacterium. ISME J. 2021, 15, 605–617. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, Y.; Lin, C.; Zhang, J.; Mai, J.; Jiang, J.; Gao, X.; Li, Y.; Zhao, G.; Zhang, L.; et al. Crosstalk between the ancestral type VII secretion system ESX-4 and other T7SS in Mycobacterium marinum. iScience 2022, 25, 103585. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Dwivedi, S.P.; Gaharwar, U.S.; Meena, R.; Rajamani, P.; Prasad, T. Recent updates on drug resistance in Mycobacterium tuberculosis. J. Appl. Microbiol. 2020, 128, 1547–1567. [Google Scholar] [CrossRef]
- Abdallah, A.M.; van Pittius, N.C.G.; DiGiuseppe Champion, P.A.; Cox, J.; Luirink, J.; Vandenbroucke-Grauls, C.M.J.E.; Appelmelk, B.J.; Bitter, W. Type VII secretion—mycobacteria show the way. Nat. Rev. Micrbiol. 2007, 5, 883–891. [Google Scholar] [CrossRef]
- Beckham, H.K.S.; Ritter, C.; Chojnowski, G.; Ziemianowicz, D.S.; Mullapudi, E.; Rettel, M.; Savitski, M.M.; Mortensen, S.A.; Kosinski, J.; Wilmanns, M. Structure of the mycobacterial ESX-5 type VII secretion system pore complex. Sci. Adv. 2021, 7, eabg9923. [Google Scholar] [CrossRef] [PubMed]
- Groschel, M.I.; Sayes, F.; Simeone, R.; Majlessi, L.; Brosch, R. ESX secretion systems: Mycobacterial evolution to counter host immunity. Nat. Rev. Microbiol. 2016, 14, 677–691. [Google Scholar] [CrossRef]
- Hayashi, J.M.; Luo, C.Y.; Mayfield, J.A.; Hsu, T.; Fukuda, T.; Walfield, A.L.; Giffen, S.R.; Leszyk, J.D.; Baer, C.E.; Bennion, O.T.; et al. Spatially distinct and metabolically active membrane domain in mycobacteria. Proc. Natl. Acad. Sci. USA 2016, 113, 5400–5405. [Google Scholar] [CrossRef]
- Cassio Barreto de Oliveira, M.; Balan, A. The ATP-Binding Cassette (ABC) Transport Systems in Mycobacterium tuberculosis: Structure, Function, and Possible Targets for Therapeutics. Biology 2020, 9, 443. [Google Scholar] [CrossRef]
- Tak, U.; Dokland, T.; Niederweis, M. Pore-forming Esx proteins mediate toxin secretion by Mycobacterium tuberculosis. Nat. Commun. 2021, 12, 394. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.V.; Nuñez, J.K.; Doudna, J.A. Biology and Applications of CRISPR Systems: Harnessing Nature’s Toolbox for Genome Engineering. Cell 2016, 164, 29–44. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.-Y.; Li, S.-S.; Ding, X.-Y.; Guo, X.-P.; Jin, Q.; Sun, Y.-C.; Hatfull, G.F. A CRISPR-Assisted Nonhomologous End-Joining Strategy for Efficient Genome Editing in Mycobacterium tuberculosis. mBio 2020, 11, e02364-19. [Google Scholar] [CrossRef] [PubMed]
- Somerville, W.; Thibert, L.; Schwartzman, K.; Behr, M.A. Extraction of Mycobacterium tuberculosis DNA: A Question of Containment. J. Clin. Microbiol. 2005, 43, 2996–2997. [Google Scholar] [CrossRef] [PubMed]
- Tullius, M.V.; Nava, S.; Horwitz, M.A.; Ehrt, S. PPE37 Is Essential for Mycobacterium tuberculosis Heme-Iron Acquisition (HIA), and a Defective PPE37 in Mycobacterium bovis BCG Prevents HIA. Infect. Immun. 2019, 87, e00518–e00540. [Google Scholar] [CrossRef] [PubMed]
- Nadolinskaia, N.I.; Zamakhaev, M.V.; Shumkov, M.S.; Armianinova, D.K.; Karpov, D.S.; Goncharenko, A.V. CRISPR Interference of Adenylate Cyclases from Mycobacterium tuberculosis. Appl. Biochem. Microbiol. 2021, 57, 421–425. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Poulton, N.C.; Chang, J.S.; Azadian, Z.A.; DeJesus, M.A.; Ruecker, N.; Zimmerman, M.D.; Eckartt, K.A.; Bosch, B.; Engelhart, C.A.; et al. CRISPRi chemical genetics and comparative genomics identify genes mediating drug potency in Mycobacterium tuberculosis. Nat. Microbiol. 2022, 7, 766–779. [Google Scholar] [CrossRef]
- Lytvynenko, I.; Paternoga, H.; Thrun, A.; Balke, A.; Müller, T.A.; Chiang, C.H.; Nagler, K.; Tsaprailis, G.; Anders, S.; Bischofs, I.; et al. Alanine Tails Signal Proteolysis in Bacterial Ribosome-Associated Quality Control. Cell 2019, 178, 76–90.e22. [Google Scholar] [CrossRef]
- Batt, S.M.; Minnikin, D.E.; Besra, G.S. The thick waxy coat of mycobacteria, a protective layer against antibiotics and the host’s immune system. Biochem. J. 2020, 477, 1983–2006. [Google Scholar] [CrossRef] [PubMed]
- Rudner, D.Z.; Losick, R. Protein subcellular localization in bacteria. Cold Spring Harb. Perspect. Biol. 2010, 2, a000307. [Google Scholar] [CrossRef] [PubMed]
- Arnold, F.M.; Weber, M.S.; Gonda, I.; Gallenito, M.J.; Adenau, S.; Egloff, P.; Zimmermann, I.; Hutter, C.A.J.; Hurlimann, L.M.; Peters, E.E.; et al. The ABC exporter IrtAB imports and reduces mycobacterial siderophores. Nature 2020, 580, 413–417. [Google Scholar] [CrossRef]
- Zhang, B.; Li, J.; Yang, X.; Wu, L.; Zhang, J.; Yang, Y.; Zhao, Y.; Zhang, L.; Yang, X.; Yang, X.; et al. Crystal Structures of Membrane Transporter MmpL3, an Anti-TB Drug Target. Cell 2019, 176, 636–648.e13. [Google Scholar] [CrossRef]
- Chim, N.; Torres, R.; Liu, Y.; Capri, J.; Batot, G.; Whitelegge, J.P.; Goulding, C.W. The Structure and Interactions of Periplasmic Domains of Crucial MmpL Membrane Proteins from Mycobacterium tuberculosis. Chem. Biol. 2015, 22, 1098–1107. [Google Scholar] [CrossRef] [PubMed]
- Hermann, C.; Karamchand, L.; Blackburn, J.M.; Soares, N.C. Cell Envelope Proteomics of Mycobacteria. J. Proteome Res. 2021, 20, 94–109. [Google Scholar] [CrossRef] [PubMed]
- Vyas, R.; Tewari, R.; Weiss, M.S.; Karthikeyan, S. Structures of ternary complexes of aspartate-semialdehyde dehydrogenase (Rv3708c) fromMycobacterium tuberculosisH37Rv. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Ahangar, M.S.; Khandokar, Y.; Nasir, N.; Vyas, R.; Biswal, B.K. HisB fromMycobacterium tuberculosis: Cloning, overexpression inMycobacterium smegmatis, purification, crystallization and preliminary X-ray crystallographic analysis. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 1451–1456. [Google Scholar] [CrossRef]
- Gouzy, A.; Poquet, Y.; Neyrolles, O. Nitrogen metabolism in Mycobacterium tuberculosis physiology and virulence. Nat. Rev. Microbiol. 2014, 12, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Guan, X.; Zhang, S.; Liu, Y.; Wang, S.; Fan, P.; Du, X.; Yan, S.; Zhang, P.; Chen, H.Y.; et al. Structural-profiling of low molecular weight RNAs by nanopore trapping/translocation using Mycobacterium smegmatis porin A. Nat. Commun. 2021, 12, 3368. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Rao, Z. Structural biology and inhibition of the Mtb cell wall glycoconjugates biosynthesis on the membrane. Curr. Opin. Struct. Biol. 2023, 82, 102670. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Wang-Kan, X.; Neuberger, A.; van Veen, H.W.; Pos, K.M.; Piddock, L.J.V.; Luisi, B.F. Multidrug efflux pumps: Structure, function and regulation. Nat. Rev. Microbiol. 2018, 16, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.A.; Ball, D.A.; Sun, M.G.; Carlsson, F.; Watkins, B.Y.; Aggarwal, N.; McCracken, J.M.; Huynh, K.K.; Brown, E.J. EccA1, a Component of the Mycobacterium marinum ESX-1 Protein Virulence Factor Secretion Pathway, Regulates Mycolic Acid Lipid Synthesis. Chem. Biol. 2012, 19, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Liu, L.; Qiu, H.; Wen, H.; He, B.; Cui, Y.; Li, S.; Zhang, X.; Zhang, L.; Tian, C.; et al. Identification and architecture of a putative secretion tube across mycobacterial outer envelope. Sci. Adv. 2021, 7, eabg5656. [Google Scholar] [CrossRef] [PubMed]
- De Siena, B.; Campolattano, N.; D’Abrosca, G.; Russo, L.; Cantillon, D.; Marasco, R.; Muscariello, L.; Waddell, S.J.; Sacco, M. Characterization of the Mycobacterial MSMEG-3762/63 Efflux Pump in Mycobacterium smegmatis Drug Efflux. Front. Microbiol. 2020, 11, 575828. [Google Scholar] [CrossRef]
- Kim, S.; Yamaoka, Y.; Ono, H.; Kim, H.; Shim, D.; Maeshima, M.; Martinoia, E.; Cahoon, E.B.; Nishida, I.; Lee, Y. AtABCA9 transporter supplies fatty acids for lipid synthesis to the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 2012, 110, 773–778. [Google Scholar] [CrossRef]
- López-Marqués, R.L.; Poulsen, L.R.; Bailly, A.; Geisler, M.; Pomorski, T.G.; Palmgren, M.G. Structure and mechanism of ATP-dependent phospholipid transporters. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2015, 1850, 461–475. [Google Scholar] [CrossRef]
- Khakhina, S.; Johnson, S.S.; Manoharlal, R.; Russo, S.B.; Blugeon, C.; Lemoine, S.; Sunshine, A.B.; Dunham, M.J.; Cowart, L.A.; Devaux, F.; et al. Control of Plasma Membrane Permeability by ABC Transporters. Eukaryot. Cell 2015, 14, 442–453. [Google Scholar] [CrossRef]
- Koen Andries, P.V.; Guillemont, H.; Hinrich, W.; Gohlmann, H.; Neefs, J.-M.; Winkler, H.; Gestel, J.V.; Timmerman, P.; Zhu, M.; Lee, E.; et al. A Diarylquinoline Drug Active on the ATP Synthase of Mycobacterium tuberculosis. Sciense 2005, 307, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Cholo, M.C.; Steel, H.C.; Fourie, P.B.; Germishuizen, W.A.; Anderson, R. Clofazimine: Current status and future prospects. J. Antimicrob. Chemother. 2011, 67, 290–298. [Google Scholar] [CrossRef] [PubMed]
- Braibant, M.; Philippe Gilot, J.C. The ATP binding cassette (ABC) transport systems of Mycobacterium tuberculosis. FEMS Micro Rev. 2000, 24, 449–467. [Google Scholar] [CrossRef] [PubMed]
- November Sankey, H.M.; Singh, P.; Rogers, J.; Reddi, A.; Steven, D.; Hartson, A.M. Role of the Mycobacterium tuberculosis ESX-4 Secretion System in Heme Iron Utilization and Pore Formation by PPE Proteins. Am. Soc. Microbiol. 2023, 8, e00573-22. [Google Scholar] [CrossRef]
- Palmer, L.D.; Skaar, E.P. Transition Metals and Virulence in Bacteria. Annu. Rev. Genet. 2016, 50, 67–91. [Google Scholar] [CrossRef]
- Taudte, N.; German, N.; Zhu, Y.-G.; Grass, G.; Rensing, C. Restoration of growth by manganese in a mutant strain of Escherichia coli lacking most known iron and manganese uptake systems. BioMetals 2016, 29, 433–450. [Google Scholar] [CrossRef] [PubMed]
- Tullius, M.V.; Harmston, C.A.; Owens, C.P.; Chim, N.; Morse, R.P.; McMath, L.M.; Iniguez, A.; Kimmey, J.M.; Sawaya, M.R.; Whitelegge, J.P.; et al. Discovery and characterization of a unique mycobacterial heme acquisition system. Proc. Natl. Acad. Sci. USA 2011, 108, 5051–5056. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Rietzschel, N.; Kwon, H.; Walter Nuno, A.B.; Hanna, D.A.; Phillips, J.D.; Raven, E.L.; Reddi, A.R.; Hamza, I. Regulation of intracellular heme trafficking revealed by subcellular reporters. Proc. Natl. Acad. Sci. USA 2016, 113, E5144–E5152. [Google Scholar] [CrossRef]
- Danilchanka, O.; Pires, D.; Anes, E.; Niederweis, M. The Mycobacterium tuberculosis Outer Membrane Channel Protein CpnT Confers Susceptibility to Toxic Molecules. Antimicrob. Agents Chemother. 2015, 59, 2328–2336. [Google Scholar] [CrossRef] [PubMed]
- Pajuelo, D.; Tak, U.; Zhang, L.; Danilchanka, O.; Tischler, A.D.; Niederweis, M. Toxin secretion and trafficking by Mycobacterium tuberculosis. Nat. Commun. 2021, 12, 6592. [Google Scholar] [CrossRef]
- Izquierdo Lafuente, B.; Ummels, R.; Kuijl, C.; Bitter, W.; Speer, A. Mycobacterium tuberculosis Toxin CpnT Is an ESX-5 Substrate and Requires Three Type VII Secretion Systems for Intracellular Secretion. mBio 2021, 12, e02920–e02983. [Google Scholar] [CrossRef]
- Clark, R.R.; Judd, J.; Lasek-Nesselquist, E.; Montgomery, S.A.; Hoffmann, J.G.; Derbyshire, K.M.; Gray, T.A. Direct cell–cell contact activates SigM to express the ESX-4 secretion system in Mycobacterium smegmatis. Proc. Natl. Acad. Sci. USA 2018, 115, E6595–E6603. [Google Scholar] [CrossRef] [PubMed]
- Umare, M.D.; Khedekar, P.B.; Chikhale, R.V. Mycobacterial Membrane Protein Large 3 (MmpL3) Inhibitors: A Promising Approach to Combat Tuberculosis. ChemMedChem 2021, 16, 3136–3148. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, P.; Zhao, B.; He, W.; Song, Z.; Pei, S.; Liu, D.; Xia, H.; Wang, S.; Ou, X.; Zheng, Y.; et al. Impact of MSMEG5257 Deletion on Mycolicibacterium smegmatis Growth. Microorganisms 2024, 12, 770. https://doi.org/10.3390/microorganisms12040770
He P, Zhao B, He W, Song Z, Pei S, Liu D, Xia H, Wang S, Ou X, Zheng Y, et al. Impact of MSMEG5257 Deletion on Mycolicibacterium smegmatis Growth. Microorganisms. 2024; 12(4):770. https://doi.org/10.3390/microorganisms12040770
Chicago/Turabian StyleHe, Ping, Bing Zhao, Wencong He, Zexuan Song, Shaojun Pei, Dongxin Liu, Hui Xia, Shengfen Wang, Xichao Ou, Yang Zheng, and et al. 2024. "Impact of MSMEG5257 Deletion on Mycolicibacterium smegmatis Growth" Microorganisms 12, no. 4: 770. https://doi.org/10.3390/microorganisms12040770




