The Diversity and Composition of Soil Microbial Communities Differ in Three Land Use Types of the Sanjiang Plain, Northeastern China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Soil Sampling Collecting
2.3. Physico-Chemical Properties Analysis
2.4. Phospholipid Fatty Acid (PLFA) Analysis
2.5. Data Analysis
- I.
- Shannon–Wiener (H): H = ∑(Pi)(lnPi);
- II.
- Simpson (D): D = 1 − Σpi2.
- * Pi = Ni/N, where Ni is the number of characteristic fatty acids in treatment; n1 is the number of individuals with the first characteristic fatty acid biomarker; n2 is the number of individuals with the second characteristic fatty acid biomarker; n is the number.
- III.
- Margalef (M): M = (S − 1)/lnN;
- IV.
- Menhinick (E): E = S/√N;
- V.
- Brillouin (B): B = N − 1 lg(N!/n1!n2!…n!).
- * where N represents the number of total characteristic fatty acids in the experiment, and S is the total number of characteristic fatty acid markers.
3. Results
3.1. Changes in Soil Physico-Chemical Properties in Different Land Use Types
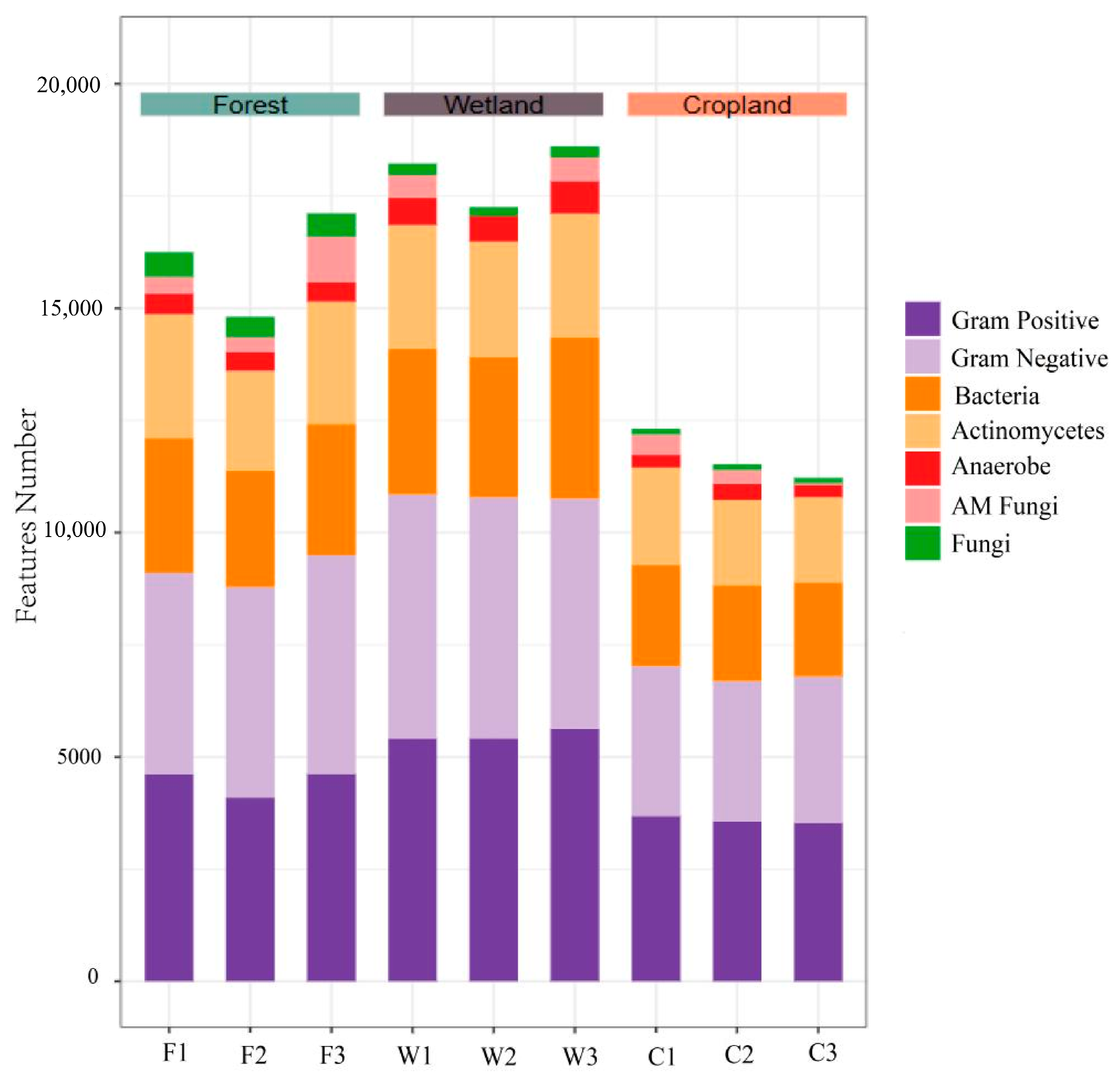
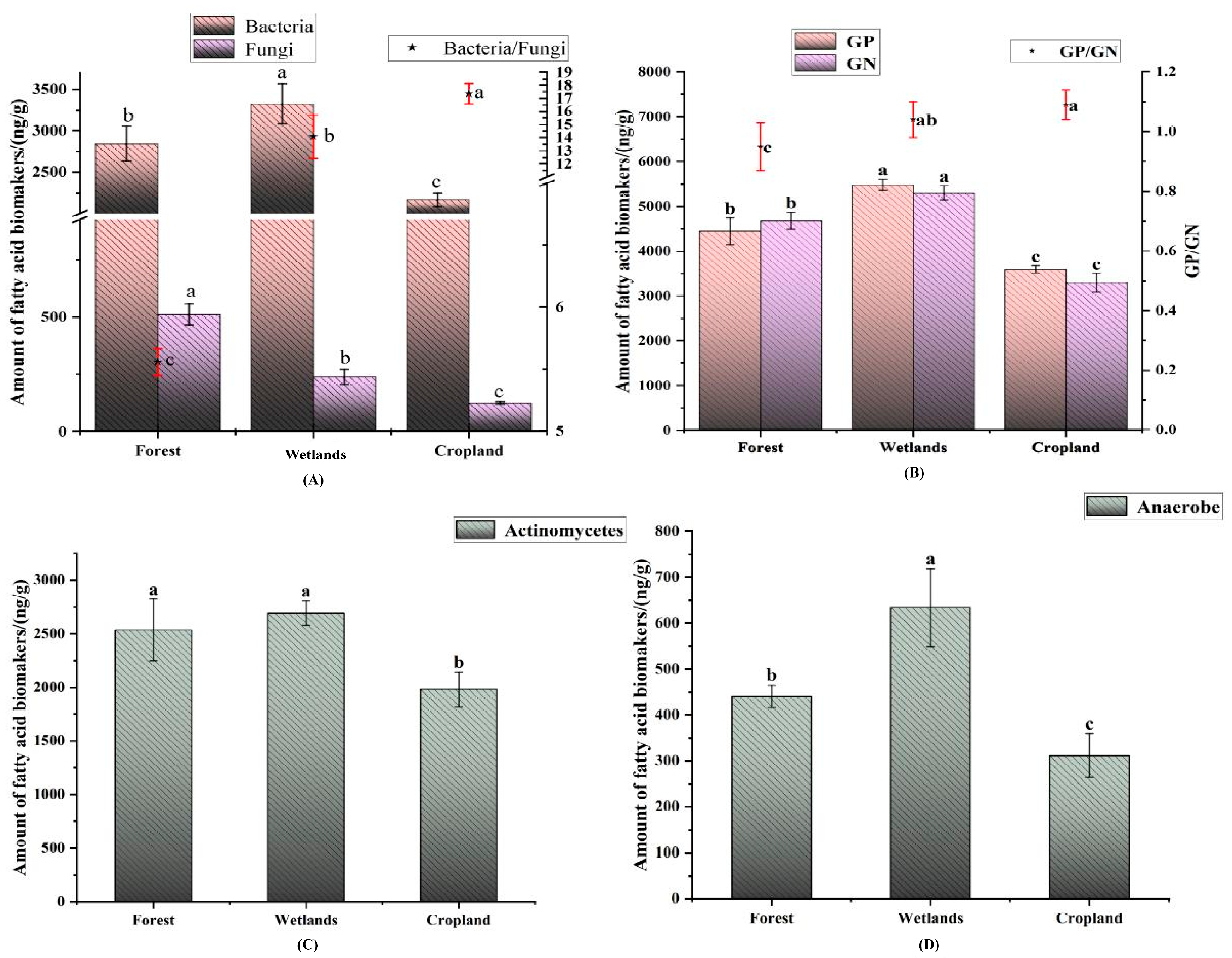
3.2. Abundance of Soil Microbial PLFA in Different Land Use Types of the Sanjiang Plain
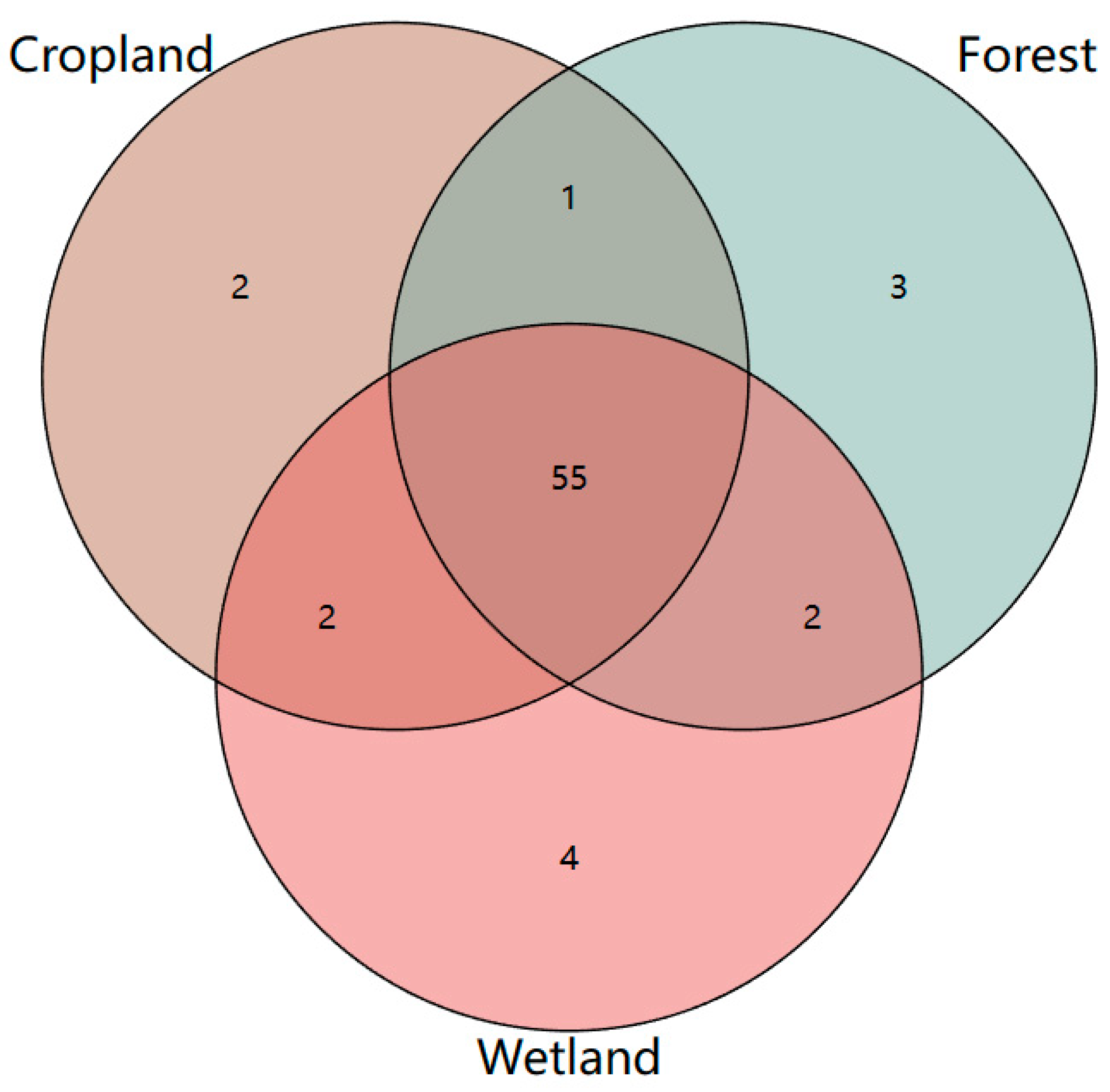
3.3. Soil Microbial Alpha Diversity Indices of the Different Land Use Types
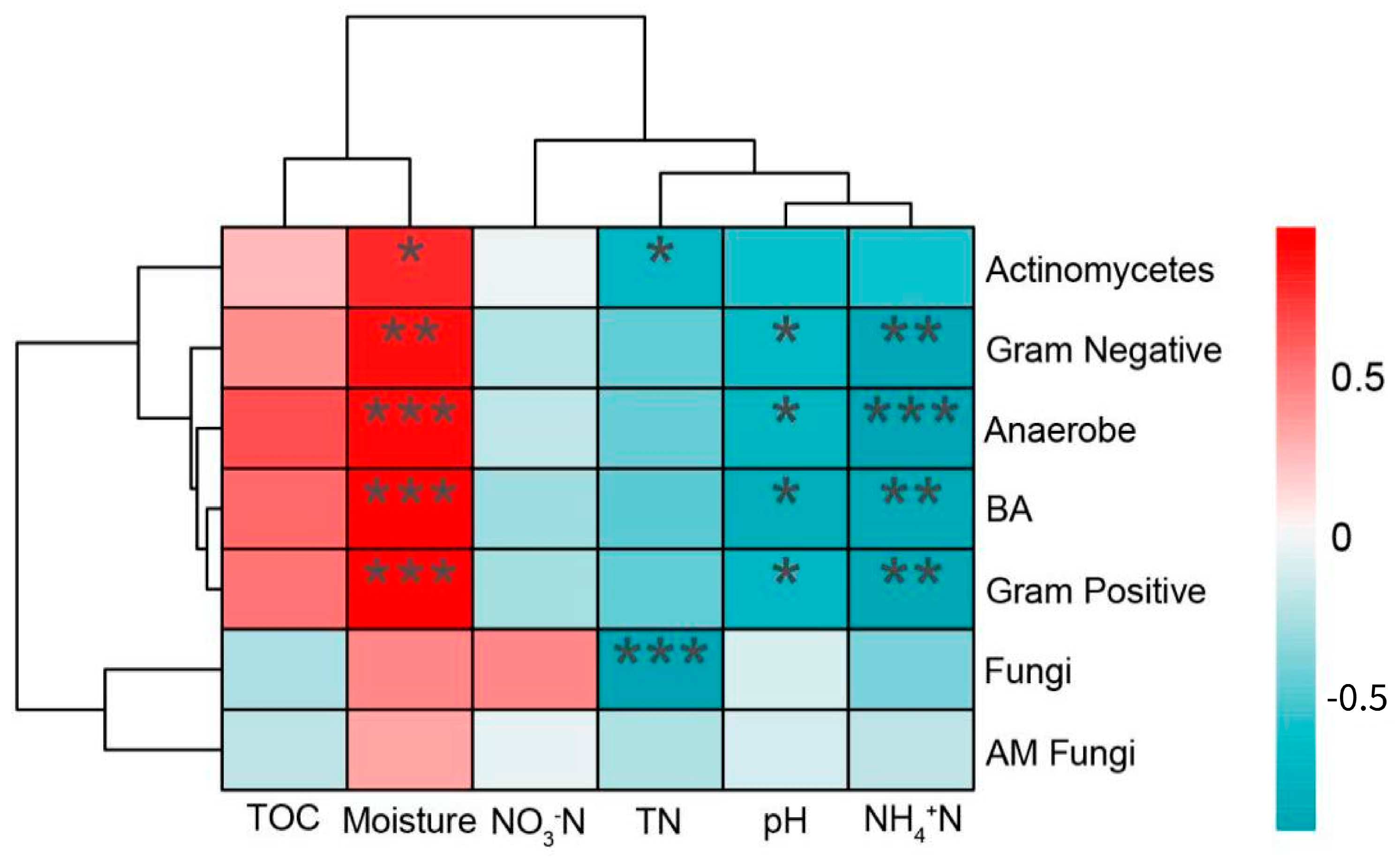
3.4. Relationship between Soil Microbial Structure and Soil Chemical Properties
4. Discussion
4.1. Changes of Soil Physicochemical Properties under Different Land use Types in Sanjiang Plain
4.2. Effects of Different Land use Types on Soil Microbial Composition in Sanjiang Plain
4.3. Effects of Different Land Use Types on Soil Microbial Diversity
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Borrelli, P.; Robinson, D.A.; Fleischer, L.R.; Lugato, E.; Ballabio, C.; Alewell, C.; Meusburger, K.; Modugno, S.; Schütt, B.; Ferro, V.; et al. An assessment of the global impact of 21st century land use change on soil erosion. Nat. Commun. 2017, 8, 2013. [Google Scholar] [CrossRef]
- Wang, X.; Xiao, X.; Xu, X.; Zou, Z.; Chen, B.; Qin, Y.; Zhang, X.; Dong, J.; Liu, D.; Pan, L.; et al. Rebound in China’s coastal wetlands following conservation and restoration. Nat. Sustain. 2021, 4, 1076–1083. [Google Scholar] [CrossRef]
- Sui, X.; Zhang, R.T.; Frey, B.; Yang, L.B.; Li, M.H.; Ni, H.W. Land use change effects on diversity of soil bacterial, acidobacterial and fungal communities in wetlands of the Sanjiang Plain, northeastern China. Sci. Rep. 2019, 9, 18535. [Google Scholar] [CrossRef]
- Turley, N.E.; Bell-Dereske, L.; Evans, S.E.; Brudvig, L.A. Agricultural land-use history and restoration impact soil microbial biodiversity. J. Appl. Ecol. 2020, 57, 852–863. [Google Scholar] [CrossRef]
- Fuhrmann, J.J.; Zuberer, D.A. 13—Carbon Transformations and Soil Organic Matter Formation. In Principles and Applications of Soil Microbiology, 3rd ed.; Gentry, T.J., Fuhrmann, J.J., Zuberer, D.A., Eds.; Elsevier: Amsterdam, The Netherlands, 2021; pp. 327–361. [Google Scholar]
- Sheng, Y.; Wang, H.; Wang, M.; Li, H.; Li, X.; Pan, F.; Chen, X.; Shen, X.; Mao, Z. Effects of soil texture on the growth of young apple trees and soil microbial community structure under replanted conditions. Hortic. Plant J. 2020, 6, 123–131. [Google Scholar] [CrossRef]
- Nannipieri, P.; Ascher, J.; Ceccherini, M.T.; Landi, L.; Pietramellara, G.; Renella, G. Microbial diversity and soil functions. Eur. J. Soil Sci. 2017, 68, 12–26. [Google Scholar] [CrossRef]
- Tiwari, S.; Singh, C.; Boudh, S.; Rai, P.K.; Gupta, V.K.; Singh, J.S. Land use change: A key ecological disturbance declines soil microbial biomass in dry tropical uplands. J. Environ. Manag. 2019, 242, 1–10. [Google Scholar] [CrossRef]
- Guo, Y.; Chen, X.; Wu, Y.; Zhang, L.; Cheng, J.; Wei, G.; Lin, Y. Natural revegetation of a semiarid habitat alters taxonomic and functional diversity of soil microbial communities. Sci. Total Environ. 2018, 635, 598–606. [Google Scholar] [CrossRef]
- Hua, H.; Sui, X.; Liu, Y.; Liu, X.; Chang, Q.; Xu, R.; Li, M.; Mu, L. Effects of land use type transformation on the structure and diversity of soil bacterial communities. Life 2024, 14, 252. [Google Scholar] [CrossRef]
- Zhu, X.; Yuan, F.; He, L.; Guo, Z.; Wang, N.; Zuo, Y.; Liu, J.; Li, K.; Wang, Y.; Sun, Y.; et al. Wetland conversion to cropland alters the microbes along soil profiles and over seasons. CATENA 2022, 214, 106282. [Google Scholar] [CrossRef]
- Liu, Y.; Ren, Z.; Qu, X.; Zhang, M.; Yu, Y.; Zhang, Y.; Peng, W. Microbial community structure and functional properties in permanently and seasonally flooded areas in Poyang Lake. Sci. Rep. 2020, 10, 4819. [Google Scholar] [CrossRef]
- Sui, X.; Zhang, R.; Yang, L.; Li, M.; Xu, N.; Liu, Y.H.; Chai, C.; Zhong, H.; Wang, J.; Ni, H.; et al. Differences in the microbial population associated with three wetland types in the Sanjiang Plain, Northeast China. Appl. Ecol. Environ. Res. 2017, 15, 79–92. [Google Scholar] [CrossRef]
- Shen, X.; Liu, B.; Jiang, M.; Lu, X. Marshland loss warms local land surface temperature in China. Geophys. Res. Lett. 2020, 47, e2020GL087648. [Google Scholar] [CrossRef]
- Mao, D.; He, X.; Wang, Z.; Tian, Y.; Xiang, H.; Yu, H.; Man, W.; Jia, M.; Ren, C.; Zheng, H. Diverse policies leading to contrasting impacts on land cover and ecosystem services in Northeast China. J. Clean. Prod. 2019, 240, 117961. [Google Scholar] [CrossRef]
- Wang, Z.; Mao, D.; Li, L.; Jia, M.; Dong, Z.; Miao, Z.; Ren, C.; Song, C. Quantifying changes in multiple ecosystem services during 1992-2012 in the Sanjiang Plain of China. Sci. Total Environ. 2015, 514, 119–130. [Google Scholar] [CrossRef]
- Yan, F.Q.; Zhang, S.W. Ecosystem service decline in response to wetland loss in the Sanjiang Plain, Northeast China. Ecol. Eng. 2019, 130, 117–121. [Google Scholar] [CrossRef]
- Yu, X.; Ding, S.; Zou, Y.; Xue, Z.; Lyu, X.; Wang, G. Review of Rapid Transformation of Floodplain Wetlands in Northeast China: Roles of Human Development and Global Environmental Change. Chin. Geogr. Sci. 2018, 28, 654–664. [Google Scholar] [CrossRef]
- Wang, S.; Xu, X.; Huang, L. Spatial and Temporal Variability of Soil Erosion in Northeast China from 2000 to 2020. Remote Sens. 2023, 15, 225. [Google Scholar] [CrossRef]
- Haibo, Y.; Le, Z.; Wei, H.; Yi, W.; Dan, W.; Shuwen, Z.; Kun, B. Cumulative effect of land use and cover changes in Naoli River basin in Sanjiang Plain on agricultural nonpoint source pollution load. Sustain. Water Resour. Manag. 2015, 1, 355–362. [Google Scholar] [CrossRef]
- Cao, H.J.; Xu, M.Y.; Zhang, R.T.; Ni, H.W. Effects of land use change on the structure and function of soil fungal community in sanjiang plain wetland. Appl. Ecol. Environ. Res. 2022, 20, 1677–1691. [Google Scholar] [CrossRef]
- Sui, X.; Zhang, R.; Frey, B.; Yang, L.; Liu, Y.; Ni, H.; Li, M.H. Soil physicochemical properties drive the variation in soil microbial communities along a forest successional series in a degraded wetland in northeastern China. Ecol. Evol. 2021, 11, 2194–2208. [Google Scholar] [CrossRef]
- Pellegrino, E.; Bosco, S.; Ciccolini, V.; Pistocchi, C.; Sabbatini, T.; Silvestri, N.; Bonari, E. Agricultural abandonment in Mediterranean reclaimed peaty soils: Long-term efects on soil chemical properties, arbuscular mycorrhizas and CO2 flux. Agric. Ecosyst. Environ. 2015, 199, 164–175. [Google Scholar] [CrossRef]
- Weng, X.H.; Wang, M.Y.; Sui, X.; Frey, B.; Liu, Y.N.; Zhang, R.T.; Ni, H.W.; Li, M.H. High Ammonium Addition Changes the Diversity and Structure of Bacterial Communities in Temperate Wetland Soils of Northeastern China. Microorganisms 2023, 11, 2033. [Google Scholar] [CrossRef]
- Bossio, D.A.; Scow, K.M. Impacts of carbon and flooding on soil microbial communities: Phospholipid fatty acid profiles and substrate utilization patterns. Microbial. Ecol. 1998, 35, 265–278. [Google Scholar] [CrossRef]
- Yao, X.; Wang, W.; Zeng, F. Application of phospholipid fatty acid method in soil microbial community analysis. Microbiol. Bull. 2016, 43, 2086–2095. [Google Scholar] [CrossRef]
- Buyer, J.S.; Vinyard, B.; Maul, J.; Selmer, K.; Lupitskyy, R.; Rice, C.; Roberts, D.P. Combined extraction method for metabolomic and PLFA analysis of soil. Appl. Soil Ecol. 2019, 135, 129–136. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Ma, J.P.; Pang, D.B.; Chen, L.; Wan, H.Y.; Chen, G.L.; Li, X.B. Phospholipid fatty acid analysis of soil microbes in typical vegetation types at different elevation on the east slope of Helan Mountain. Acta Ecol. Sin. 2022, 42, 5045–5505. [Google Scholar]
- Jinlei, C.; Ling, F.; Xiang, K. Composition, structure and regional characteristics of two forest communities in the central subtropics. For. Sci. 2019, 55, 159–172. [Google Scholar]
- Dănescu, A.; Albrecht, A.T.; Bauhus, J. Structural diversity promotes productivity of mixed, uneven-aged forests in southwestern Germany. Oecologia 2016, 182, 319–333. [Google Scholar] [CrossRef]
- Wang, M.; Weng, X.; Zhang, R.; Yang, L.; Liu, Y.; Sui, X. The Diversity and Composition of Soil Microbial Community Differ in Three Typical Wetland Types of the Sanjiang Plain, Northeastern China. Sustainability 2022, 14, 14394. [Google Scholar] [CrossRef]
- Tian, G.; Badejo, M.A.; Okoh, A.I.; Ishida, F.; Kolawole, G.O.; Hayashi, Y.; Salako, F.K. Effects of residue quality and climate on plant residue decomposition and nutrient release along the transect from humid forest to Sahel of West Africa. Biogeochemistry 2007, 86, 217–229. [Google Scholar] [CrossRef]
- Novo, P.; Vesna, T. Significance of harvest residues in sustainable management of arable land i. Decomposition of harvest residues. Arch. Tech. Sci. 2022, 1, 49–56. [Google Scholar] [CrossRef]
- Chunguang, W.; Haixing, L.; Tijiu, C.; Xiaoxin, S. Variation of soil carbon and nitrogen storage in a natural restoration chronosequence of reclaimed temperate marshes. Glob. Ecol. Conserv. 2021, 27, e01589. [Google Scholar] [CrossRef]
- Yu, S.; Ehrenfeld, J.G. The effects of changes in soil moisture on nitrogen cycling in acid wetland types of the New Jersey Pinelands (USA). Soil Biol. Biochem. 2009, 41, 2394–2405. [Google Scholar] [CrossRef]
- Bossio, D.A.; Girvan, M.S.; Verchot, L.; Bullimore, J.; Borelli, T.; Albrecht, A. Soil microbial community response to land use change in an agricultural landscape of western Kenya. Microb. Ecol. 2005, 49, 50–62. [Google Scholar] [CrossRef]
- Gartzia-Bengoetxea, N.; Kandeler, E.; de Arano, I.M.; Arias-González, A. Soil microbial functional activity is governed by a combination of tree species composition and soil properties in temperate forests. Appl. Soil Ecol. 2016, 100, 57–64. [Google Scholar] [CrossRef]
- Clairmont, L.K.; Stevens, K.J.; Slawson, R.M. Site-specific differences in microbial community structure and function within the rhizosphere and rhizoplane of wetland plants is plant species dependent. Rhizosphere 2019, 9, 56–68. [Google Scholar] [CrossRef]
- Choi, H.; Geronimo, F.K.; Jeon, M.; Kim, L.H. Evaluation of bacterial community in constructed wetlands treating different sources of wastewater. Ecol. Eng. 2022, 182, 106703. [Google Scholar] [CrossRef]
- Wang, M.; Sun, M.; Zhao, Y.; Shi, Y.; Su, S.; Wang, S.; Zhou, Y.; Chen, L. Seasonal changes of soil microbiota and its association with environmental factors in coal mining subsidence area. AMB Express 2023, 13, 147. [Google Scholar] [CrossRef]
- Liu, Z.-D.; Song, Y.-Y.; Ma, X.-Y.; Yuan, J.-B.; Lou, Y.-J.; Yang, C.; Tang, H.-R.; Song, C.-C. Deep soil microbial carbon metabolic function is important but often neglected: A study on the Songnen Plain reed wetland, Northeast China. Fundam. Res. 2023, 3, 833–843. [Google Scholar] [CrossRef]
- Lynn, T.M.; Liu, Q.; Hu, Y.; Yuan, H.; Wu, X.; Khai, A.A.; Ge, T. Influence of land use on bacterial and archaeal diversity and community structures in three natural ecosystems and one agricultural soil. Arch. Microbiol. 2017, 199, 711–721. [Google Scholar] [CrossRef]
- Suleiman, A.K.A.; Manoeli, L.; Boldo, J.T.; Pereira, M.G.; Roesch, L.F.W. Shifts in soil bacterial community after eight years of land-use change. Syst. Appl. Microbiol. 2013, 36, 137–144. [Google Scholar] [CrossRef]


| Microbial Group | Peak Name |
|---|---|
| Bacteria | 12:0, 13:0, 14:0, 15:0, 16:0, 18:0, 22:0, 24:0 |
| Gram-Positive | 11:0 iso, 11:0 anteiso, 12:0 iso, 12:0 anteiso, 13:0 iso, 13:0 anteiso, 14:1 iso ω7c, 14:0 iso, 14:0 anteiso, 15:1 iso ω9c, 15:1 iso ω6c, 15:1 anteiso ω9c, 15:0 iso, 15:0 anteiso, 16:0 iso, 16:0 anteiso, 17:1 iso ω9c, 17:0 iso, 17:0 anteiso, 18:0 iso, 19:0 iso, 19:0 anteiso, 20:0 iso, 22:0 is0 |
| Gram-Negative | 13:1 ω5c, 13:1 ω4c, 13:1 ω3c, 12:0 2OH, 14:1 ω7c, 14:1 ω5c 15:1 ω9c, 15:1 ω8c, 15:1 ω7c, 15:1 ω6c, 15:1 ω5c, 14:0 2OH, 16:1 ω9c, 16:1 ω7c 16:1 ω6c, 16:1 ω4c, 16:1 ω3c, 17:1 ω9c, 17:1 ω8c, 17:1 ω7c, 17:1 ω6c, 17:0 cyclo ω7c, 17:1 ω5c, 17:1 ω4c, 17:1 ω3c, 16:0 2OH, 18:1 ω8c, 18:1 ω7c, 18:1 ω6c, 18:1 ω5c, 18:1 ω3c, 19:1 ω9c, 19:1 ω8c, 19:1 ω7c, 19:1 ω6c, 19:0cyclo ω9c, 19:0 cyclo ω7c, 19:0 cyclo ω6c, 20:1 ω9c, 20:1 ω8c, 20:1 ω6c, 20:1 ω4c, 20:0 cyclo ω6c, 21:1 ω9c, 21:1 ω8c, 21:1 ω6c, 21:1 ω5c, 21:1 ω4c, 21:1 ω3c, 22:1 ω9c, 22:1 ω8c, 22:1 ω6c 22:1 ω5c, 22:1 ω3c, 22:0 cyclo ω6c, 24:1 ω9c, 24:1 w7c |
| Actinomycetes | 16:0 10-methyl, 17:1 ω7c 10-methyl, 17:0 10-methyl, 18:1 ω7c10-methyl, 18:0 10-methyl, 19:1 ω7c 10-methyl, 20:0 10-methyl |
| Fungi | 18:1ω9c, 18:2 ω6c, 23:0 |
| Protozoa | 19:3ω6c, 20:3ω6c, 20:4ω6c, 20:5ω3c |
| AM Fungi | 16:1 ω5c |
| Anaerobic | 13:0 DMA, 15:0 DMA, 16:2 DMA, 16:1 w9c DMA, 16:1 w7c DMA, 17:0 DMA, 18:2 DMA, 18:1 w9c DMA, 18:1 w7c DMA, 18:0 DMA |
| Site | pH | TN (g/kg) | TOC (g/kg) | NH4+N (mg/kg) | NO3−N (mg/kg) | SWC (%) |
|---|---|---|---|---|---|---|
| Forest | 7.49 ± 0.35 a | 1.89 ± 0.40 c | 19.22 ± 1.99 b | 16.85 ± 1.83 b | 0.27 ± 0.05 a | 0.23 ± 0.00 b |
| Wetland | 5.61 ± 0.37 b | 3.56 ± 0.39 b | 40.63 ± 1.03 a | 10.65 ± 0.92 b | 0.15 ± 0.02 a | 0.43 ± 0.13 a |
| Cropland | 5.95 ± 0.12 b | 4.63 ± 0.56 a | 20.34 ± 1.67 b | 55.53 ± 5.62 a | 0.22 ± 0.12 a | 0.16 ± 0.02 b |
| Diversity Index | Forest | Wetland | Cropland |
|---|---|---|---|
| Shannon–Wiener (H) | 3.29 ± 0.07 a | 3.21 ± 0.02 a | 2.23 ± 0.08 b |
| Simpson (D) | 0.95 ± 0.01 a | 0.94 ± 0.00 a | 0.78 ± 0.07 b |
| Margalef (M) | 4.68 ± 0.11 a | 4.64 ± 0.23 a | 3.34 ± 0.09 b |
| Menhinick (E) | 0.35 ± 0.01 a | 0.35 ± 0.02 a | 0.28 ± 0.04 b |
| Brillouin (B) | 3.23 ± 0.05 a | 3.12 ± 0.06 a | 2.65 ± 0.05 b |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.; Wang, M.; Gao, X.; Zhao, W.; Miao, P.; Liu, Y.; Zhang, R.; Wang, X.; Sui, X.; Li, M.-H. The Diversity and Composition of Soil Microbial Communities Differ in Three Land Use Types of the Sanjiang Plain, Northeastern China. Microorganisms 2024, 12, 780. https://doi.org/10.3390/microorganisms12040780
Wang S, Wang M, Gao X, Zhao W, Miao P, Liu Y, Zhang R, Wang X, Sui X, Li M-H. The Diversity and Composition of Soil Microbial Communities Differ in Three Land Use Types of the Sanjiang Plain, Northeastern China. Microorganisms. 2024; 12(4):780. https://doi.org/10.3390/microorganisms12040780
Chicago/Turabian StyleWang, Shenzheng, Mingyu Wang, Xin Gao, Wenqi Zhao, Puwen Miao, Yingnan Liu, Rongtao Zhang, Xin Wang, Xin Sui, and Mai-He Li. 2024. "The Diversity and Composition of Soil Microbial Communities Differ in Three Land Use Types of the Sanjiang Plain, Northeastern China" Microorganisms 12, no. 4: 780. https://doi.org/10.3390/microorganisms12040780
APA StyleWang, S., Wang, M., Gao, X., Zhao, W., Miao, P., Liu, Y., Zhang, R., Wang, X., Sui, X., & Li, M.-H. (2024). The Diversity and Composition of Soil Microbial Communities Differ in Three Land Use Types of the Sanjiang Plain, Northeastern China. Microorganisms, 12(4), 780. https://doi.org/10.3390/microorganisms12040780







