Current Advances in the Functional Diversity and Mechanisms Underlying Endophyte–Plant Interactions
Abstract
:1. Introduction
2. Systemic and Non-Systemic Endophytes
2.1. Definition of Systemic and Non-Systemic Endophytes
2.2. Comparative Genomics of Systemic Endophytes and Non-Systemic Endophytes
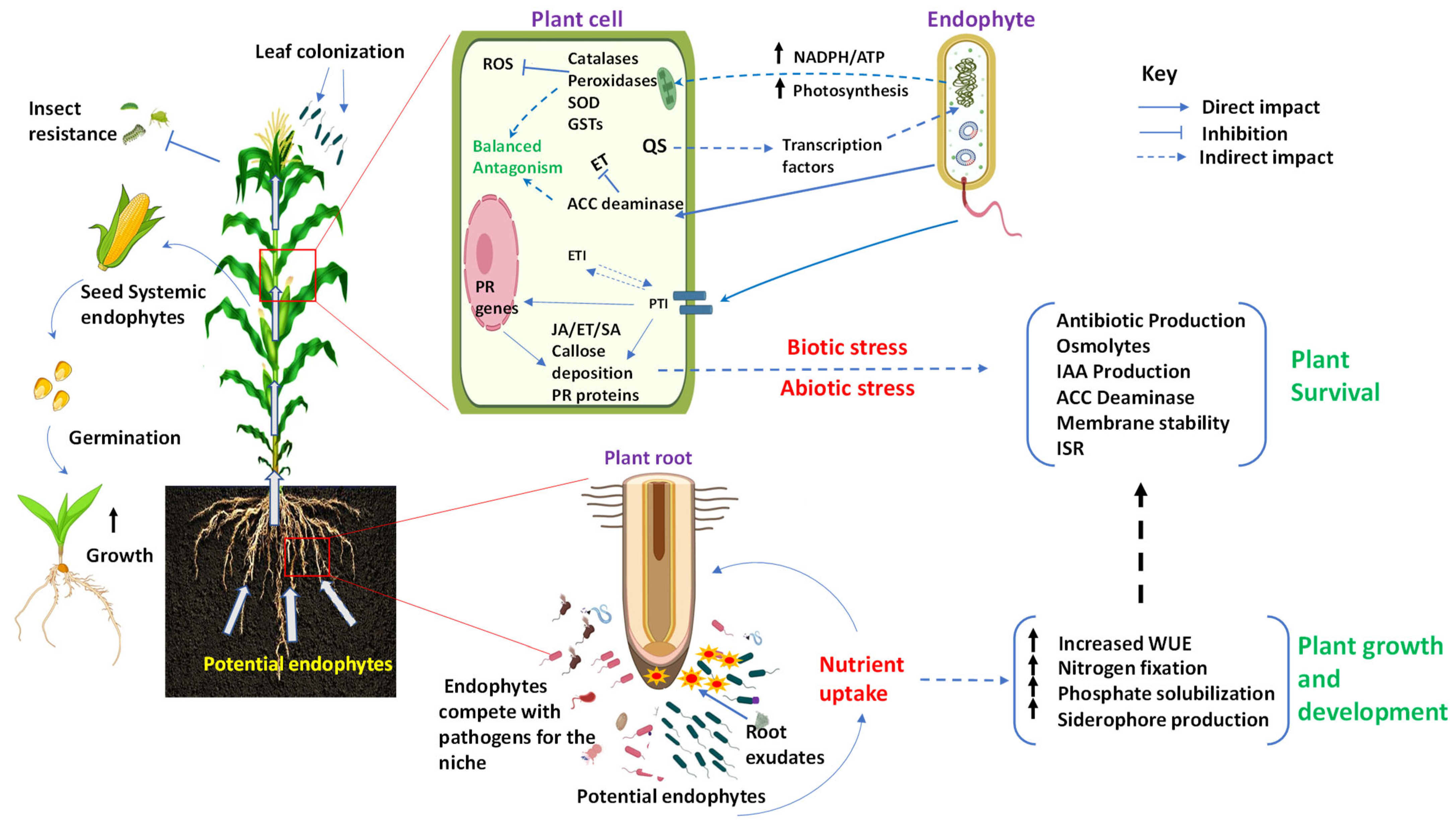
3. Impact of Endophytes on Host Plants
3.1. Promoting Plant Growth
3.1.1. Enhancing Nutrient Availability
3.1.2. Promoting the Biosynthesis of Phytohormones
3.2. Enhancing Stress Tolerance and Disease Resistance
3.2.1. Biotic Stresses
3.2.2. Abiotic Stress
4. Regulation of Endophytes by Host Plant
4.1. The Recruitment of Endophytes Regulated by Plant
4.1.1. Rhizosphere Metabolites from Host Plant
4.1.2. The Vertical Transmission of Endophytes from Plant
4.2. The Colonization and Growth of Endophytes Regulated by the Plant
4.2.1. Regulation of Plant Immune Response during Endophyte Colonization
4.2.2. The Nutrient Assimilation and Hormone Metabolism of Plant
4.2.3. Host Tissue Specificity of Endophytes
4.3. The Community Structure of Endophytes Regulated by The Plant
4.3.1. Physical Barrier Formation Pathways in Plants
4.3.2. Innate Immune Pathways in Plants
4.3.3. Metabolic Pathways Involved in Plant Secondary Metabolite Biosynthesis
4.3.4. Metabolic Pathway of Hormones
4.4. The Potential to Confer Novel Functions upon Endophytes
4.5. The Interconversion of Non-Systemic Endophytes across Different Life Cycles
4.6. Plant Host Genetic Loci Regulating Endophytes
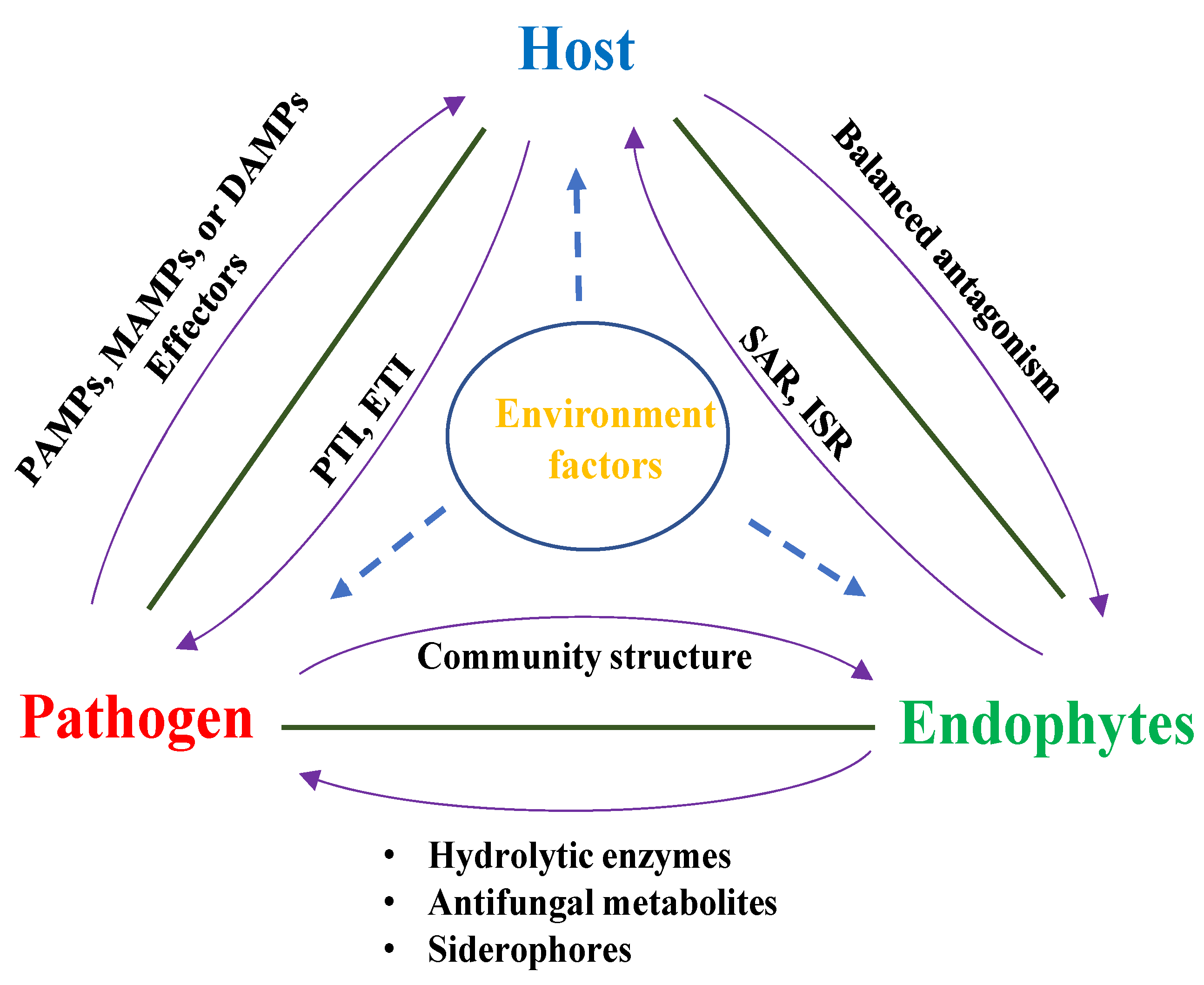
5. Host–Endophyte–Pathogen Interaction
5.1. Host–Endophyte–Pathogen Triangle Concept
5.2. The Endophyte Functions as an Extension of the Plant’s Immune System
6. Prospects of Endophytes in Sustainable Agricultural Development
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Turner, T.R.; James, E.K.; Poole, P.S. The Plant Microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, R.; Redman, R. More than 400 Million Years of Evolution and Some Plants Still Can’t Make It on Their Own: Plant Stress Tolerance via Fungal Symbiosis. J. Exp. Bot. 2008, 59, 1109–1114. [Google Scholar] [CrossRef] [PubMed]
- Wani, Z.A.; Ashraf, N.; Mohiuddin, T.; Riyaz-Ul-Hassan, S. Plant-Endophyte Symbiosis, an Ecological Perspective. Appl. Microbiol. Biotechnol. 2015, 99, 2955–2965. [Google Scholar] [CrossRef]
- del Carmen Orozco-Mosqueda, M.; Santoyo, G. Plant-Microbial Endophytes Interactions: Scrutinizing Their Beneficial Mechanisms from Genomic Explorations. Curr. Plant Biol. 2021, 25, 100189. [Google Scholar]
- Santoyo, G.; Moreno-Hagelsieb, G.; del Carmen Orozco-Mosqueda, M.; Glick, B.R. Plant Growth-Promoting Bacterial Endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Anand, U.; Pal, T.; Yadav, N.; Singh, V.K.; Tripathi, V.; Choudhary, K.K.; Shukla, A.K.; Sunita, K.; Kumar, A.; Bontempi, E. Current Scenario and Future Prospects of Endophytic Microbes: Promising Candidates for Abiotic and Biotic Stress Management for Agricultural and Environmental Sustainability. Microb. Ecol. 2023, 86, 1455–1486. [Google Scholar] [CrossRef] [PubMed]
- Mahdi, L.K.; Miyauchi, S.; Uhlmann, C.; Garrido-Oter, R.; Langen, G.; Wawra, S.; Niu, Y.; Guan, R.; Robertson-Albertyn, S.; Bulgarelli, D. The Fungal Root Endophyte Serendipita Vermifera Displays Inter-Kingdom Synergistic Beneficial Effects with the Microbiota in Arabidopsis Thaliana and Barley. ISME J. 2022, 16, 876–889. [Google Scholar] [CrossRef] [PubMed]
- Zamioudis, C.; Pieterse, C.M. Modulation of Host Immunity by Beneficial Microbes. Mol. Plant-Microbe Interact. 2012, 25, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, S.; Bhatt, A. Microbial Endophytes: Emerging Trends and Biotechnological Applications. Curr. Microbiol. 2023, 80, 249. [Google Scholar]
- Oukala, N.; Aissat, K.; Pastor, V. Bacterial Endophytes: The Hidden Actor in Plant Immune Responses against Biotic Stress. Plants 2021, 10, 1012. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, Y.-X.; Zhang, N.; Hu, B.; Jin, T.; Xu, H.; Qin, Y.; Yan, P.; Zhang, X.; Guo, X. NRT1. 1B Is Associated with Root Microbiota Composition and Nitrogen Use in Field-Grown Rice. Nat. Biotechnol. 2019, 37, 676–684. [Google Scholar] [CrossRef] [PubMed]
- Hassani, M.A.; Durán, P.; Hacquard, S. Microbial Interactions within the Plant Holobiont. Microbiome 2018, 6, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.K.; Meyer, T.E. Evolutionary Analysis by Whole-Genome Comparisons. J. Bacteriol. 2002, 184, 2260–2272. [Google Scholar] [CrossRef]
- Schardl, C.L.; Leuchtmann, A.; Spiering, M.J. Symbioses of Grasses with Seedborne Fungal Endophytes. Annu. Rev. Plant Biol. 2004, 55, 315–340. [Google Scholar] [CrossRef] [PubMed]
- Henry, L.P.; Bergelson, J. Evolutionary Implications of Host Genetic Control for Engineering Beneficial Microbiomes. Curr. Opin. Syst. Biol. 2023, 34, 100455. [Google Scholar] [CrossRef] [PubMed]
- Eida, A.A.; Ziegler, M.; Lafi, F.F.; Michell, C.T.; Voolstra, C.R.; Hirt, H.; Saad, M.M. Desert Plant Bacteria Reveal Host Influence and Beneficial Plant Growth Properties. PLoS ONE 2018, 13, e0208223. [Google Scholar] [CrossRef]
- Matsumoto, H.; Fan, X.; Wang, Y.; Kusstatscher, P.; Duan, J.; Wu, S.; Chen, S.; Qiao, K.; Wang, Y.; Ma, B. Bacterial Seed Endophyte Shapes Disease Resistance in Rice. Nat. Plants 2021, 7, 60–72. [Google Scholar] [CrossRef] [PubMed]
- Mengistu, A.A. Endophytes: Colonization, Behaviour, and Their Role in Defense Mechanism. Int. J. Microbiol. 2020, 2020, 6927219. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Van Themaat, E.V.L.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [PubMed]
- Truyens, S.; Weyens, N.; Cuypers, A.; Vangronsveld, J. Bacterial Seed Endophytes: Genera, Vertical Transmission and Interaction with Plants. Environ. Microbiol. Rep. 2015, 7, 40–50. [Google Scholar] [CrossRef]
- Lekshmi, R.; Sora, S.; Anith, K.; Soniya, E. Root Colonization by the Endophytic Fungus Piriformospora Indica Shortens the Juvenile Phase of Piper Nigrum L. by Fine Tuning the Floral Promotion Pathways. Front. Plant Sci. 2022, 13, 954693. [Google Scholar] [CrossRef] [PubMed]
- Nie, P.; Li, X.; Wang, S.; Guo, J.; Zhao, H.; Niu, D. Induced Systemic Resistance against Botrytis Cinerea by Bacillus Cereus AR156 through a JA/ET-and NPR1-Dependent Signaling Pathway and Activates PAMP-Triggered Immunity in Arabidopsis. Front. Plant Sci. 2017, 8, 247629. [Google Scholar] [CrossRef] [PubMed]
- Bai, B.; Liu, W.; Qiu, X.; Zhang, J.; Zhang, J.; Bai, Y. The Root Microbiome: Community Assembly and Its Contributions to Plant Fitness. J. Integr. Plant Biol. 2022, 64, 230–243. [Google Scholar] [CrossRef]
- Trdá, L.; Fernandez, O.; Boutrot, F.; Héloir, M.; Kelloniemi, J.; Daire, X.; Adrian, M.; Clément, C.; Zipfel, C.; Dorey, S. The Grapevine Flagellin Receptor VvFLS2 Differentially Recognizes Flagellin-derived Epitopes from the Endophytic Growth-promoting Bacterium Burkholderia Phytofirmans and Plant Pathogenic Bacteria. New Phytol. 2014, 201, 1371–1384. [Google Scholar] [CrossRef] [PubMed]
- Velásquez, A.C.; Huguet-Tapia, J.C.; He, S.Y. Shared in Planta Population and Transcriptomic Features of Nonpathogenic Members of Endophytic Phyllosphere Microbiota. Proc. Natl. Acad. Sci. USA 2022, 119, e2114460119. [Google Scholar] [CrossRef]
- Karpinets, T.V.; Park, B.H.; Syed, M.H.; Klotz, M.G.; Uberbacher, E.C. Metabolic Environments and Genomic Features Associated with Pathogenic and Mutualistic Interactions between Bacteria and Plants. Mol. Plant-Microbe Interact. 2014, 27, 664–677. [Google Scholar] [CrossRef] [PubMed]
- Saikkonen, K.; Faeth, S.H.; Helander, M.; Sullivan, T.J. Fungal endophytes: A continuum of interactions with host plants. Annu. Rev. Ecol. Syst. 1998, 29, 319–343. [Google Scholar] [CrossRef]
- Schulz, B.; Boyle, C. The Endophytic Continuum. Mycol. Res. 2005, 109, 661–686. [Google Scholar] [CrossRef] [PubMed]
- Khare, E.; Mishra, J.; Arora, N.K. Multifaceted interactions between endophytes and plant: Developments and prospects. Front. Microbiol. 2018, 9, 411604. [Google Scholar] [CrossRef] [PubMed]
- Pathak, P.; Rai, V.K.; Can, H.; Singh, S.K.; Kumar, D.; Bhardwaj, N.; Kumar, A. Plant-endophyte interaction during biotic stress management. Plants 2022, 11, 2203. [Google Scholar] [CrossRef]
- Sena, L.; Mica, E.; Valè, G.; Vaccino, P.; Pecchioni, N. Exploring the potential of endophyte-plant interactions for improving crop sustainable yields in a changing climate. Front. Plant Sci. 2024, 15, 1349401. [Google Scholar] [CrossRef]
- Chitnis, V.R.; Suryanarayanan, T.S.; Nataraja, K.N.; Prasad, S.R.; Oelmüller, R.; Shaanker, R.U. Fungal endophyte-mediated crop improvement: The way ahead. Front. Plant Sci. 2020, 11, 561007. [Google Scholar] [CrossRef]
- Watts, D.; Palombo, E.A.; Jaimes Castillo, A.; Zaferanloo, B. Endophytes in agriculture: Potential to improve yields and tolerances of agricultural crops. Microorganisms 2023, 11, 1276. [Google Scholar] [CrossRef] [PubMed]
- White, J.F.; Kingsley, K.L.; Zhang, Q.; Verma, R.; Obi, N.; Dvinskikh, S.; Kowalski, K.P. Endophytic microbes and their potential applications in crop management. Pest Manag. Sci. 2019, 75, 2558–2565. [Google Scholar] [CrossRef] [PubMed]
- Botella, L.; Diez, J.J. Phylogenic Diversity of Fungal Endophytes in Spanish Stands of Pinus Halepensis. Fungal Divers. 2011, 47, 9–18. [Google Scholar]
- Herre, E.; Knowlton, N.; Mueller, U.; Rehner, S. Patterns of Ecological Transmission and Evolutionary Association. Trends Ecol. Evol. 1999, 14, 49–53. [Google Scholar] [CrossRef] [PubMed]
- Soliman, S.S.; Trobacher, C.P.; Tsao, R.; Greenwood, J.S.; Raizada, M.N. A Fungal Endophyte Induces Transcription of Genes Encoding a Redundant Fungicide Pathway in Its Host Plant. BMC Plant Biol. 2013, 13, 93. [Google Scholar] [CrossRef]
- Christensen, M.J.; Bennett, R.J.; Ansari, H.A.; Koga, H.; Johnson, R.D.; Bryan, G.T.; Simpson, W.R.; Koolaard, J.P.; Nickless, E.M.; Voisey, C.R. Epichloë Endophytes Grow by Intercalary Hyphal Extension in Elongating Grass Leaves. Fungal Genet. Biol. 2008, 45, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Higgins, K.L.; Arnold, A.E.; Coley, P.D.; Kursar, T.A. Communities of Fungal Endophytes in Tropical Forest Grasses: Highly Diverse Host-and Habitat Generalists Characterized by Strong Spatial Structure. Fungal Ecol. 2014, 8, 1–11. [Google Scholar] [CrossRef]
- Załuga, J.; Stragier, P.; Baeyen, S.; Haegeman, A.; Van Vaerenbergh, J.; Maes, M.; De Vos, P. Comparative Genome Analysis of Pathogenic and Non-Pathogenic Clavibacter Strains Reveals Adaptations to Their Lifestyle. BMC Genom. 2014, 15, 392. [Google Scholar] [CrossRef] [PubMed]
- Mitter, B.; Petric, A.; Shin, M.W.; Chain, P.S.; Hauberg-Lotte, L.; Reinhold-Hurek, B.; Nowak, J.; Sessitsch, A. Comparative Genome Analysis of Burkholderia Phytofirmans PsJN Reveals a Wide Spectrum of Endophytic Lifestyles Based on Interaction Strategies with Host Plants. Front. Plant Sci. 2013, 4, 120. [Google Scholar] [CrossRef] [PubMed]
- Bertalan, M.; Albano, R.; de Pádua, V.; Rouws, L.; Rojas, C.; Hemerly, A.; Teixeira, K.; Schwab, S.; Araujo, J.; Oliveira, A. Complete Genome Sequence of the Sugarcane Nitrogen-Fixing Endophyte Gluconacetobacter Diazotrophicus Pal5. BMC Genom. 2009, 10, 450. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Zhu, Y.; Chang, H.; Wang, C.; Yang, J.; Shi, J.; Gao, J.; Yang, W.; Lan, L.; Wang, Y. An SHR–SCR Module Specifies Legume Cortical Cell Fate to Enable Nodulation. Nature 2021, 589, 586–590. [Google Scholar] [CrossRef] [PubMed]
- Santi, C.; Bogusz, D.; Franche, C. Biological Nitrogen Fixation in Non-Legume Plants. Ann. Bot. 2013, 111, 743–767. [Google Scholar] [CrossRef] [PubMed]
- Schulte, C.C.; Borah, K.; Wheatley, R.M.; Terpolilli, J.J.; Saalbach, G.; Crang, N.; de Groot, D.H.; Ratcliffe, R.G.; Kruger, N.J.; Papachristodoulou, A. Metabolic Control of Nitrogen Fixation in Rhizobium-Legume Symbioses. Sci. Adv. 2021, 7, eabh2433. [Google Scholar] [CrossRef]
- Chaudhary, R.; Kumar, V.; Gupta, S.; Naik, B.; Prasad, R.; Mishra, S.; Saris, P.E.J.; Kumar, V. Finger Millet (Eleusine coracana) Plant–Endophyte Dynamics: Plant Growth, Nutrient Uptake, and Zinc Biofortification. Microorganisms 2023, 11, 973. [Google Scholar] [CrossRef] [PubMed]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial Endophyte Colonization and Distribution within Plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef] [PubMed]
- Oteino, N.; Lally, R.D.; Kiwanuka, S.; Lloyd, A.; Ryan, D.; Germaine, K.J.; Dowling, D.N. Plant Growth Promotion Induced by Phosphate Solubilizing Endophytic Pseudomonas Isolates. Front. Microbiol. 2015, 6, 745. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Lou, K.; Li, C. Growth and Photosynthetic Efficiency Promotion of Sugar Beet (Beta Vulgaris L.) by Endophytic Bacteria. Photosynth. Res. 2010, 105, 5–13. [Google Scholar] [CrossRef] [PubMed]
- Almuhayawi, M.S.; Abdel-Mawgoud, M.; Al Jaouni, S.K.; Almuhayawi, S.M.; Alruhaili, M.H.; Selim, S.; AbdElgawad, H. Bacterial Endophytes as a Promising Approach to Enhance the Growth and Accumulation of Bioactive Metabolites of Three Species of Chenopodium Sprouts. Plants 2021, 10, 2745. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.-M.; Radhakrishnan, R.; You, Y.-H.; Joo, G.-J.; Lee, I.-J.; Lee, K.-E.; Kim, J.-H. Phosphate Solubilizing Bacillus Megaterium Mj1212 Regulates Endogenous Plant Carbohydrates and Amino Acids Contents to Promote Mustard Plant Growth. Indian J. Microbiol. 2014, 54, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Spaepen, S.; Vanderleyden, J. Auxin and Plant-Microbe Interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a001438. [Google Scholar] [CrossRef] [PubMed]
- Zúñiga, A.; Poupin, M.J.; Donoso, R.; Ledger, T.; Guiliani, N.; Gutiérrez, R.A.; González, B. Quorum Sensing and Indole-3-Acetic Acid Degradation Play a Role in Colonization and Plant Growth Promotion of Arabidopsis Thaliana by Burkholderia Phytofirmans PsJN. Mol. Plant-Microbe Interact. 2013, 26, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, R.; Waqas, M.; Khan, A.L.; Asaf, S.; Khan, M.A.; Kang, S.-M.; Yun, B.-W.; Lee, I.-J. Seed-Borne Endophytic Bacillus Amyloliquefaciens RWL-1 Produces Gibberellins and Regulates Endogenous Phytohormones of Oryza sativa. Plant Physiol. Biochem. 2016, 106, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Bhore, S.J.; Ravichantar, N.; Loh, C.Y. Screening of Endophytic Bacteria Isolated from Leaves of Sambung Nyawa [Gynura Procumbens (Lour.) Merr.] for Cytokinin-like Compounds. Bioinformation 2010, 5, 191. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xiong, K.; Wen, W.; Li, L.; Xu, D. Functional Endophytes Regulating Plant Secondary Metabolism: Current Status, Prospects and Applications. Int. J. Mol. Sci. 2023, 24, 1153. [Google Scholar] [CrossRef] [PubMed]
- Van Loon, L.C.; Bakker, P.A.; van der Heijdt, W.H.; Wendehenne, D.; Pugin, A. Early Responses of Tobacco Suspension Cells to Rhizobacterial Elicitors of Induced Systemic Resistance. Mol. Plant-Microbe Interact. 2008, 21, 1609–1621. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Hao, H.; Lu, X.; Zhao, X.; Wang, Y.; Zhang, Y.; Xie, Z.; Wang, R. Transcriptome Profiling of Genes Involved in Induced Systemic Salt Tolerance Conferred by Bacillus Amyloliquefaciens FZB42 in Arabidopsis Thaliana. Sci. Rep. 2017, 7, 10795. [Google Scholar] [CrossRef]
- Ek-Ramos, M.J.; Gomez-Flores, R.; Orozco-Flores, A.A.; Rodríguez-Padilla, C.; González-Ochoa, G.; Tamez-Guerra, P. Bioactive Products from Plant-Endophytic Gram-Positive Bacteria. Front. Microbiol. 2019, 10, 433349. [Google Scholar] [CrossRef] [PubMed]
- Tymon, L.S.; Morgan, P.; Gundersen, B.; Inglis, D.A. Potential of Endophytic Fungi Collected from Cucurbita Pepo Roots Grown under Three Different Agricultural Mulches as Antagonistic Endophytes to Verticillium Dahliae in Western Washington. Microbiol. Res. 2020, 240, 126535. [Google Scholar] [CrossRef] [PubMed]
- Kupferschmied, P.; Maurhofer, M.; Keel, C. Promise for Plant Pest Control: Root-Associated Pseudomonads with Insecticidal Activities. Front. Plant Sci. 2013, 4, 55361. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Fei, Y.J.; Wu, Y.B.; Luo, D.L.; Chen, M.; Sun, K.; Dai, C.C. Endophytic fungus reshapes spikelet microbiome to reduce mycotoxin produced by Fusarium proliferatum through altering rice metabolites. J. Agric. Food Chem. 2023, 71, 11350–11364. [Google Scholar] [CrossRef] [PubMed]
- Miljaković, D.; Marinković, J.; Balešević-Tubić, S. The Significance of Bacillus Spp. in Disease Suppression and Growth Promotion of Field and Vegetable Crops. Microorganisms 2020, 8, 1037. [Google Scholar] [CrossRef] [PubMed]
- Olowe, O.M.; Akanmu, A.O.; Asemoloye, M.D. Exploration of Microbial Stimulants for Induction of Systemic Resistance in Plant Disease Management. Ann. Appl. Biol. 2020, 177, 282–293. [Google Scholar] [CrossRef]
- Glick, B.R. Bacteria with ACC Deaminase Can Promote Plant Growth and Help to Feed the World. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Ravanbakhsh, M.; Kowalchuk, G.A.; Jousset, A. Root-Associated Microorganisms Reprogram Plant Life History along the Growth–Stress Resistance Tradeoff. ISME J. 2019, 13, 3093–3101. [Google Scholar] [CrossRef] [PubMed]
- de Bruijn, F.J. Molecular Microbial Ecology of the Rhizosphere; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013. [Google Scholar]
- Ali, A.H.; Radwan, U.; El-Zayat, S.; El-Sayed, M.A. The Role of the Endophytic Fungus, Thermomyces Lanuginosus, on Mitigation of Heat Stress to Its Host Desert Plant Cullen Plicata. Biol. Futur. 2019, 70, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xie, Z.; Zhang, X.; Lang, D.; Zhang, X. Growth-Promoting Bacteria Alleviates Drought Stress of G. Uralensis through Improving Photosynthesis Characteristics and Water Status. J. Plant Interact. 2019, 14, 580–589. [Google Scholar] [CrossRef]
- Ismail, A.H.; Mehmood, A.; Qadir, M.; Husna, A.I.; Hamayun, M.; Khan, N. Thermal Stress Alleviating Potential of Endophytic Fungus Rhizopus Oryzae Inoculated to Sunflower (Helianthus Annuus L.) and Soybean (Glycine Max L.). Pak. J. Bot 2020, 52, 1857–1865. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Asaf, S.; Khan, A.; Adhikari, A.; Jan, R.; Ali, S.; Imran, M.; Kim, K.; Lee, I. Plant Growth-promoting Endophytic Bacteria Augment Growth and Salinity Tolerance in Rice Plants. Plant Biol. 2020, 22, 850–862. [Google Scholar] [CrossRef] [PubMed]
- Azizi, M.; Fard, E.M.; Ghabooli, M. Piriformospora Indica Affect Drought Tolerance by Regulation of Genes Expression and Some Morphophysiological Parameters in Tomato (Solanum Lycopersicum L.). Sci. Hortic. 2021, 287, 110260. [Google Scholar] [CrossRef]
- Bouzouina, M.; Kouadria, R.; Lotmani, B. Fungal Endophytes Alleviate Salt Stress in Wheat in Terms of Growth, Ion Homeostasis and Osmoregulation. J. Appl. Microbiol. 2021, 130, 913–925. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.R.; Dai, L.; Xu, G.F.; Wang, H.S. A Strain of Phoma Species Improves Drought Tolerance of Pinus Tabulaeformis. Sci. Rep. 2021, 11, 7637. [Google Scholar] [CrossRef] [PubMed]
- Ravi, R.K.; Valli, P.P.S.; Muthukumar, T. Physiological Characterization of Root Endophytic Fusarium Haematococcum for Hydrolytic Enzyme Production, Nutrient Solubilization and Salinity Tolerance. Biocatal. Agric. Biotechnol. 2022, 43, 102392. [Google Scholar]
- Zhang, Y.; Tian, Z.; Xi, Y.; Wang, X.; Chen, S.; He, M.; Chen, Y.; Guo, Y. Improvement of Salt Tolerance of Arabidopsis Thaliana Seedlings Inoculated with Endophytic Bacillus Cereus KP120. J. Plant Interact. 2022, 17, 884–893. [Google Scholar] [CrossRef]
- Castrillo, G.; Teixeira, P.J.P.L.; Paredes, S.H.; Law, T.F.; De Lorenzo, L.; Feltcher, M.E.; Finkel, O.M.; Breakfield, N.W.; Mieczkowski, P.; Jones, C.D. Root Microbiota Drive Direct Integration of Phosphate Stress and Immunity. Nature 2017, 543, 513–518. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Navarro, D.N.; Dardanelli, M.S.; Ruíz-Saínz, J.E. Attachment of Bacteria to the Roots of Higher Plants. FEMS Microbiol. Lett. 2007, 272, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Bacilio-Jiménez, M.; Aguilar-Flores, S.; Ventura-Zapata, E.; Pérez-Campos, E.; Bouquelet, S.; Zenteno, E. Chemical Characterization of Root Exudates from Rice (Oryza Sativa) and Their Effects on the Chemotactic Response of Endophytic Bacteria. Plant Soil 2003, 249, 271–277. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E. Revealing Structure and Assembly Cues for Arabidopsis Root-Inhabiting Bacterial Microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Kawasaki, A.; Donn, S.; Ryan, P.R.; Mathesius, U.; Devilla, R.; Jones, A.; Watt, M. Microbiome and Exudates of the Root and Rhizosphere of Brachypodium Distachyon, a Model for Wheat. PLoS ONE 2016, 11, e0164533. [Google Scholar] [CrossRef]
- Yeoh, Y.K.; Dennis, P.G.; Paungfoo-Lonhienne, C.; Weber, L.; Brackin, R.; Ragan, M.A.; Schmidt, S.; Hugenholtz, P. Evolutionary Conservation of a Core Root Microbiome across Plant Phyla along a Tropical Soil Chronosequence. Nat. Commun. 2017, 8, 215. [Google Scholar] [CrossRef] [PubMed]
- Nguema-Ona, E.; Vicré-Gibouin, M.; Cannesan, M.-A.; Driouich, A. Arabinogalactan Proteins in Root–Microbe Interactions. Trends Plant Sci. 2013, 18, 440–449. [Google Scholar] [CrossRef]
- Kost, T.; Stopnisek, N.; Agnoli, K.; Eberl, L.; Weisskopf, L. Oxalotrophy, a Widespread Trait of Plant-Associated Burkholderia Species, Is Involved in Successful Root Colonization of Lupin and Maize by Burkholderia Phytofirmans. Front. Microbiol. 2014, 4, 421. [Google Scholar] [CrossRef]
- Ma, L.; Wang, W.-Q.; Shi, R.; Zhang, X.-M.; Li, X.; Yang, Y.-S.; Mo, M.H. Effects of Organic Acids on the Chemotaxis Profiles and Biocontrol Traits of Antagonistic Bacterial Endophytes against Root-Rot Disease in Panax Notoginseng. Antonie Van Leeuwenhoek 2021, 114, 1771–1789. [Google Scholar] [CrossRef] [PubMed]
- Barret, M.; Briand, M.; Bonneau, S.; Préveaux, A.; Valière, S.; Bouchez, O.; Hunault, G.; Simoneau, P.; Jacques, M.-A. Emergence Shapes the Structure of the Seed Microbiota. Appl. Environ. Microbiol. 2015, 81, 1257–1266. [Google Scholar] [CrossRef] [PubMed]
- Bziuk, N.; Maccario, L.; Straube, B.; Wehner, G.; Sørensen, S.J.; Schikora, A.; Smalla, K. The Treasure inside Barley Seeds: Microbial Diversity and Plant Beneficial Bacteria. Environ. Microbiome 2021, 16, 20. [Google Scholar] [CrossRef]
- Gagic, M.; Faville, M.J.; Zhang, W.; Forester, N.T.; Rolston, M.P.; Johnson, R.D.; Voisey, C.R. Seed transmission of Epichloë endophytes in Lolium perenne is heavily influenced by host genetics. Front. Plant Sci. 2018, 9, 1580. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zuo, S.; Xu, L.; Zou, Y.; Song, W. Study on Diversity of Endophytic Bacterial Communities in Seeds of Hybrid Maize and Their Parental Lines. Arch. Microbiol. 2012, 194, 1001–1012. [Google Scholar] [PubMed]
- Cregger, M.; Veach, A.; Yang, Z.; Crouch, M.; Vilgalys, R.; Tuskan, G.; Schadt, C. The Populus Holobiont: Dissecting the Effects of Plant Niches and Genotype on the Microbiome. Microbiome 2018, 6, 31. [Google Scholar] [CrossRef]
- Wei, Z.; Gu, Y.; Friman, V.-P.; Kowalchuk, G.A.; Xu, Y.; Shen, Q.; Jousset, A. Initial Soil Microbiome Composition and Functioning Predetermine Future Plant Health. Sci. Adv. 2019, 5, eaaw0759. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.A.; Santos-Medellín, C.M.; Liechty, Z.S.; Nguyen, B.; Lurie, E.; Eason, S.; Phillips, G.; Sundaresan, V. Compositional Shifts in Root-Associated Bacterial and Archaeal Microbiota Track the Plant Life Cycle in Field-Grown Rice. PLoS Biol. 2018, 16, e2003862. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.R.; Lundberg, D.S.; Del Rio, T.G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host Genotype and Age Shape the Leaf and Root Microbiomes of a Wild Perennial Plant. Nat. Commun. 2016, 7, 12151. [Google Scholar] [CrossRef] [PubMed]
- Johnston-Monje, D.; Raizada, M.N. Conservation and Diversity of Seed Associated Endophytes in Zea across Boundaries of Evolution, Ethnography and Ecology. PLoS ONE 2011, 6, e20396. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; del Rio, T.G. Defining the Core Arabidopsis Thaliana Root Microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef]
- Jin, T.; Wang, Y.; Huang, Y.; Xu, J.; Zhang, P.; Wang, N.; Liu, X.; Chu, H.; Liu, G.; Jiang, H. Taxonomic Structure and Functional Association of Foxtail Millet Root Microbiome. Gigascience 2017, 6, 1–12. [Google Scholar] [CrossRef]
- Hamonts, K.; Trivedi, P.; Garg, A.; Janitz, C.; Grinyer, J.; Holford, P.; Botha, F.C.; Anderson, I.C.; Singh, B.K. Field Study Reveals Core Plant Microbiota and Relative Importance of Their Drivers. Environ. Microbiol. 2018, 20, 124–140. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, Y.; Zhang, P.; Trivedi, P.; Riera, N.; Wang, Y.; Liu, X.; Fan, G.; Tang, J.; Coletta-Filho, H.D. The Structure and Function of the Global Citrus Rhizosphere Microbiome. Nat. Commun. 2018, 9, 4894. [Google Scholar] [CrossRef] [PubMed]
- Bloemberg, G.V.; Wijfjes, A.H.; Lamers, G.E.; Stuurman, N.; Lugtenberg, B.J. Simultaneous Imaging of Pseudomonas Fluorescens WCS365 Populations Expressing Three Different Autofluorescent Proteins in the Rhizosphere: New Perspectives for Studying Microbial Communities. Mol. Plant-Microbe Interact. 2000, 13, 1170–1176. [Google Scholar] [CrossRef] [PubMed]
- Gamalero, E.; Lingua, G.; Berta, G.; Lemanceau, P.P. Methods for Studying Root Colonization by Introduced Beneficial Bacteria. Agronomie 2003, 23, 407–418. [Google Scholar] [CrossRef]
- Compant, S.; Clément, C.; Sessitsch, A. Plant Growth-Promoting Bacteria in the Rhizo-and Endosphere of Plants: Their Role, Colonization, Mechanisms Involved and Prospects for Utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The Plant Immune System. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef] [PubMed]
- Dodds, P.N.; Rathjen, J.P. Plant Immunity: Towards an Integrated View of Plant–Pathogen Interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.-M.; He, S.Y.; Xin, X.-F. Pattern-Recognition Receptors Are Required for NLR-Mediated Plant Immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef] [PubMed]
- Ngou, B.P.M.; Ahn, H.-K.; Ding, P.; Jones, J.D. Mutual Potentiation of Plant Immunity by Cell-Surface and Intracellular Receptors. Nature 2021, 592, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Plett, J.M.; Martin, F.M. Know Your Enemy, Embrace Your Friend: Using Omics to Understand How Plants Respond Differently to Pathogenic and Mutualistic Microorganisms. Plant J. 2018, 93, 729–746. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S.; Kracher, B.; Hiruma, K.; Münch, P.C.; Garrido-Oter, R.; Thon, M.R.; Weimann, A.; Damm, U.; Dallery, J.-F.; Hainaut, M. Survival Trade-Offs in Plant Roots during Colonization by Closely Related Beneficial and Pathogenic Fungi. Nat. Commun. 2016, 7, 11362. [Google Scholar] [CrossRef]
- Zhang, S.; Wei, W.; Zhu, W.; Yu, Y.; Chen, P. De Novo Analysis of Wolfiporia Cocos Transcriptome to Reveal the Differentially Expressed Carbohydrate-Active Enzymes (CAZymes) Genes during the Early Stage of Sclerotial Growth. Front. Microbiol. 2016, 7, 178337. [Google Scholar] [CrossRef] [PubMed]
- Tisserant, E.; Malbreil, M.; Kuo, A.; Kohler, A.; Symeonidi, A.; Balestrini, R.; Charron, P.; Duensing, N.; Frei dit Frey, N.; Gianinazzi-Pearson, V. Genome of an Arbuscular Mycorrhizal Fungus Provides Insight into the Oldest Plant Symbiosis. Proc. Natl. Acad. Sci. USA 2013, 110, 20117–20122. [Google Scholar] [CrossRef] [PubMed]
- Kohler, A.; Kuo, A.; Nagy, L.G.; Morin, E.; Barry, K.W.; Buscot, F.; Canbäck, B.; Choi, C.; Cichocki, N.; Clum, A. Convergent Losses of Decay Mechanisms and Rapid Turnover of Symbiosis Genes in Mycorrhizal Mutualists. Nat. Genet. 2015, 47, 410–415. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.; Kohler, A.; Murat, C.; Veneault-Fourrey, C.; Hibbett, D.S. Unearthing the Roots of Ectomycorrhizal Symbioses. Nat. Rev. Microbiol. 2016, 14, 760–773. [Google Scholar] [CrossRef] [PubMed]
- Cord-Landwehr, S.; Melcher, R.L.; Kolkenbrock, S.; Moerschbacher, B.M. A Chitin Deacetylase from the Endophytic Fungus Pestalotiopsis Sp. Efficiently Inactivates the Elicitor Activity of Chitin Oligomers in Rice Cells. Sci. Rep. 2016, 6, 38018. [Google Scholar] [CrossRef] [PubMed]
- Bonfante, P.; Genre, A. Mechanisms Underlying Beneficial Plant–Fungus Interactions in Mycorrhizal Symbiosis. Nat. Commun. 2010, 1, 48. [Google Scholar] [CrossRef] [PubMed]
- Hiruma, K.; Gerlach, N.; Sacristán, S.; Nakano, R.T.; Hacquard, S.; Kracher, B.; Neumann, U.; Ramírez, D.; Bucher, M.; O’Connell, R.J. Root Endophyte Colletotrichum Tofieldiae Confers Plant Fitness Benefits That Are Phosphate Status Dependent. Cell 2016, 165, 464–474. [Google Scholar] [CrossRef] [PubMed]
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; Glavina del Rio, T.; Jones, C.D.; Tringe, S.G. Salicylic Acid Modulates Colonization of the Root Microbiome by Specific Bacterial Taxa. Science 2015, 349, 860–864. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Nester, E.W. Indoleacetic Acid, a Product of Transferred DNA, Inhibits Vir Gene Expression and Growth of Agrobacterium Tumefaciens C58. Proc. Natl. Acad. Sci. USA 2006, 103, 4658–4662. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Yang, S.; Meng, L.; Wang, B.-G. The Plant Hormone Abscisic Acid Regulates the Growth and Metabolism of Endophytic Fungus Aspergillus Nidulans. Sci. Rep. 2018, 8, 6504. [Google Scholar] [CrossRef] [PubMed]
- Du, W.; Yao, Z.; Li, J.; Sun, C.; Xia, J.; Wang, B.; Shi, D.; Ren, L. Diversity and Antimicrobial Activity of Endophytic Fungi Isolated from Securinega Suffruticosa in the Yellow River Delta. PLoS ONE 2020, 15, e0229589. [Google Scholar] [CrossRef]
- Bouffaud, M.; Poirier, M.; Muller, D.; Moënne-Loccoz, Y. Root Microbiome Relates to Plant Host Evolution in Maize and Other P Oaceae. Environ. Microbiol. 2014, 16, 2804–2814. [Google Scholar] [CrossRef]
- Schlaeppi, K.; Dombrowski, N.; Oter, R.G.; Ver Loren van Themaat, E.; Schulze-Lefert, P. Quantitative Divergence of the Bacterial Root Microbiota in Arabidopsis Thaliana Relatives. Proc. Natl. Acad. Sci. USA 2014, 111, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Reisberg, E.E.; Hildebrandt, U.; Riederer, M.; Hentschel, U. Distinct Phyllosphere Bacterial Communities on Arabidopsis Wax Mutant Leaves. PLoS ONE 2013, 8, e78613. [Google Scholar] [CrossRef] [PubMed]
- Bodenhausen, N.; Bortfeld-Miller, M.; Ackermann, M.; Vorholt, J.A. A Synthetic Community Approach Reveals Plant Genotypes Affecting the Phyllosphere Microbiota. PLoS Genet. 2014, 10, e1004283. [Google Scholar] [CrossRef]
- Lü, S.; Zhao, H.; Des Marais, D.L.; Parsons, E.P.; Wen, X.; Xu, X.; Bangarusamy, D.K.; Wang, G.; Rowland, O.; Juenger, T. Arabidopsis ECERIFERUM9 Involvement in Cuticle Formation and Maintenance of Plant Water Status. Plant Physiol. 2012, 159, 930–944. [Google Scholar] [CrossRef]
- Salas-González, I.; Reyt, G.; Flis, P.; Custódio, V.; Gopaulchan, D.; Bakhoum, N.; Dew, T.P.; Suresh, K.; Franke, R.B.; Dangl, J.L. Coordination between Microbiota and Root Endodermis Supports Plant Mineral Nutrient Homeostasis. Science 2021, 371, eabd0695. [Google Scholar] [CrossRef] [PubMed]
- Oldstone-Jackson, C.; Huang, F.; Bergelson, J. Microbe-Associated Molecular Pattern Recognition Receptors Have Little Effect on Endophytic Arabidopsis Thaliana Microbiome Assembly in the Field. Front. Plant Sci. 2023, 14, 1276472. [Google Scholar] [CrossRef]
- Chen, T.; Nomura, K.; Wang, X.; Sohrabi, R.; Xu, J.; Yao, L.; Paasch, B.C.; Ma, L.; Kremer, J.; Cheng, Y. A Plant Genetic Network for Preventing Dysbiosis in the Phyllosphere. Nature 2020, 580, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T. Assembly and Ecological Function of the Root Microbiome across Angiosperm Plant Species. Proc. Natl. Acad. Sci. USA 2018, 115, E1157–E1165. [Google Scholar] [CrossRef] [PubMed]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D. Hormone Crosstalk in Plant Disease and Defense: More than Just Jasmonate-Salicylate Antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef] [PubMed]
- Boachon, B.; Burdloff, Y.; Ruan, J.-X.; Rojo, R.; Junker, R.R.; Vincent, B.; Nicolè, F.; Bringel, F.; Lesot, A.; Henry, L. A Promiscuous CYP706A3 Reduces Terpene Volatile Emission from Arabidopsis Flowers, Affecting Florivores and the Floral Microbiome. Plant Cell 2019, 31, 2947–2972. [Google Scholar] [CrossRef]
- Brachi, B.; Filiault, D.; Whitehurst, H.; Darme, P.; Le Gars, P.; Le Mentec, M.; Morton, T.C.; Kerdaffrec, E.; Rabanal, F.; Anastasio, A. Plant Genetic Effects on Microbial Hubs Impact Host Fitness in Repeated Field Trials. Proc. Natl. Acad. Sci. USA 2022, 119, e2201285119. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Singh, S.K.; Peng, L.; Kaushal, R.; Vílchez, J.I.; Shao, C.; Wu, X.; Zheng, S.; Morcillo, R.J.; Paré, P.W. Flavonoid-Attracted Aeromonas Sp. from the Arabidopsis Root Microbiome Enhances Plant Dehydration Resistance. ISME J. 2022, 16, 2622–2632. [Google Scholar] [CrossRef] [PubMed]
- Harbort, C.J.; Hashimoto, M.; Inoue, H.; Niu, Y.; Guan, R.; Rombolà, A.D.; Kopriva, S.; Voges, M.J.; Sattely, E.S.; Garrido-Oter, R. Root-Secreted Coumarins and the Microbiota Interact to Improve Iron Nutrition in Arabidopsis. Cell Host Microbe 2020, 28, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.C.; Jiang, T.; Liu, Y.-X.; Bai, Y.-C.; Reed, J.; Qu, B.; Goossens, A.; Nützmann, H.-W.; Bai, Y.; Osbourn, A. A Specialized Metabolic Network Selectively Modulates Arabidopsis Root Microbiota. Science 2019, 364, eaau6389. [Google Scholar] [CrossRef]
- Kudjordjie, E.N.; Sapkota, R.; Steffensen, S.K.; Fomsgaard, I.S.; Nicolaisen, M. Maize Synthesized Benzoxazinoids Affect the Host Associated Microbiome. Microbiome 2019, 7, 59. [Google Scholar] [CrossRef] [PubMed]
- Lopes, L.D.; Wang, P.; Futrell, S.L.; Schachtman, D.P. Sugars and Jasmonic Acid Concentration in Root Exudates Affect Maize Rhizosphere Bacterial Communities. Appl. Environ. Microbiol. 2022, 88, e00971-22. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Teplitski, M.; Robinson, J.B.; Bauer, W.D. Production of Substances by Medicago Truncatula That Affect Bacterial Quorum Sensing. Mol. Plant-Microbe Interact. 2003, 16, 827–834. [Google Scholar] [CrossRef] [PubMed]
- Roman-Reyna, V.; Pinili, D.; Borja, F.N.; Quibod, I.L.; Groen, S.C.; Mulyaningsih, E.S.; Rachmat, A.; Slamet-Loedin, I.H.; Alexandrov, N.; Mauleon, R. The Rice Leaf Microbiome Has a Conserved Community Structure Controlled by Complex Host-Microbe Interactions. BioRxiv 2019, 615278. [Google Scholar] [CrossRef]
- Takeda, N.; Handa, Y.; Tsuzuki, S.; Kojima, M.; Sakakibara, H.; Kawaguchi, M. Gibberellins Interfere with Symbiosis Signaling and Gene Expression and Alter Colonization by Arbuscular Mycorrhizal Fungi in Lotus Japonicus. Plant Physiol. 2015, 167, 545–557. [Google Scholar] [CrossRef] [PubMed]
- Thrall, P.H.; Hochberg, M.E.; Burdon, J.J.; Bever, J.D. Coevolution of Symbiotic Mutualists and Parasites in a Community Context. Trends Ecol. Evol. 2007, 22, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Bae, H. Horizontal Gene Transfer and Endophytes: An Implication for the Acquisition of Novel Traits. Plants 2020, 9, 305. [Google Scholar] [CrossRef] [PubMed]
- Mc Ginty, S.E.; Rankin, D.J.; Brown, S.P. Horizontal Gene Transfer and the Evolution of Bacterial Cooperation. Evolution 2011, 65, 21–32. [Google Scholar] [CrossRef]
- Gu, Y.; Wang, Y.; Sun, Y.; Zhao, K.; Xiang, Q.; Yu, X.; Zhang, X.; Chen, Q. Genetic Diversity and Characterization of Arsenic-Resistant Endophytic Bacteria Isolated from Pteris Vittata, an Arsenic Hyperaccumulator. BMC Microbiol. 2018, 18, 42. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Zhao, J.; Tang, N.; Sun, H.; Huang, J. Horizontal Gene Transfer from Bacteria and Plants to the Arbuscular Mycorrhizal Fungus Rhizophagus Irregularis. Front. Plant Sci. 2018, 9, 353100. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sun, S.; Ge, W.; Zhao, L.; Hou, B.; Wang, K.; Lyu, Z.; Chen, L.; Xu, S.; Guo, J. Horizontal Gene Transfer of Fhb7 from Fungus Underlies Fusarium Head Blight Resistance in Wheat. Science 2020, 368, eaba5435. [Google Scholar] [CrossRef] [PubMed]
- Sudhakar, T.; Dash, S.; Rao, R.; Srinivasan, R.; Zacharia, S.; Atmanand, M.; Subramaniam, B.; Nayak, S. Do Endophytic Fungi Possess Pathway Genes for Plant Secondary Metabolites. Curr. Sci. 2013, 104, 178. [Google Scholar]
- Stierle, A.; Strobel, G.; Stierle, D. Taxol and Taxane Production by Taxomyces Andreanae, an Endophytic Fungus of Pacific Yew. Science 1993, 260, 214–216. [Google Scholar] [CrossRef] [PubMed]
- Moricca, S.; Ragazzi, A. Fungal Endophytes in Mediterranean Oak Forests: A Lesson from Discula Quercina. Phytopathology 2008, 98, 380–386. [Google Scholar] [CrossRef] [PubMed]
- Cui, R.; Lu, X.; Chen, X.; Malik, W.A.; Wang, D.; Wang, J.; Wang, S.; Guo, L.; Chen, C.; Wang, X. A Novel Raffinose Biological Pathway Is Observed by Symbionts of Cotton≡ Verticillium Dahliae to Improve Salt Tolerance Genetically on Cotton. J. Agron. Crop Sci. 2021, 207, 956–969. [Google Scholar] [CrossRef]
- Freeman, S.; Rodriguez, R.J. Genetic Conversion of a Fungal Plant Pathogen to a Nonpathogenic, Endophytic Mutualist. Science 1993, 260, 75–78. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Xie, J.; Fu, Y.; Cheng, J.; Qu, Z.; Zhao, Z.; Cheng, S.; Chen, T.; Li, B.; Wang, Q. A 2-Kb Mycovirus Converts a Pathogenic Fungus into a Beneficial Endophyte for Brassica Protection and Yield Enhancement. Mol. Plant 2020, 13, 1420–1433. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Caddell, D.F.; Xu, G.; Dahlen, L.; Washington, L.; Yang, J.; Coleman-Derr, D. Genome Wide Association Study Reveals Plant Loci Controlling Heritability of the Rhizosphere Microbiome. ISME J. 2021, 15, 3181–3194. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–Microbiome Interactions: From Community Assembly to Plant Health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Hewezi, T.; Lebeis, S.L.; Pantalone, V.; Grewal, P.S.; Staton, M.E. Soil Indigenous Microbiome and Plant Genotypes Cooperatively Modify Soybean Rhizosphere Microbiome Assembly. BMC Microbiol. 2019, 19, 201. [Google Scholar] [CrossRef]
- Robertson-Albertyn, S.; Alegria Terrazas, R.; Janiak, A.; Szarejko, I.; Hedley, P.E.; Bulgarelli, D. Root Hair Mutations Displace the Barley Rhizosphere Microbiota. Front. Plant Sci. 2017, 8, 266759. [Google Scholar] [CrossRef] [PubMed]
- Zgadzaj, R.; Garrido-Oter, R.; Jensen, D.B.; Koprivova, A.; Schulze-Lefert, P.; Radutoiu, S. Root Nodule Symbiosis in Lotus Japonicus Drives the Establishment of Distinctive Rhizosphere, Root, and Nodule Bacterial Communities. Proc. Natl. Acad. Sci. USA 2016, 113, E7996–E8005. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Thingholm, L.B.; Skiecevičienė, J.; Rausch, P.; Kummen, M.; Hov, J.R.; Degenhardt, F.; Heinsen, F.-A.; Rühlemann, M.C.; Szymczak, S. Genome-Wide Association Analysis Identifies Variation in Vitamin D Receptor and Other Host Factors Influencing the Gut Microbiota. Nat. Genet. 2016, 48, 1396–1406. [Google Scholar] [CrossRef] [PubMed]
- Horton, M.W.; Bodenhausen, N.; Beilsmith, K.; Meng, D.; Muegge, B.D.; Subramanian, S.; Vetter, M.M.; Vilhjálmsson, B.J.; Nordborg, M.; Gordon, J.I. Genome-Wide Association Study of Arabidopsis Thaliana Leaf Microbial Community. Nat. Commun. 2014, 5, 5320. [Google Scholar] [CrossRef] [PubMed]
- Wallace, J.G.; Kremling, K.A.; Kovar, L.L.; Buckler, E.S. Quantitative Genetics of the Maize Leaf Microbiome. Phytobiomes J. 2018, 2, 208–224. [Google Scholar] [CrossRef]
- Bergelson, J.; Mittelstrass, J.; Horton, M.W. Characterizing Both Bacteria and Fungi Improves Understanding of the Arabidopsis Root Microbiome. Sci. Rep. 2019, 9, 24. [Google Scholar] [CrossRef]
- VanWallendael, A.; Benucci, G.M.N.; da Costa, P.B.; Fraser, L.; Sreedasyam, A.; Fritschi, F.; Juenger, T.E.; Lovell, J.T.; Bonito, G.; Lowry, D.B. Host Genotype Controls Ecological Change in the Leaf Fungal Microbiome. PLoS Biol. 2022, 20, e3001681. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zhang, Q.; Li, B.; Jin, Y.; Jiang, L.; Wu, R. Network Mapping of Root–Microbe Interactions in Arabidopsis Thaliana. NPJ Biofilms Microbiomes 2021, 7, 72. [Google Scholar] [CrossRef] [PubMed]
- Balint-Kurti, P.; Simmons, S.J.; Blum, J.E.; Ballaré, C.L.; Stapleton, A.E. Maize Leaf Epiphytic Bacteria Diversity Patterns Are Genetically Correlated with Resistance to Fungal Pathogen Infection. Mol. Plant-Microbe Interact. 2010, 23, 473–484. [Google Scholar] [CrossRef] [PubMed]
- Wippel, K.; Tao, K.; Niu, Y.; Zgadzaj, R.; Kiel, N.; Guan, R.; Dahms, E.; Zhang, P.; Jensen, D.B.; Logemann, E. Host Preference and Invasiveness of Commensal Bacteria in the Lotus and Arabidopsis Root Microbiota. Nat. Microbiol. 2021, 6, 1150–1162. [Google Scholar] [CrossRef] [PubMed]
- Lv, S.; Yang, Y.; Yu, G.; Peng, L.; Zheng, S.; Singh, S.K.; Vílchez, J.I.; Kaushal, R.; Zi, H.; Yi, D. Dysfunction of Histone Demethylase IBM1 in Arabidopsis Causes Autoimmunity and Reshapes the Root Microbiome. ISME J. 2022, 16, 2513–2524. [Google Scholar] [CrossRef] [PubMed]
- Kaushal, R.; Peng, L.; Singh, S.K.; Zhang, M.; Zhang, X.; Vílchez, J.I.; Wang, Z.; He, D.; Yang, Y.; Lv, S. Dicer-like Proteins Influence Arabidopsis Root Microbiota Independent of RNA-Directed DNA Methylation. Microbiome 2021, 9, 57. [Google Scholar] [CrossRef] [PubMed]
- Roussin-Léveillée, C.; Rossi, C.; Castroverde, C.; Peter, M. The plant disease triangle facing climate change: A molecular perspective. Trends Plant Sci. 2024; ahead of print. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.T.; Thireault, C.A.; Paasch, B.C.; Zhang, L.; He, S.Y. Roles of Microbiota in Autoimmunity in Arabidopsis. bioRxiv 2023. [Google Scholar] [CrossRef]
- Carrión, V.J.; Perez-Jaramillo, J.; Cordovez, V.; Tracanna, V.; De Hollander, M.; Ruiz-Buck, D.; Mendes, L.W.; van Ijcken, W.F.; Gomez-Exposito, R.; Elsayed, S.S. Pathogen-Induced Activation of Disease-Suppressive Functions in the Endophytic Root Microbiome. Science 2019, 366, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Mao, Z.; Zhang, W.; Wu, C.; Feng, H.; Peng, Y.; Shahid, H.; Cui, Z.; Ding, P.; Shan, T. Diversity and Antibacterial Activity of Fungal Endophytes from Eucalyptus Exserta. BMC Microbiol. 2021, 21, 155. [Google Scholar] [CrossRef]
- Partida-Martinez, L.P.; Hertweck, C. Pathogenic Fungus Harbours Endosymbiotic Bacteria for Toxin Production. Nature 2005, 437, 884–888. [Google Scholar] [CrossRef] [PubMed]
- Busby, P.E.; Peay, K.G.; Newcombe, G. Common Foliar Fungi of Populus Trichocarpa Modify Melampsora Rust Disease Severity. New Phytol. 2016, 209, 1681–1692. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.K.; Dara, S.K. Fungal and Bacterial Endophytes as Microbial Control Agents for Plant-Parasitic Nematodes. Int. J. Environ. Res. Public Health 2021, 18, 4269. [Google Scholar] [CrossRef] [PubMed]
- Bu, Z.; Li, W.; Liu, X.; Liu, Y.; Gao, Y.; Pei, G.; Zhuo, R.; Cui, K.; Qin, Z.; Zheng, H. The Rice Endophyte-Derived α-Mannosidase ShAM1 Degrades Host Cell Walls to Activate DAMP-Triggered Immunity against Disease. Microbiol. Spectr. 2023, 11, e04824-22. [Google Scholar] [CrossRef] [PubMed]
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.-T.; Weigel, D.; Kemen, E.M. Microbial Hub Taxa Link Host and Abiotic Factors to Plant Microbiome Variation. PLoS Biol. 2016, 14, e1002352. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Yang, T.; Friman, V.-P.; Xu, Y.; Shen, Q.; Jousset, A. Trophic Network Architecture of Root-Associated Bacterial Communities Determines Pathogen Invasion and Plant Health. Nat. Commun. 2015, 6, 8413. [Google Scholar] [CrossRef]
- Hu, J.; Wei, Z.; Friman, V.-P.; Gu, S.; Wang, X.; Eisenhauer, N.; Yang, T.; Ma, J.; Shen, Q.; Xu, Y. Probiotic Diversity Enhances Rhizosphere Microbiome Function and Plant Disease Suppression. MBio 2016, 7, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Innerebner, G.; Knief, C.; Vorholt, J.A. Protection of Arabidopsis Thaliana against Leaf-Pathogenic Pseudomonas Syringae by Sphingomonas Strains in a Controlled Model System. Appl. Environ. Microbiol. 2011, 77, 3202–3210. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Shukla, P. Molecular Modeling and Docking of Microbial Inulinases towards Perceptive Enzyme–Substrate Interactions. Indian J. Microbiol. 2012, 52, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Asaf, S.; Khan, A.L.; Khan, M.A.; Al-Harrasi, A.; Lee, I.-J. Complete Genome Sequencing and Analysis of Endophytic Sphingomonas Sp. LK11 and Its Potential in Plant Growth. 3 Biotech 2018, 8, 389. [Google Scholar] [CrossRef] [PubMed]
- Robinson, R.J.; Fraaije, B.A.; Clark, I.M.; Jackson, R.W.; Hirsch, P.R.; Mauchline, T.H. Endophytic Bacterial Community Composition in Wheat (Triticum Aestivum) Is Determined by Plant Tissue Type, Developmental Stage and Soil Nutrient Availability. Plant Soil 2016, 405, 381–396. [Google Scholar] [CrossRef]
- Li, Z.; Bai, X.; Jiao, S.; Li, Y.; Li, P.; Yang, Y.; Zhang, H.; Wei, G. A Simplified Synthetic Community Rescues Astragalus Mongholicus from Root Rot Disease by Activating Plant-Induced Systemic Resistance. Microbiome 2021, 9, 217. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, J.; Liu, F.; Liang, J.; Zhao, P.; Tsui, C.K.; Cai, L. Cross-Kingdom Synthetic Microbiota Supports Tomato Suppression of Fusarium Wilt Disease. Nat. Commun. 2022, 13, 7890. [Google Scholar] [CrossRef] [PubMed]
- De Souza, R.S.C.; Armanhi, J.S.L.; Arruda, P. From Microbiome to Traits: Designing Synthetic Microbial Communities for Improved Crop Resiliency. Front. Plant Sci. 2020, 11, 553605. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, C.; Onyino, J.; Gao, X. Current Advances in the Functional Diversity and Mechanisms Underlying Endophyte–Plant Interactions. Microorganisms 2024, 12, 779. https://doi.org/10.3390/microorganisms12040779
Zhao C, Onyino J, Gao X. Current Advances in the Functional Diversity and Mechanisms Underlying Endophyte–Plant Interactions. Microorganisms. 2024; 12(4):779. https://doi.org/10.3390/microorganisms12040779
Chicago/Turabian StyleZhao, Caihong, Johnmark Onyino, and Xiquan Gao. 2024. "Current Advances in the Functional Diversity and Mechanisms Underlying Endophyte–Plant Interactions" Microorganisms 12, no. 4: 779. https://doi.org/10.3390/microorganisms12040779
APA StyleZhao, C., Onyino, J., & Gao, X. (2024). Current Advances in the Functional Diversity and Mechanisms Underlying Endophyte–Plant Interactions. Microorganisms, 12(4), 779. https://doi.org/10.3390/microorganisms12040779






