Randomised, Placebo-Controlled Investigation of the Impact of Probiotic Consumption on Gut Microbiota Diversity and the Faecal Metabolome in Seniors
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Compliance
2.3. Questionnaires
2.4. Sample Collection and Pretreatment
2.5. Gut Microbiota Characterisation
2.6. Faecal Metabolite Analysis
2.7. Statistical Analyses
3. Results
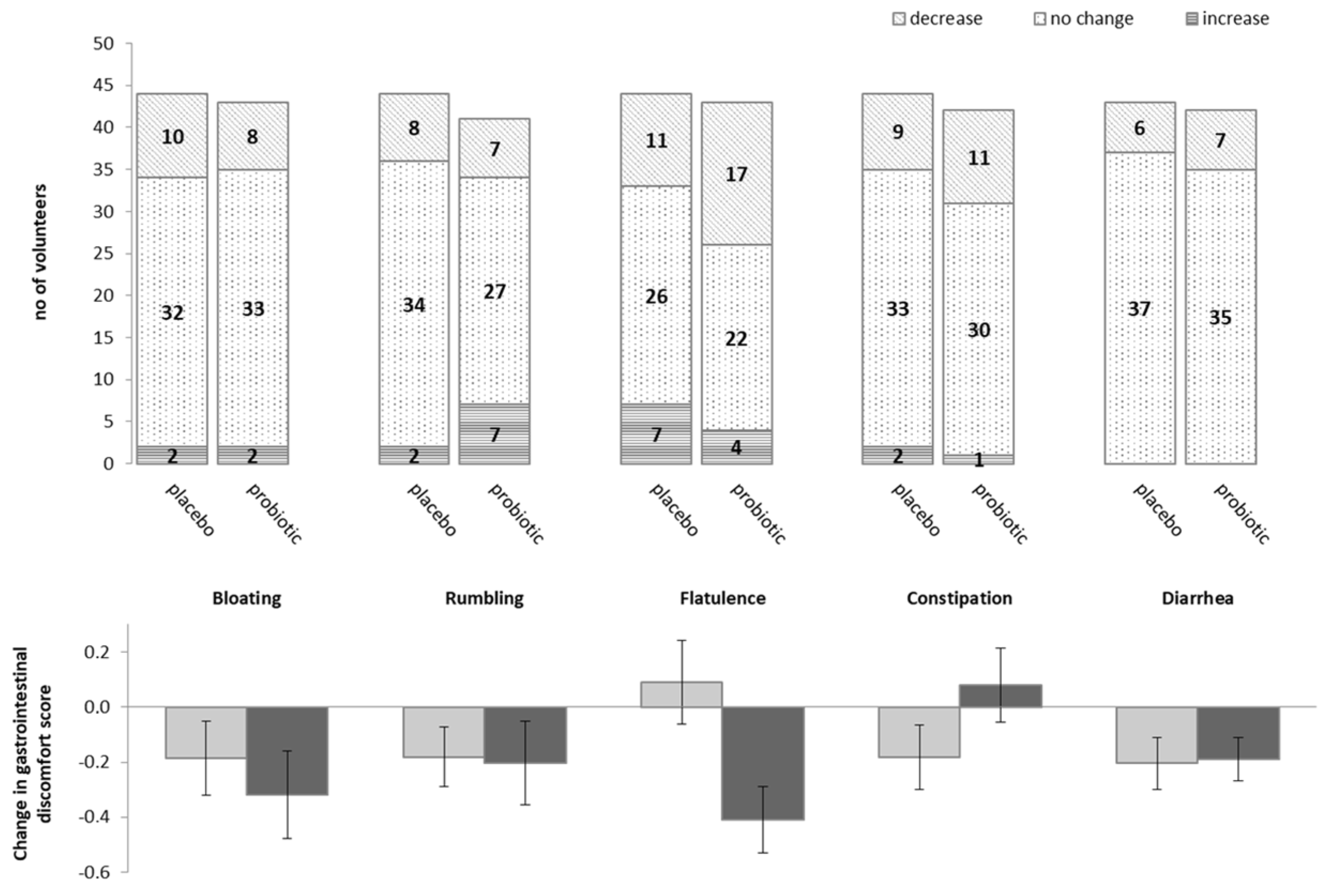
3.1. Impact of Probiotics on Self-Reported Gastro-Intestinal Discomfort
3.2. C. difficile
3.3. Faecal Microbiota of Danish Elderly
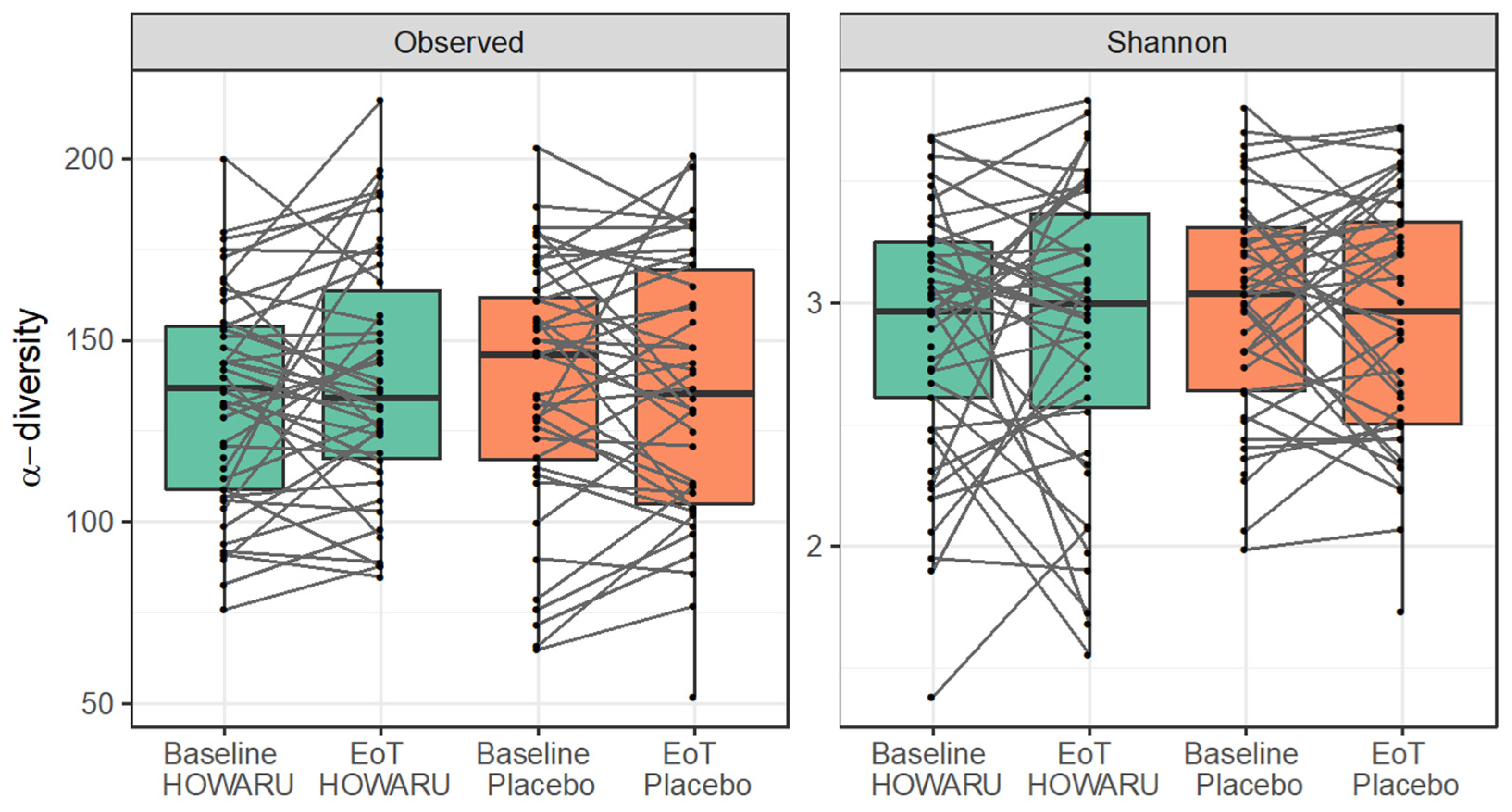
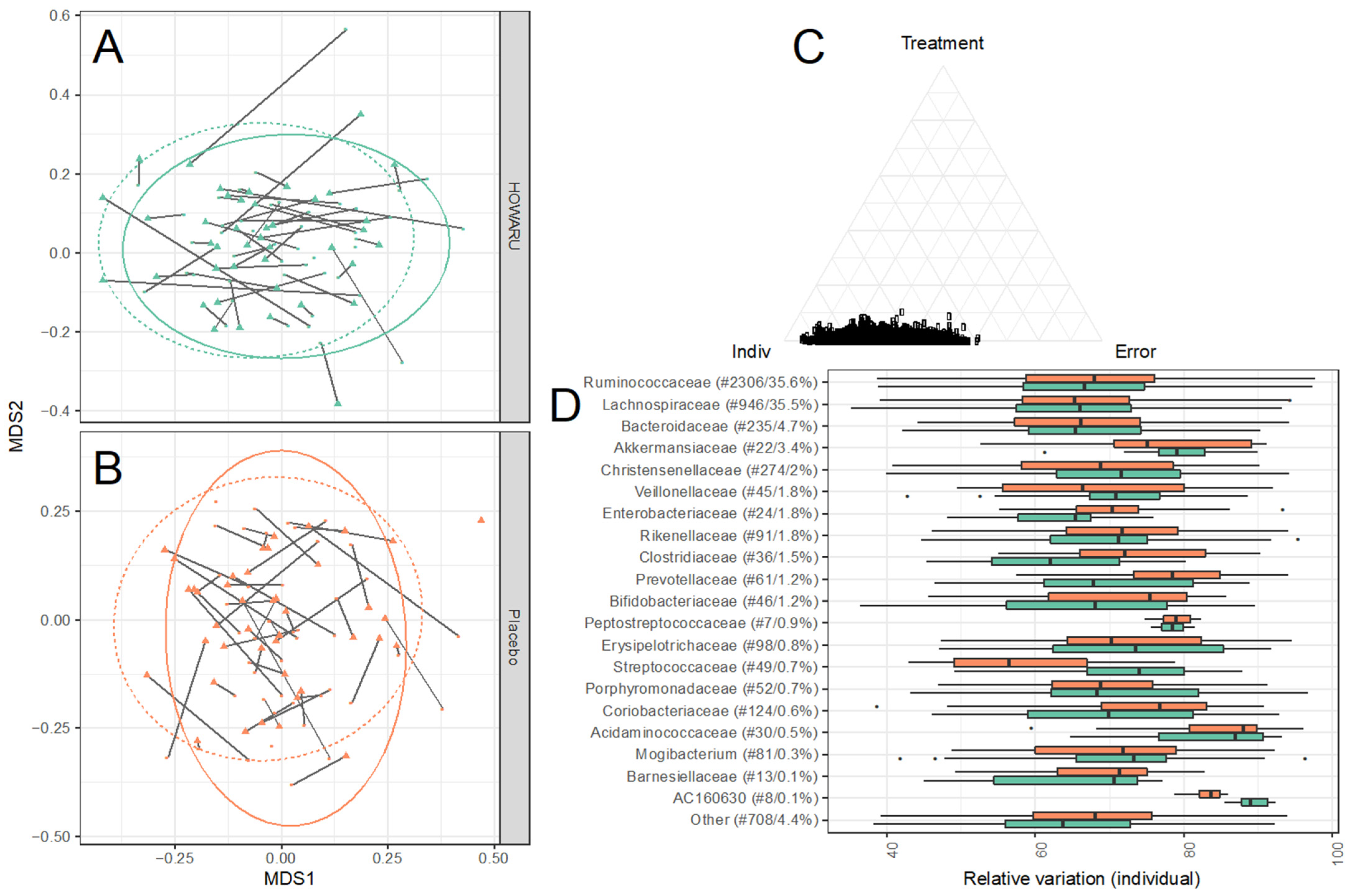
3.4. Effects of Intervention on Microbial Diversity and Composition
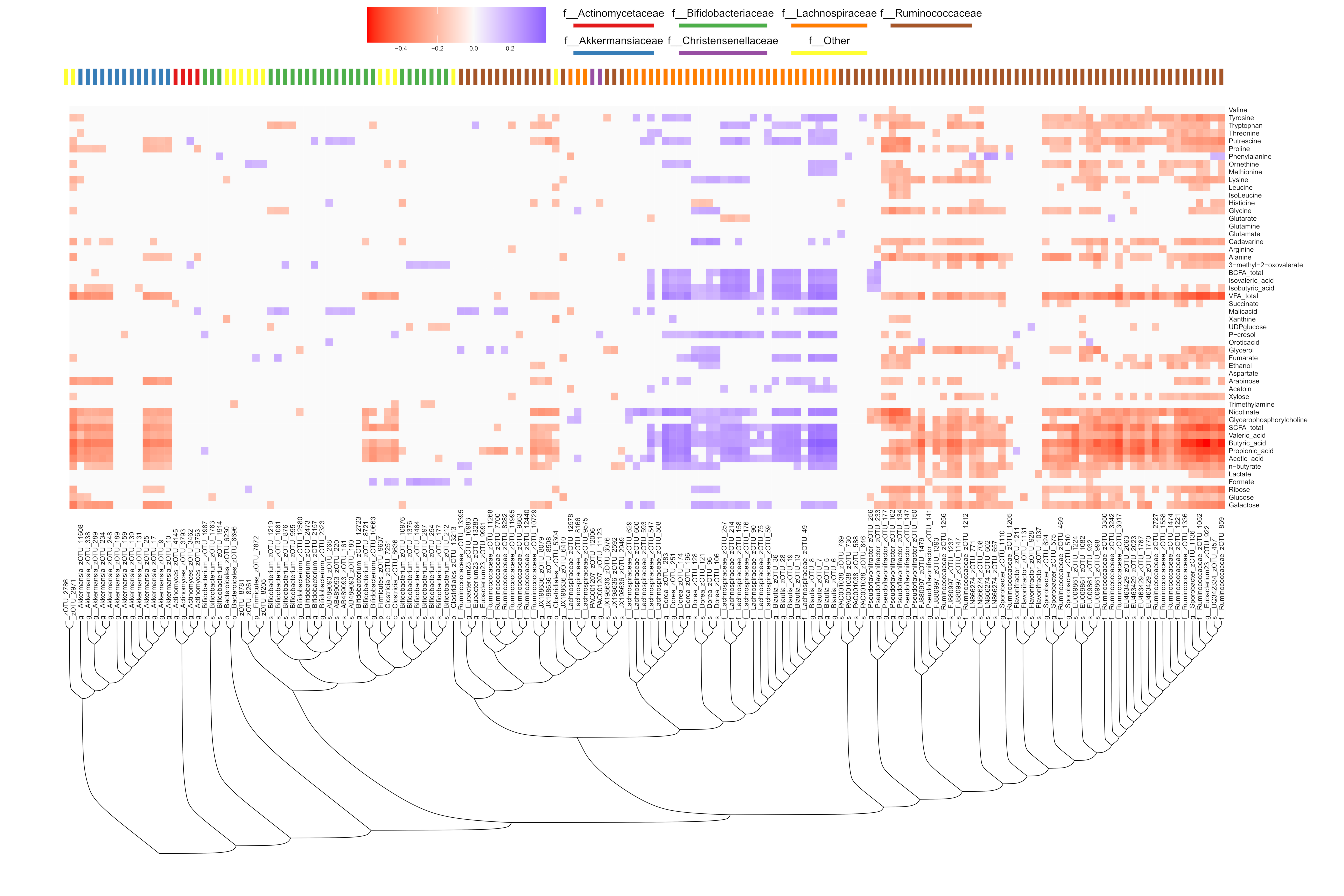
3.5. Effects of Probiotics on Metabolite Composition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khan, S.S.; Singer, B.D.; Vaughan, D.E. Molecular and physiological manifestations and measurement of aging in humans. Aging Cell 2017, 16, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Mariat, D.; Firmesse, O.; Levenez, F.; Guimarăes, V.D.; Sokol, H.; Doré, J.; Corthier, G.; Furet, J.P. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009, 9, 123. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Cusack, S.í.; O’Sullivan, O.; Greene-Diniz, R.; de Weerd, H.; Flannery, E.; Marchesi, J.R.; Falush, D.; Dinan, T.; Fitzgerald, G.; et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. USA 2011, 108, 4586–4591. [Google Scholar] [CrossRef] [PubMed]
- Claesson, M.J.; Jeffery, I.B.; Conde, S.; Power, S.E.; ‘Connor, E.M.; Cusack, S.; Harris, H.M.B.; Coakley, M.; Lakshminarayanan, B.; ‘Sullivan, O.; et al. Gut microbiota composition correlates with diet and health in the elderly. Nature 2012, 488, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Van Tongeren, S.P.; Slaets, J.P.; Harmsen, H.J.; Welling, G.W. Fecal microbiota composition and frailty. Appl. Environ. Microbiol. 2005, 71, 6438–6442. [Google Scholar] [CrossRef] [PubMed]
- Tiihonen, K.; Tynkkynen, S.; Ouwehand, A.; Ahlroos, T.; Rautonen, N. The effect of ageing with and without non-steroidal anti-inflammatory drugs on gastrointestinal microbiology and immunology. Br. J. Nutr. 2008, 100, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.A.; Jeffery, I.B.; Beaumont, M.; Bell, J.T.; Clark, A.G.; Ley, R.E.; O’Toole, P.W.; Spector, T.D.; Steves, C.J. Signatures of early frailty in the gut microbiota. Genome Med. 2016, 8, 8. [Google Scholar] [CrossRef] [PubMed]
- An, R.; Wilms, E.; Masclee, A.A.M.; Smidt, H.; Zoetendal, E.G.; Jonkers, D. Age-dependent changes in GI physiology and microbiota: Time to reconsider? J. Gut 2018, 67, 2213–2222. [Google Scholar] [CrossRef] [PubMed]
- Castro-Mejía, J.L.; Khakimov, B.; Krych, Ł.; Bülow, J.; Bechshøft, R.L.; Højfeldt, G.; Mertz, K.H.; Garne, E.S.; Schacht, S.R.; Ahmad, H.F.; et al. Physical fitness in community-dwelling older adults is linked to dietary intake, gut microbiota, and metabolomic signatures. Aging Cell 2020, 19, e13105. [Google Scholar] [CrossRef]
- Ghosh, T.S.; Rampelli, S.; Jeffery, I.B.; Santoro, A.; Neto, M.; Capri, M.; Giampieri, E.; Jennings, A.; Candela, M.; Turroni, S.; et al. Mediterranean diet intervention alters the gut microbiome in older people reducing frailty and improving health status: The NU-AGE 1-year dietary intervention across five European countries. Gut 2020, 69, 1218–1228. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.; Prasad, J.; Gill, H.; Stevenson, L.; Gopal, P. Impact of consumption of different levels of Bifidobacterium lactis HN019 on the intestinal microflora of elderly human subjects. J. Nutr. Health Aging 2007, 11, 26–31. [Google Scholar] [PubMed]
- Lahtinen, S.J.; Forssten, S.; Aakko, J.; Granlund, L.; Rautonen, N.; Salminen, S.; Viitanen, M.; Ouwehand, A.C. Probiotic cheese containing Lactobacillus rhamnosus HN001 and Lactobacillus acidophilus NCFM® modifies subpopulations of fecal lactobacilli and Clostridium difficile in the elderly. Age 2012, 34, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, N.B.; Bryrup, T.; Allin, K.H.; Nielsen, T.; Hansen, T.H.; Pedersen, O. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: A systematic review of randomized controlled trials. Genome Med. 2016, 8, 52. [Google Scholar] [CrossRef] [PubMed]
- Eloe-Fadrosh, E.A.; Brady, A.; Crabtree, J.; Drabek, E.F.; Ma, B.; Mahurkar, A.; Ravel, J.; Haverkamp, M.; Fiorino, A.-M.; Botelho, C.; et al. Functional Dynamics of the Gut Microbiome in Elderly People during Probiotic Consumption. mBio 2015, 6, e00231-00215. [Google Scholar] [CrossRef]
- Rampelli, S.; Candela, M.; Severgnini, M.; Biagi, E.; Turroni, S.; Roselli, M.; Carnevali, P.; Donini, L.; Brigidi, P. A probiotics-containing biscuit modulates the intestinal microbiota in the elderly. J. Nutr. Health Aging 2013, 17, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Kwok, L.Y.; Wang, L.; Zhang, J.; Guo, Z.; Zhang, H. A pilot study on the effect of Lactobacillus casei Zhang on intestinal microbiota parameters in Chinese subjects of different age. Benef. Microbes 2014, 5, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Senan, S.; Prajapati, J.B.; Joshi, C.G.; Sreeja, V.; Gohel, M.K.; Trivedi, S.; Patel, R.M.; Pandya, H.; Singh, U.S.; Phatak, A.; et al. Geriatric Respondents and Non-Respondents to Probiotic Intervention Can be Differentiated by Inherent Gut Microbiome Composition. Front. Microbiol. 2015, 6, 944. [Google Scholar] [CrossRef]
- Hutchinson, A.N.; Bergh, C.; Kruger, K.; Sűsserová, M.; Allen, J.; Améen, S.; Tingö, L. The Effect of Probiotics on Health Outcomes in the Elderly: A Systematic Review of Randomized, Placebo-Controlled Studies. Microorganisms 2021, 9, 1344. [Google Scholar] [CrossRef]
- Miller, L.E.; Lehtoranta, L.; Lehtinen, M.J. The Effect of Bifidobacterium animalis ssp. lactis HN019 on Cellular Immune Function in Healthy Elderly Subjects: Systematic Review and Meta-Analysis. Nutrients 2017, 9, 191. [Google Scholar] [CrossRef]
- Šola, K.F.; Vladimir-Knežević, S.; Hrabač, P.; Mucalo, I.; Saso, L.; Verbanac, D. The effect of multistrain probiotics on functional constipation in the elderly: A randomized controlled trial. Eur. J. Clin. Nutr. 2022, 76, 1675–1681. [Google Scholar] [CrossRef] [PubMed]
- Barker, A.K.; Duster, M.; Valentine, S.; Hess, T.; Archbald-Pannone, L.; Guerrant, R.; Safdar, N. A randomized controlled trial of probiotics for Clostridium difficile infection in adults (PICO). J. Antimicrob. Chemother. 2017, 72, 3177–3180. [Google Scholar] [CrossRef] [PubMed]
- Ouwehand, A.C.; DongLian, C.; Weijian, X.; Stewart, M.; Ni, J.; Stewart, T.; Miller, L.E. Probiotics reduce symptoms of antibiotic use in a hospital setting: A randomized dose response study. Vaccine 2014, 32, 458–463. [Google Scholar] [CrossRef] [PubMed]
- Hickson, M.; D’Souza, A.L.; Muthu, N.; Rogers, T.R.; Want, S.; Rajkumar, C.; Bulpitt, C.J. Use of probiotic Lactobacillus preparation to prevent diarrhoea associated with antibiotics: Randomised double blind placebo controlled trial. BMJ Clin. Res. Ed. 2007, 335, 80. [Google Scholar] [CrossRef]
- Tremaroli, V.; Backhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, T.S.; Shanahan, F.; O’Toole, P.W. The gut microbiome as a modulator of healthy ageing. Nat. Rev. Gastroenterol. Hepatol. 2022, 19, 565–584. [Google Scholar] [CrossRef] [PubMed]
- Woodmansey, E.J.; McMurdo, M.E.T.; Macfarlane, G.T.; Macfarlane, S. Comparison of Compositions and Metabolic Activities of Fecal Microbiotas in Young Adults and in Antibiotic-Treated and Non-Antibiotic-Treated Elderly Subjects. Appl. Environ. Microbiol. 2004, 70, 6113–6122. [Google Scholar] [CrossRef] [PubMed]
- Gill, C.I.R.; Heavey, P.; McConville, E.; Bradbury, I.; Fässler, C.; Mueller, S.; Cresci, A.; Dore, J.; Norin, E.; Rowland, I. Effect of Fecal Water on an In Vitro Model of Colonic Mucosal Barrier Function. Nutr. Cancer 2007, 57, 59–65. [Google Scholar] [CrossRef]
- Yadav, H.; Lee, J.-H.; Lloyd, J.; Walter, P.; Rane, S.G. Beneficial metabolic effects of a probiotic via butyrate-induced GLP-1 hormone secretion. J. Biol. Chem. 2013, 288, 25088–25097. [Google Scholar] [CrossRef]
- Nagpal, R.; Wang, S.; Ahmadi, S.; Hayes, J.; Gagliano, J.; Subashchandrabose, S.; Kitzman, D.W.; Becton, T.; Read, R.; Yadav, H. Human-origin probiotic cocktail increases short-chain fatty acid production via modulation of mice and human gut microbiome. Sci. Rep. 2018, 8, 12649. [Google Scholar] [CrossRef]
- Lessa, F.C.; Mu, Y.; Bamberg, W.M.; Beldavs, Z.G.; Dumyati, G.K.; Dunn, J.R.; Farley, M.M.; Holzbauer, S.M.; Meek, J.I.; Phipps, E.C.; et al. Burden of Clostridium difficile Infection in the United States. N. Engl. J. Med. 2015, 372, 825–834. [Google Scholar] [CrossRef]
- Dumic, I.; Nordin, T.; Jecmenica, M.; Stojkovic Lalosevic, M.; Milosavljevic, T.; Milovanovic, T. Gastrointestinal Tract Disorders in Older Age. Can. J. Gastroenterol. Hepatol. 2019, 2019, 6757524. [Google Scholar] [CrossRef]
- Olsen, M.A.; Stwalley, D.; Demont, C.; Dubberke, E.R. Increasing Age Has Limited Impact on Risk of Clostridium difficile Infection in an Elderly Population. Open Forum Infect. Dis. 2018, 5, ofy160. [Google Scholar] [CrossRef] [PubMed]
- Rea, M.C.; O’Sullivan, O.; Shanahan, F.; O’Toole, P.W.; Stanton, C.; Ross, R.P.; Hill, C. Clostridium difficile Carriage in Elderly Subjects and Associated Changes in the Intestinal Microbiota. J. Clin. Microbiol. 2012, 50, 867–875. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, E.; Kato, H.; Kita, H.; Karasawa, T.; Maegawa, T.; Koino, Y.; Matsumoto, K.; Takada, T.; Nomoto, K.; Tanaka, R.; et al. Clostridium difficile colonization in healthy adults: Transient colonization and correlation with enterococcal colonization. J. Med. Microbiol. 2004, 53, 167–172. [Google Scholar] [CrossRef]
- Van Zanten, G.C.; Krych, L.; Röytiö, H.; Forssten, S.; Lahtinen, S.J.; Al-Soud, W.A.; Sørensen, S.; Svensson, B.; Jespersen, L.; Jakobsen, M. Synbiotic Lactobacillus acidophilus NCFM and cellobiose does not affect human gut bacterial diversity but increases abundance of lactobacilli, bifidobacteria and branched-chain fatty acids: A randomized, double-blinded cross-over trial. FEMS Microbiol. Ecol. 2014, 90, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Lehtinen, M.J.; Hibberd, A.A.; Mannikko, S.; Yeung, N.; Kauko, T.; Forssten, S.; Lehtoranta, L.; Lahtinen, S.J.; Stahl, B.; Lyra, A.; et al. Nasal microbiota clusters associate with inflammatory response, viral load, and symptom severity in experimental rhinovirus challenge. Sci. Rep. 2018, 8, 11411. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Ouwehand, A.C.; Tiihonen, K.; Saarinen, M.; Putaala, H.; Rautonen, N. Influence of a combination of Lactobacillus acidophilus NCFM and lactitol on healthy elderly: Intestinal and immune parameters. Br. J. Nutr. 2009, 101, 367–375. [Google Scholar] [CrossRef]
- Hao, J.; Liebeke, M.; Astle, W.; De Iorio, M.; Bundy, J.G.; Ebbels, T.M. Bayesian deconvolution and quantification of metabolites in complex 1D NMR spectra using BATMAN. Nat. Protoc. 2014, 9, 1416–1427. [Google Scholar] [CrossRef]
- Pérez Martínez, G.; Bäuerl, C.; Collado, M.C. Understanding gut microbiota in elderly’s health will enable intervention through probiotics. Benef. Microbes 2014, 5, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Nobaek, S.; Johansson, M.L.; Molin, G.; Ahrné, S.; Jeppsson, B. Alteration of intestinal microflora is associated with reduction in abdominal bloating and pain in patients with irritable bowel syndrome. Am. J. Gastroenterol. 2000, 95, 1231–1238. [Google Scholar] [CrossRef] [PubMed]
- Drouault-Holowacz, S.; Bieuvelet, S.; Burckel, A.; Cazaubiel, M.; Dray, X.; Marteau, P. A double blind randomized controlled trial of a probiotic combination in 100 patients with irritable bowel syndrome. Gastroenterol. Clin. Biol. 2008, 32, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Waller, P.A.; Gopal, P.K.; Leyer, G.J.; Ouwehand, A.C.; Reifer, C.; Stewart, M.E.; Miller, L.E. Dose-response effect of Bifidobacterium lactis HN019 on whole gut transit time and functional gastrointestinal symptoms in adults. Scand. J. Gastroenterol. 2011, 46, 1057–1064. [Google Scholar] [CrossRef] [PubMed]
- Yeun, Y.; Lee, J. Effect of a double-coated probiotic formulation on functional constipation in the elderly: A randomized, double blind, controlled study. Arch. Pharm. Res. 2015, 38, 1345–1350. [Google Scholar] [CrossRef] [PubMed]
- Van den Nieuwboer, M.; Klomp-Hogeterp, A.; Verdoorn, S.; Metsemakers-Brameijer, L.; Vriend, T.M.; Claassen, E.; Larsen, O.F. Improving the bowel habits of elderly residents in a nursing home using probiotic fermented milk. Benef. Microbes 2015, 6, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Ringel-Kulka, T.; Palsson, O.S.; Maier, D.; Carroll, I.; Galanko, J.A.; Leyer, G.; Ringel, Y. Probiotic bacteria Lactobacillus acidophilus NCFM and Bifidobacterium lactis Bi-07 versus placebo for the symptoms of bloating in patients with functional bowel disorders: A double-blind study. J. Clin. Gastroenterol. 2011, 45, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Biagi, E.; Franceschi, C.; Rampelli, S.; Severgnini, M.; Ostan, R.; Turroni, S.; Consolandi, C.; Quercia, S.; Scurti, M.; Monti, D.; et al. Gut Microbiota and Extreme Longevity. Curr. Biol. 2016, 26, 1480–1485. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Suda, W.; Kim, S.; Oshima, K.; Fukuda, S.; Ohno, H.; Morita, H.; Hattori, M. Robustness of gut microbiota of healthy adults in response to probiotic intervention revealed by high-throughput pyrosequencing. DNA Res. 2013, 20, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.T.; Maw, A.; Tmanova, L.L.; Pino, A.; Ancy, K.; Crawford, C.V.; Simon, M.S.; Evans, A.T. Timely Use of Probiotics in Hospitalized Adults Prevents Clostridium difficile Infection: A Systematic Review with Meta-Regression Analysis. Gastroenterology 2017, 152, 1889–1900.e1889. [Google Scholar] [CrossRef]
- Miyajima, F.; Roberts, P.; Swale, A.; Price, V.; Jones, M.; Horan, M.; Beeching, N.; Brazier, J.; Parry, C.; Pendleton, N.; et al. Characterisation and carriage ratio of Clostridium difficile strains isolated from a community-dwelling elderly population in the United Kingdom. PLoS ONE 2011, 6, e22804. [Google Scholar] [CrossRef] [PubMed]
- Ryan, J.; Murphy, C.; Twomey, C.; Paul Ross, R.; Rea, M.C.; MacSharry, J.; Sheil, B.; Shanahan, F. Asymptomatic carriage of Clostridium difficile in an Irish continuing care institution for the elderly: Prevalence and characteristics. Ir. J. Med. Sci. 2010, 179, 245–250. [Google Scholar] [CrossRef] [PubMed]
- Windey, K.; De Preter, V.; Verbeke, K. Relevance of protein fermentation to gut health. Mol. Nutr. Food Res. 2012, 56, 184–196. [Google Scholar] [CrossRef] [PubMed]
- Rios-Covian, D.; González, S.; Nogacka, A.M.; Arboleya, S.; Salazar, N.; Gueimonde, M.; de los Reyes-Gavilán, C.G. An Overview on Fecal Branched Short-Chain Fatty Acids Along Human Life and as Related with Body Mass Index: Associated Dietary and Anthropometric Factors. Front. Microbiol. 2020, 11, 973. [Google Scholar] [CrossRef] [PubMed]
- Abdugheni, R.; Wang, W.Z.; Wang, Y.J.; Du, M.X.; Liu, F.L.; Zhou, N.; Jiang, C.Y.; Wang, C.Y.; Wu, L.; Ma, J.; et al. Metabolite profiling of human-originated Lachnospiraceae at the strain level. iMeta 2022, 1, e58. [Google Scholar] [CrossRef]
- Brasili, E.; Mengheri, E.; Tomassini, A.; Capuani, G.; Roselli, M.; Finamore, A.; Sciubba, F.; Marini, F.; Miccheli, A. Lactobacillus acidophilus La5 and Bifidobacterium lactis Bb12 Induce Different Age-Related Metabolic Profiles Revealed by 1H-NMR Spectroscopy in Urine and Feces of Mice. J. Nutr. 2013, 143, 1549–1557. [Google Scholar] [CrossRef]
- Vemuri, R.; Shinde, T.; Gundamaraju, R.; Gondalia, S.V.; Karpe, A.V.; Beale, D.J.; Martoni, C.J.; Eri, R. Lactobacillus acidophilus DDS-1 Modulates the Gut Microbiota and Improves Metabolic Profiles in Aging Mice. Nutrients 2018, 10, 1255. [Google Scholar] [CrossRef]


| Placebo n = 45 | Probiotics n = 45 | p-Value | ||
|---|---|---|---|---|
| Age (years) | Median [IQR] | 77 (76–80) | 79 (76–82) | 0.15 |
| Sex | Male | 18 (40%) | 15 (33%) | 0.66 |
| Female | 27 (60%) | 30 (67%) | ||
| Medication # | Yes | 28 (62%) | 28 (62%) | 1.00 |
| No | 17 (38%) | 17 (38%) | ||
| Defecation frequency | Less than every second day | 2 (5%) | 0 (0%) | 0.42 |
| Every second day | 4 (10%) | 5 (12%) | ||
| Once daily | 19 (45%) | 26 (61%) | ||
| 1–2 times daily | 7 (17%) | 5 (12%) | ||
| 2–3 times daily | 9 (21%) | 5 (12%) | ||
| More than 3 times daily | 1 (2%) | 2 (5%) | ||
| Stool consistency | Very hard | 4 (9%) | 2 (4%) | 0.27 |
| Hard | 7 (16%) | 11 (24%) | ||
| Neither hard nor soft | 23 (52%) | 18 (40%) | ||
| Soft | 10 (23%) | 11 (24%) | ||
| Very soft | 0 (0%) | 3 (7%) | ||
| Bloating | No discomfort | 34 (77%) | 35 (78%) | 0.87 |
| Mild to moderate discomfort | 8 (18%) | 7 (16%) | ||
| Strong to very strong discomfort | 2 (5%) | 3 (7%) | ||
| Rumbling | No discomfort | 34 (77%) | 35 (81%) | 0.82 |
| Mild to moderate discomfort | 8 (18%) | 7 (16%) | ||
| Strong to very strong discomfort | 2 (5%) | 1 (2%) | ||
| Flatulence | No discomfort | 31 (69%) | 18 (41%) | 0.03 |
| Mild to moderate discomfort | 9 (20%) | 16 (36%) | ||
| Strong to very strong discomfort | 5 (11%) | 10 (23%) | ||
| Constipation | No discomfort | 35 (80%) | 33 (75%) | 0.36 |
| Mild to moderate discomfort | 6 (14%) | 10 (23%) | ||
| Strong to very strong discomfort | 3 (7%) | 1 (2%) | ||
| Diarrhoea | No discomfort | 36 (84%) | 37 (84%) | 0.246 |
| Mild to moderate discomfort | 4 (9%) | 1 (2%) | ||
| Strong to very strong discomfort | 3 (7%) | 6 (14%) | ||
| Intestinal pain, after meal | No discomfort | 41 (91%) | 43 (96%) | 0.68 |
| Mild to moderate discomfort | 4 (9%) | 2 (4%) | ||
| Strong to very strong discomfort | 0 (0%) | 0 (0%) | ||
| Intestinal pain, general | No discomfort | 45 (100%) | 44 (100%) | 1.00 |
| Mild to moderate discomfort | 0 (0%) | 0 (0%) | ||
| Strong to very strong discomfort | 0 (0%) | 0 (0%) |
| Phylum | Class | Order | Family | Genus | Species | No. of zOTUs | No. of Samples | No. of Samples (%) | Maximum Reads (%) | Mean Reads (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Actinobacteria | Coriobacteriia | Coriobacteriales | Coriobacteriaceae | 35 | 95 | 97 | 0.4 | 0.0 | ||
| Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | Bacteroides | 78 | 98 | 100 | 19.7 | 2.2 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Rikenellaceae | Alistipes | Alistipes | 52 | 98 | 100 | 6.6 | 0.9 |
| Bacteroidetes | Bacteroidia | Bacteroidales | Bacteroidaceae | Bacteroides | 148 | 95 | 97 | 15.2 | 1.2 | |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | 725 | 98 | 100 | 45.3 | 23.0 | ||
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Faecalibacterium | 115 | 98 | 100 | 35.8 | 7.8 | |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Blautia | 73 | 98 | 100 | 53.6 | 6.8 | |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | 928 | 98 | 100 | 12.8 | 4.5 | ||
| Firmicutes | Clostridia | Clostridiales | 367 | 98 | 100 | 6.7 | 2.8 | |||
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Sporobacter | 148 | 98 | 100 | 8.3 | 1.9 | |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Oscillibacter | 63 | 98 | 100 | 15.9 | 1.8 | |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Dorea | Dorea | 13 | 98 | 100 | 3.9 | 0.9 |
| Firmicutes | 97 | 98 | 100 | 3.5 | 0.5 | |||||
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Lachnospira | 12 | 98 | 100 | 8.4 | 0.4 | |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Falcatimonas | Falcatimonas | 4 | 98 | 100 | 2.3 | 0.4 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Pseudoflavonifractor | 60 | 98 | 100 | 1.6 | 0.3 | |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Agathobaculum | 25 | 98 | 100 | 3.6 | 0.3 | |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Ruthenibacterium | Ruthenibacterium | 4 | 98 | 100 | 1.2 | 0.1 |
| Firmicutes | Clostridia | Clostridiales | Mogibacterium | PAC001168 | 14 | 98 | 100 | 0.6 | 0.1 | |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Subdoligranulum | DQ800172 | 4 | 97 | 99 | 13.1 | 1.7 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Faecalibacterium | GL538271 | 4 | 97 | 99 | 10.8 | 1.4 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Subdoligranulum | 11 | 97 | 99 | 5.7 | 1.1 | |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | 4 | 97 | 99 | 5.4 | 0.7 | ||
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Pseudoflavonifractor | LN866274 | 4 | 97 | 99 | 0.8 | 0.1 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | PAC000195 | 1 | 97 | 99 | 0.2 | 0.1 | |
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae | Clostridium | 23 | 96 | 98 | 14.2 | 1.3 | |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Oscillibacter | PAC001129 | 2 | 96 | 98 | 6.9 | 0.8 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | PAC000672 | 9 | 96 | 98 | 1.8 | 0.3 | |
| Firmicutes | Clostridia | Clostridiales | Christensenellaceae | PAC001207 | 50 | 96 | 98 | 1.6 | 0.3 | |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Ruminococcus | 48 | 96 | 98 | 1.5 | 0.2 | |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | PAC000195 | PAC000195 | 1 | 96 | 98 | 0.3 | 0.1 |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Eubacterium23 | 36 | 95 | 97 | 43.7 | 2.7 | |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Paludicola | 48 | 95 | 97 | 0.9 | 0.1 | |
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Subdoligranulum | DQ793991 | 20 | 95 | 97 | 0.5 | 0.1 |
| Firmicutes | Clostridia | Clostridiales | Christensenellaceae | PAC001207 | EU472329 | 4 | 94 | 96 | 2.7 | 0.3 |
| Firmicutes | Clostridia | Clostridiales | Christensenellaceae | 44 | 94 | 96 | 0.5 | 0.1 | ||
| Firmicutes | Clostridia | Clostridiales | Ruminococcaceae | Oscillibacter | JPJG | 6 | 93 | 95 | 5.5 | 0.8 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Streptococcus | 35 | 93 | 95 | 11.8 | 0.4 | |
| Firmicutes | Clostridia | Clostridiales | Peptostreptococcaceae | Clostridioides | Clostridium | 3 | 93 | 95 | 1.6 | 0.1 |
| Firmicutes | Clostridia | Clostridiales | Lachnospiraceae | Shuttleworthia | 4 | 93 | 95 | 0.3 | 0.1 | |
| Firmicutes | Clostridia | Clostridiales | Mogibacterium | 15 | 93 | 95 | 0.3 | 0.0 | ||
| Unclassified | 168 | 98 | 100 | 3.5 | 0.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van Zanten, G.C.; Madsen, A.L.; Yde, C.C.; Krych, L.; Yeung, N.; Saarinen, M.T.; Kot, W.; Jensen, H.M.; Rasmussen, M.A.; Ouwehand, A.C.; et al. Randomised, Placebo-Controlled Investigation of the Impact of Probiotic Consumption on Gut Microbiota Diversity and the Faecal Metabolome in Seniors. Microorganisms 2024, 12, 796. https://doi.org/10.3390/microorganisms12040796
van Zanten GC, Madsen AL, Yde CC, Krych L, Yeung N, Saarinen MT, Kot W, Jensen HM, Rasmussen MA, Ouwehand AC, et al. Randomised, Placebo-Controlled Investigation of the Impact of Probiotic Consumption on Gut Microbiota Diversity and the Faecal Metabolome in Seniors. Microorganisms. 2024; 12(4):796. https://doi.org/10.3390/microorganisms12040796
Chicago/Turabian Stylevan Zanten, Gabriella C., Anne Lundager Madsen, Christian C. Yde, Lukasz Krych, Nicolas Yeung, Markku T. Saarinen, Witold Kot, Henrik Max Jensen, Morten A. Rasmussen, Arthur C. Ouwehand, and et al. 2024. "Randomised, Placebo-Controlled Investigation of the Impact of Probiotic Consumption on Gut Microbiota Diversity and the Faecal Metabolome in Seniors" Microorganisms 12, no. 4: 796. https://doi.org/10.3390/microorganisms12040796
APA Stylevan Zanten, G. C., Madsen, A. L., Yde, C. C., Krych, L., Yeung, N., Saarinen, M. T., Kot, W., Jensen, H. M., Rasmussen, M. A., Ouwehand, A. C., & Nielsen, D. S. (2024). Randomised, Placebo-Controlled Investigation of the Impact of Probiotic Consumption on Gut Microbiota Diversity and the Faecal Metabolome in Seniors. Microorganisms, 12(4), 796. https://doi.org/10.3390/microorganisms12040796






