Rhizospheric Bacteria of Cover Legumes from Acidic Soils Are Capable of Solubilizing Different Inorganic Phosphates
Abstract
:1. Introduction
2. Materials and Methods
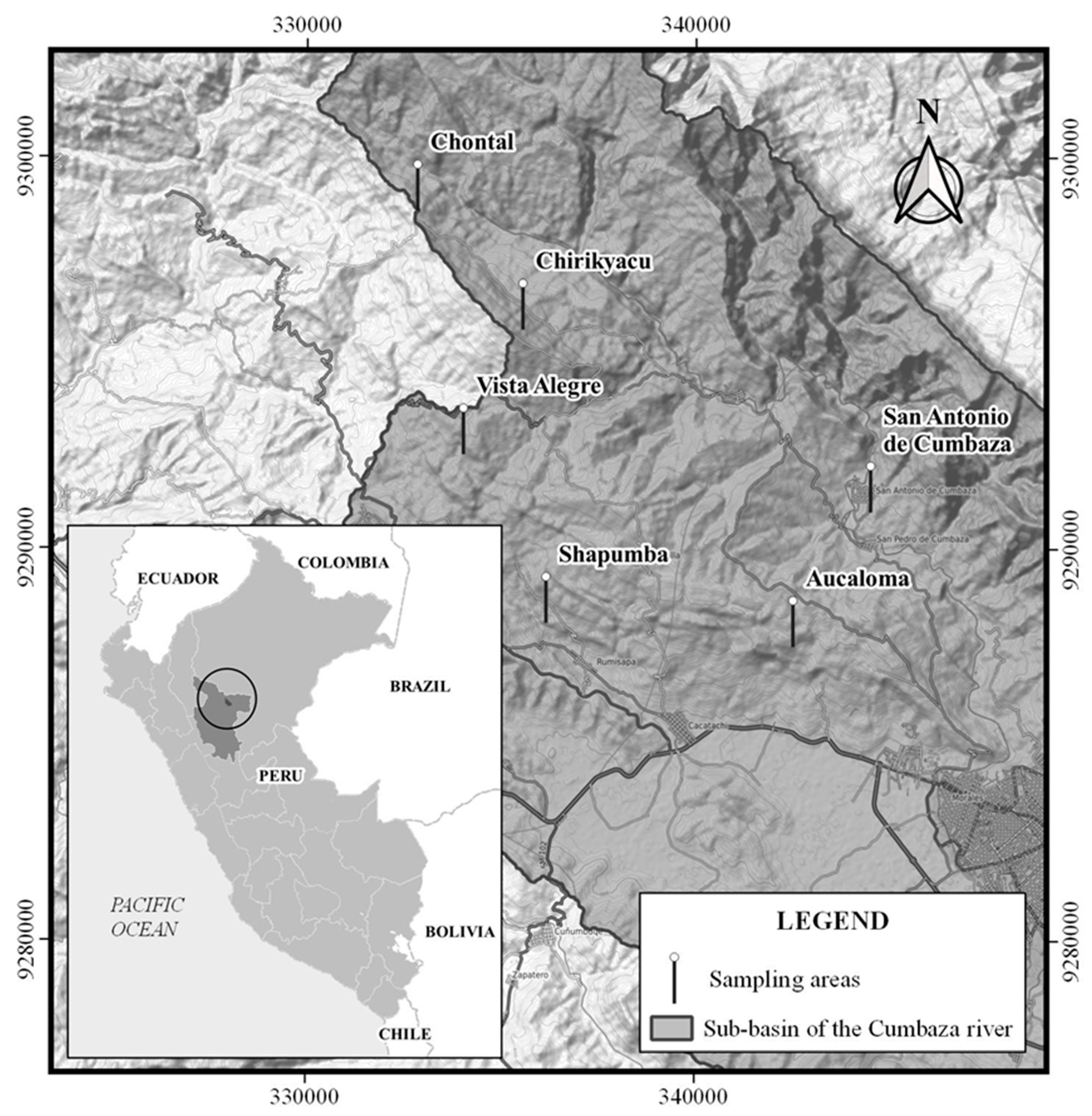
2.1. Study Area and Soil Sampling
2.2. Greenhouse Trial
2.3. Isolation of Bacteria from Rhizospheric Soils
2.4. Evaluation of Phosphate Solubilization by Rhizospheric Bacteria
2.5. Molecular Characterization of Phosphate-Solubilizing Bacteria
2.6. Research Design and Statistical Analysis
3. Results
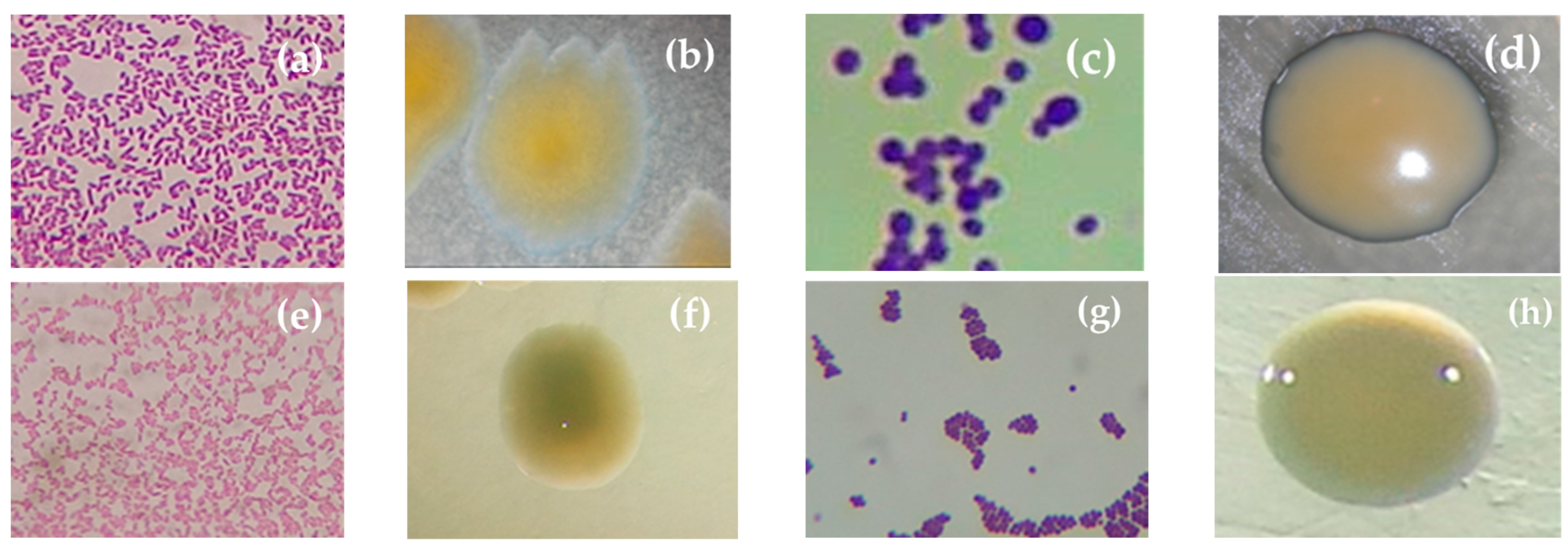
3.1. Isolation and Morphocolonial Characterization of Rhizospheric Bacteria
3.2. Solubilization of AlPO4 and FePO4 by Rhizospheric Bacteria
3.2.1. Solubilization of AlPO4 by Rhizospheric Bacteria
3.2.2. Solubilization of FePO4 by Rhizospheric Bacteria
3.3. Molecular Characterization of Phosphate-Solubilizing Bacteria
4. Discussion
4.1. Isolation and Morphocolonial Characterization of Rhizospheric Bacteria
4.2. Solubilization of AlPO4 by Rhizospheric Bacteria
4.3. Solubilization of FePO4 by Rhizospheric Bacteria
4.4. Molecular Characterization of Rhizospheric Bacteria
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ríos-Ruiz, W.F.; Torres-Chávez, E.E.; Torres-Delgado, J.; Rojas-García, J.C.; Bedmar, E.J.; Valdez-Nuñez, R.A. Inoculation of bacterial consortium increases rice yield (Oryza sativa L.) reducing applications of nitrogen fertilizer in San Martin region, Peru. Rhizosphere 2020, 14, 100200. [Google Scholar] [CrossRef]
- Rios-Ruiz, W.F.; Tuanama-Reátegui, C.; Huamán-Córdova, G.; Valdez-Nuñez, R.A. Co-Inoculation of Endophytes Bacillus siamensis TUR07-02b and Priestia megaterium SMBH14-02 Promotes Growth in Rice with Low Doses of Nitrogen Fertilizer. Plants 2023, 12, 524. [Google Scholar] [CrossRef]
- Souza, V.S.; Santos, D.D.C.; Ferreira, J.G.; de Souza, S.O.; Gonçalo, T.P.; de Sousa, J.V.A.; Cherubin, M.R. Cover crop diversity for sustainable agriculture: Insights from the Cerrado biome. Soil Use Manag. 2024, 40, e13014. [Google Scholar] [CrossRef]
- Li, W.; Liu, Y.; Duan, J.; Liu, G.; Nie, X.; Li, Z. Leguminous cover orchard improves soil quality, nutrient preservation capacity, and aggregate stoichiometric balance: A 22-year homogeneous experimental site. Agric. Ecosyst. Environ. 2024, 363, 108876. [Google Scholar] [CrossRef]
- Jing, T.; Li, J.; He, Y.; Shankar, A.; Saxena, A.; Tiwari, A.; Awasthi, M.K. Role of calcium nutrition in plant physiology: Advances in research and insights into acidic soil conditions—A comprehensive review. Plant Physiol. Biochem. 2024, 210, 108602. [Google Scholar] [CrossRef] [PubMed]
- Pang, F.; Li, Q.; Solanki, M.K.; Wang, Z.; Xing, Y.X.; Dong, D.F. Soil phosphorus transformation and plant uptake driven by phosphate-solubilizing microorganisms. Front. Microbiol. 2024, 15, 1383813. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.D.; Pan, G.; Lu, X.; Qi, W. Phosphorus solubilizing microorganisms: Potential promoters of agricultural and environmental engineering. Front. Bioeng. Biotechnol. 2023, 11, 1–5. [Google Scholar] [CrossRef]
- Pradhan, M.; Das, R.; Sahoo, R.; Adak, T.; Pradhan, C.; Mohanty, S. Comparative P solubilizing efficiencies of five acid soil bacteria incubated with calcium, aluminium and iron phosphates. Indian J. Biochem. Biophys. 2022, 59, 947–955. [Google Scholar] [CrossRef]
- Wan, W.; Tan, J.; Wang, Y.; Qin, Y.; He, H.; Wu, H.; He, D. Responses of the rhizosphere bacterial community in acidic crop soil to pH: Changes in diversity, composition, interaction, and function. Sci. Total Environ. 2020, 700, 134418. [Google Scholar] [CrossRef]
- Janati, W.; Mikou, K.; El Ghadraoui, L.; Errachidi, F. Growth stimulation of two legumes (Vicia faba and Pisum sativum) using phosphate-solubilizing bacteria inoculation. Front. Microbiol. 2023, 14, 1212702. [Google Scholar] [CrossRef]
- Lang, M.; Christie, P.; Zhang, J.; Li, X. Long-term phosphorus application to a maize monoculture influences the soil microbial community and its feedback effects on maize seedling biomass. Appl. Soil Ecol. 2018, 128, 12–22. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, D.; Xiong, Z.; Wang, Z.; Gao, M. Changes in rhizosphere phosphorus fractions and phosphate-mineralizing microbial populations in acid soil as influenced by organic acid exudation. Soil Till. Res. 2023, 225. [Google Scholar] [CrossRef]
- Kiprotich, K.; Muoma, J.; Omayio, D.O.; Ndombi, T.S.; Wekesa, C. Molecular characterization and mineralizing potential of phosphorus solubilizing bacteria colonizing common bean (Phaseolus vulgaris L.) rhizosphere in Western Kenya. Int. J. Microbiol. 2023, 2023, 6668097. [Google Scholar] [CrossRef] [PubMed]
- Ducousso-Détrez, A.; Lahrach, Z.; Fontaine, J.; Lounès-Hadj Sahraoui, A.; Hijri, M. Cultural techniques capture diverse phosphate-solubilizing bacteria in rock phosphate-enriched habitats. Front. Microbiol. 2024, 15, 1280848. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, Z.; Ahmad, M.; Raza, M.A.; Hilger, T.; Rasche, F. Phosphate-Solubilizing Bacillus sp. Modulate Soil Exoenzyme Activities and Improve Wheat Growth. Microb. Ecol. 2024, 87, 31. Available online: https://link.springer.com/article/10.1007/s00248-023-02340-5 (accessed on 25 April 2024). [CrossRef] [PubMed]
- Wendimu, A.; Yoseph, T.; Ayalew, T. Ditching phosphatic fertilizers for phosphate-solubilizing biofertilizers: A step towards sustainable agriculture and environmental health. Sustainability 2023, 15, 1713. [Google Scholar] [CrossRef]
- Instituto Nacional de Estadística e Informática (INEI). Superficie Agrícola por Usos de la Tierra, Según Ámbito Geográfico, 2021–2022. Available online: https://www.inei.gob.pe/media/MenuRecursivo/indices_tematicos/13_11.xlsx (accessed on 12 April 2024).
- Ríos-Ruiz, W.F.; Barrios-López, L.; Rojas-García, J.C.; Valdez-Nuñez, R.A. Mycotrophic capacity and diversity of native arbuscular mycorrhizal fungi isolated from degraded soils. Sci. Agrop. 2019, 10, 99–108. [Google Scholar] [CrossRef]
- Ministerio de ambiente (MINAM)-Sistema Nacional de Información Ambiental (SINIA). Zonificación Ecológica y Económica de la Sub Cuenca del Cumbaza. 2010. Available online: https://sinia.minam.gob.pe/sites/default/files/siar-sanmartin/archivos/public/docs/93.pdf (accessed on 26 April 2024).
- Valdez-Nuñez, R.A.; Ríos-Ruiz, W.F.; Ormeño-Orrillo, E.; Torres-Chávez, E.E.; Torres-Delgado, J. Caracterización genética de bacterias endofíticas de arroz (Oryza sativa L.) con actividad antimicrobiana contra Burkholderia glumae. Rev. Argic. Microbiol. 2020, 52, 315–327. [Google Scholar] [CrossRef]
- Sylvester-Bradley, R.; Asakawa, N.; Torraca, S.L.; Magalhães, F.M.M.; Oliveira, L.A.; Pereira, R.M. Quantitative Survey of Phosphate-Solubilizing Microorganisms in the Rhizosphere of Grasses and Forage Legumes in Amazonia. Acta Amaz. 1982, 12, 15–22. Available online: https://www.cabidigitallibrary.org/doi/full/10.5555/19841985608 (accessed on 12 April 2024). [CrossRef]
- Panda, B.; Rahman, H.; Panda, J. Phosphate solubilizing bacteria from the acidic soils of Eastern Himalayan region and their antagonistic effect on fungal pathogens. Rhizosphere 2016, 2, 62–71. [Google Scholar] [CrossRef]
- Marra, L.M.; Oliveira, S.M.D.; Soares, C.R.F.S.; Moreira, F.M.D.S. Solubilisation of inorganic phosphates by inoculant strains from tropical legumes. Sci. Agric. 2011, 68, 603–609. [Google Scholar] [CrossRef]
- Murphy, J.; Riley, J.P. A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Kim, O.S.; Cho, Y.J.; Lee, K.; Yoon, S.H.; Kim, M.; Na, H.; Park, S.C.; Jeon, Y.S.; Lee, J.H.; Yi, H.; et al. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 2012, 62, 716–721. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Chun, J. PHYDIT. Molecular Sequence Editor for Phylogeny. Version 3. 1. 2001. Available online: http://plaza.snu.ac.kr/~jchun/phydit/ (accessed on 12 April 2024).
- Sharma, P.; Sharma, M.M.M.; Malik, A.; Vashisth, M.; Singh, D.; Kumar, R.; Pandey, V. Rhizosphere, Rhizosphere Biology, and Rhizospheric Engineering. In Plant Growth-Promoting Microbes for Sustainable Biotic and Abiotic Stress Management; Springer: Cham, Switzerland, 2021; pp. 577–624. Available online: https://link.springer.com/chapter/10.1007/978-3-030-66587-6_21 (accessed on 18 April 2024).
- Aliyat, F.Z.; Maldani, M.; El Guilli, M.; Nassiri, L.; Ibijbijen, J. Phosphate-solubilizing bacteria isolated from phosphate solid sludge and their ability to solubilize three inorganic phosphate forms: Calcium, iron, and aluminum phosphates. Microorganisms 2022, 10, 980. [Google Scholar] [CrossRef]
- Shrivastava, M.; Srivastava, P.C.; D’Souza, S.F. Phosphate-Solubilizing Microbes: Diversity and Phosphates Solubilization Mechanism. In Role of Rhizospheric Microbes in Soil; Meena, V., Ed.; Springer: Singapore, 2018; Volume 2, pp. 137–165. [Google Scholar] [CrossRef]
- Silva, L.I.D.; Pereira, M.C.; Carvalho, A.M.X.D.; Buttrós, V.H.; Pasqual, M.; Dória, J. Phosphorus-solubilizing microorganisms: A key to sustainable agriculture. Agriculture 2023, 13, 462. [Google Scholar] [CrossRef]
- Chen, M.; Luo, X.; Jiang, L.; Dong, R.; Siddique, K.H.; He, J. Legume crops use a phosphorus-mobilising strategy to adapt to low plant-available phosphorus in acidic soil in southwest China. Plant Soil Environ. 2023, 69, 471–479. [Google Scholar] [CrossRef]
- Lazali, M.; Drevon, J.J. Mechanisms and adaptation strategies of tolerance to phosphorus deficiency in legumes. Communic. Soil Sci. Plant Anal. 2021, 52, 1469–1483. [Google Scholar] [CrossRef]
- Mendoza-Arroyo, G.E.; Chan-Bacab, M.J.; Aguila-Ramírez, R.N.; Ortega-Morales, B.O.; Canche Solis, R.E.; Chab-Ruiz, A.O.; Camacho-Chab, J.C. Inorganic phosphate solubilization by a novel isolated bacterial strain Enterobacter sp. ITCB-09 and its application potential as biofertilizer. Agriculture 2020, 10, 383. [Google Scholar] [CrossRef]
- Kang, S.M.; Khan, M.A.; Hamayun, M.; Kim, L.R.; Kwon, E.H.; Kang, Y.S.; Lee, I.J. Phosphate-solubilizing Enterobacter ludwigii AFFR02 and Bacillus megaterium Mj1212 rescues alfalfa’s growth under post-drought stress. Agriculture 2021, 11, 485. [Google Scholar] [CrossRef]
- Soares, A.S.; Nascimento, V.L.; De Oliveira, E.E.; Jumbo, L.V.; Dos Santos, G.R.; Queiroz, L.L.; de Souza Aguiar, R.W. Pseudomonas aeruginosa and Bacillus cereus isolated from Brazilian Cerrado soil act as phosphate-solubilizing bacteria. Curr. Microbiol. 2023, 80, 146. [Google Scholar] [CrossRef] [PubMed]
- de Araújo, V.L.V.P.; Junior, M.A.L.; de Souza Júnior, V.S.; de Araújo Filho, J.C.; Fracetto, F.J.C.; Andreote, F.D.; Fracetto, G.G.M. Bacteria from tropical semiarid temporary ponds promote maize growth under hydric stress. Microbiol. Res. 2020, 240, 126564. [Google Scholar] [CrossRef]



| Areas | Vigna unguiculata | Canavalia ensiformis | Crotalaria juncea |
|---|---|---|---|
| Chirikyacu | - | Sfcv-001-01 | Sfcr-003-02 |
| - | Sfcv-017-03 | - | |
| Vista Alegre | Sfc-054-02 | - | - |
| Sfc-070-04 | - | - | |
| Chontal | Sfc-159-01 | Sfcv-041-01-2 | Sfcr-043-02 |
| - | Sfcv-041-9 | - | |
| San Antonio de Cumbaza | - | Sfcv-129-01 | Sfcr-123-01 |
| Shapumba | - | Sfcv-098-02 | Sfcr-116-01 |
| Aucaloma | Sfc-093-04 | Sfcv-082-11 | - |
| - | Sfcv-089-01 | - |
| Strains | Microscopic Features | Macroscopic Features | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Gram Staining | Bacteria Shape | Colony Diameter (mm) | Color | Surface | Elevation | Colony Shape | Margin | Consistency | |
| Sfc-054-02 | − | bacillus | 2.17 | cream | smooth | P | Ci | En | creamy |
| Sfc-070-04 | − | bacillus | 1.04 | cream | smooth | P | Ci | En | creamy |
| Sfc-159-01 | − | bacillus | 2.15 | cream | smooth | P | Ci | En | creamy |
| Sfc-093-04 | + | coccus | 0.99 | orange | smooth | P | Ci | En | creamy |
| Sfcv-001-01 | − | bacillus | 0.82 | cream | smooth | C | Ci | En | creamy |
| Sfcv-017-03 | − | bacillus | 0.9 | cream | smooth | P | Ci | En | creamy |
| Sfcv-041-01-2 | − | bacillus | 3.86 | brown | smooth | P | Fu | L | creamy |
| Sfcv-041-9 | − | bacillus | 1.39 | cream | smooth | P | Ci | En | creamy |
| Sfcv-129-01 | − | bacillus | 0.87 | cream | smooth | P | Ci | En | creamy |
| Sfcv-098-02 | + | coccus | 1.08 | orange-yellow | smooth | P | Ci | En | creamy |
| Sfcv-082-11 | − | bacillus | 1.48 | cream | smooth | P | Ci | En | creamy |
| Sfcv-089-01 | − | bacillus | 1.67 | cream | smooth | P | Ci | En | creamy |
| Sfcr-003-02 | + | coccus | 0.38 | cream | smooth | P | Ci | En | creamy |
| Sfcr-043-02 | − | bacillus | 1.05 | cream | smooth | P | Ci | En | creamy |
| Sfcr-123-01 | − | bacillus | 0.82 | cream | smooth | P | Ci | En | creamy |
| Sfcr-116-01 | − | bacillus | 1.26 | cream | smooth | P | Ci | En | creamy |
| Strains | AlPO4 | FePO4 | ||
|---|---|---|---|---|
| pH | Solubilized P (mg L−1) | pH | Solubilized P (mg L−1) | |
| CIAT 899 | 4.86 (±0.02) b | NPS | 4.26 (±0.01) c | NPS |
| Sfc-054-02 | 4.18 (±0.08) efg | 15.81 (±0.77) cde | 4.08 (±0.05) cd | 28.97 (±1.80) cd |
| Sfc-070-04 | 4.41 (±0.02) d | NPS | 3.29 (±0.04) e | 17.22 (±1.18) e |
| Sfc-093-04 | 5.23 (±0.14) a | 26.50 (±1.58) b | 4.02 (±0.01) cd | NPS |
| Sfc-159-01 | 4.34 (±0.02) de | 17.09 (±2.37) cd | 3.73 (±0.01) cde | 37.31 (±0.90) a |
| Sfcv-001-01 | 4.81 (±0.02) bc | NPS | 3.85 (±0.01) cde | 25.98 (±0.33) d |
| Sfcv-017-03 | 4.08 (±0.07) fg | 9.40 (±1.76) ef | 3.86 (±0.00) cde | 32.18 (±1.07) bc |
| Sfcv-041-01-2 | 5.21 (±0.00) a | 55.98 (±6.27) a | 5.37 (±0.02) b | 15.36 (±1.86) e |
| Sfcv-041-9 | 4.03 (±0.06) g | 6.41 (±0.61) fg | 3.83 (±0.01) cde | 34.96 (±1.32) ab |
| Sfcv-082-11 | 4.88 (±0.05) b | NPS | 3.86 (±0.03) cde | 29.19 (±1.47) cd |
| Sfcv-089-01 | 4.80 (±0.03) bc | NPS | 3.95 (±0.04) cd | 30.68 (±2.08) c |
| Sfcv-098-02 | 4.63 (±0.03) c | 22.65 (±0.55) bc | 3.89 (±0.02) cd | NPS |
| Sfcv-129-01 | 4.14 (±0.03) fg | 5.98 (±1.47) fg | 4.08 (±0.01) cd | NPS |
| Sfcr-003-02 | 4.37 (±0.11) d | 12.61 (±0.92) def | 4.24 (±0.67) c | 26.50 (±1.52) d |
| Sfcr-043-02 | 4.25 (±0.08) def | NPS | 3.54 (±0.01) de | 32.61 (±0.51) bc |
| Sfcr-123-01 | 4.34 (±0.02) de | NPS | 3.51 (±0.01) de | 30.47 (±1.09) c |
| Sfcr-116-01 | 4.41 (±0.03) d | NPS | 3.56 (±0.00) de | 30.90 (±0.39) c |
| CV (%) | 2.28 | 3.42 | 6.25 | 2.37 |
| Strains | Place of Origin | Host Legume | Solubilizing Phosphate | Most Related Species | Similarity (%) | Identified Strain/Accession Number in GenBank |
|---|---|---|---|---|---|---|
| Sfcv-041-01-2 | Chontal | Canavalia ensiformis | AlPO4 | Pseudomonas aeruginosa JCM 5962T/ Pseudonomas paraeruginosa PA7T | 99.93 | Pseudomonas sp. Sfcv-041-01-2/PP319610 |
| Sfc-093-04 | Aucaloma | Vigna unguiculata | AlPO4 | Staphylococcus saprophyticus ATCC 15305T | 100 | Staphylococcus saprophyticus Sfc-093-04/PP319607 |
| Sfcv-098-02 | Shapumba | Canavalia ensiformis | AlPO4 | Staphylococcus saprophyticus ATCC 15305T | 100 | Staphylococcus saprophyticus Sfcv-098-02/PP319608 |
| Sfcr-043-02 | Chontal | Crotalaria juncea | FePO4 | Enterobacter Sichuanensis WCHECL1597T/Enterobacter roggenkampii DSM 16690T/ Enterobacter quasiroggenkampii WCHECL 1060T/Enterobacter vonholyi E13T | 99.86 | Enterobacter sp. Sfcr-043-02T/PP319606 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ríos-Ruiz, W.F.; Casique-Huamanguli, R.D.; Valdez-Nuñez, R.A.; Rojas-García, J.C.; Calixto-García, A.R.; Ríos-Reátegui, F.; Pompa-Vásquez, D.F.; Padilla-Santa-Cruz, E. Rhizospheric Bacteria of Cover Legumes from Acidic Soils Are Capable of Solubilizing Different Inorganic Phosphates. Microorganisms 2024, 12, 1101. https://doi.org/10.3390/microorganisms12061101
Ríos-Ruiz WF, Casique-Huamanguli RD, Valdez-Nuñez RA, Rojas-García JC, Calixto-García AR, Ríos-Reátegui F, Pompa-Vásquez DF, Padilla-Santa-Cruz E. Rhizospheric Bacteria of Cover Legumes from Acidic Soils Are Capable of Solubilizing Different Inorganic Phosphates. Microorganisms. 2024; 12(6):1101. https://doi.org/10.3390/microorganisms12061101
Chicago/Turabian StyleRíos-Ruiz, Winston F., Roy D. Casique-Huamanguli, Renzo A. Valdez-Nuñez, Jose C. Rojas-García, Anderson R. Calixto-García, Franz Ríos-Reátegui, Danny F. Pompa-Vásquez, and Euler Padilla-Santa-Cruz. 2024. "Rhizospheric Bacteria of Cover Legumes from Acidic Soils Are Capable of Solubilizing Different Inorganic Phosphates" Microorganisms 12, no. 6: 1101. https://doi.org/10.3390/microorganisms12061101





