Transcriptomic Signatures of Zika Virus Infection in Patients and a Cell Culture Model
Abstract
1. Introduction
2. Materials and Methods
2.1. A549 Cells
2.2. Zika Virus Infections
2.3. RNA-Seq of Mock and ZIKV Infected A549 Cells
2.4. RNA-Seq Analysis
2.5. Differential Gene Expression
2.6. Alternative Splicing
2.7. RT-qPCR Analysis
2.8. Alternative Splicing (AS) PCR Analysis
3. Results
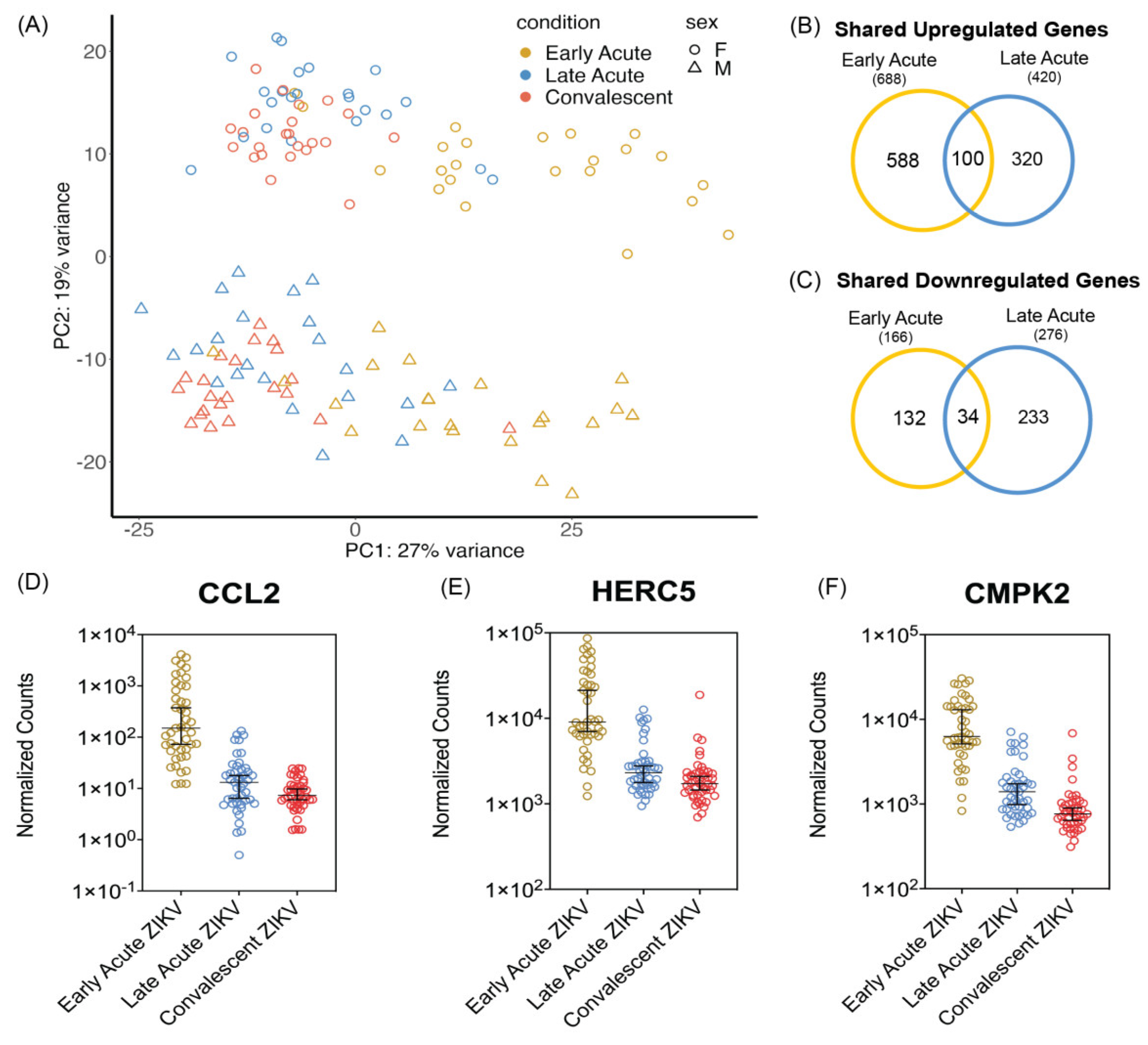
3.1. PBMCs Isolated from ZIKV Infected Patients Show Differential Transcriptomic Profiles during Early and Late Acute Stages of Infections
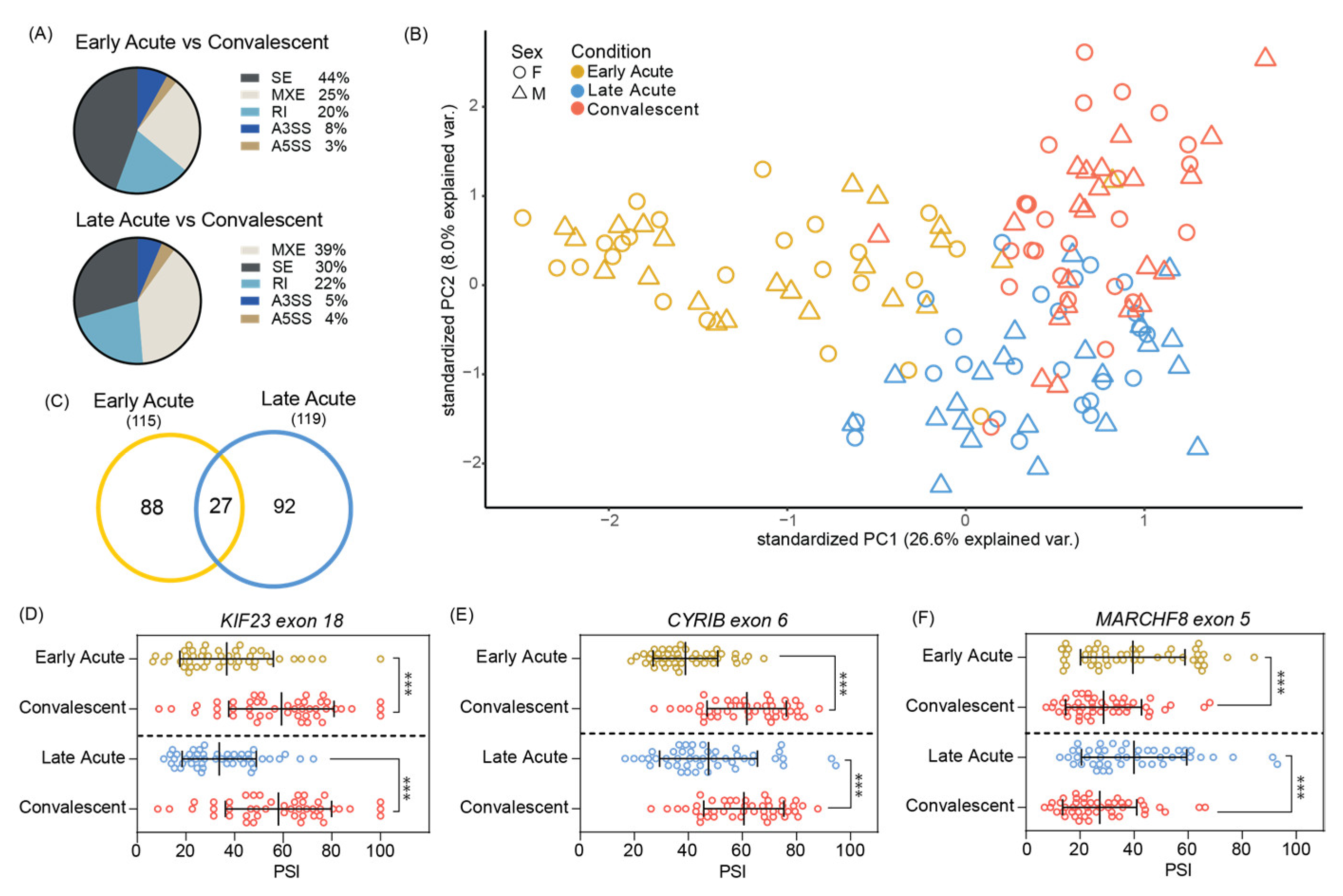
3.2. Significant Skipped Exon Alternative Splicing Events Found in Acute ZIKV Infected Pediatric PMBCs
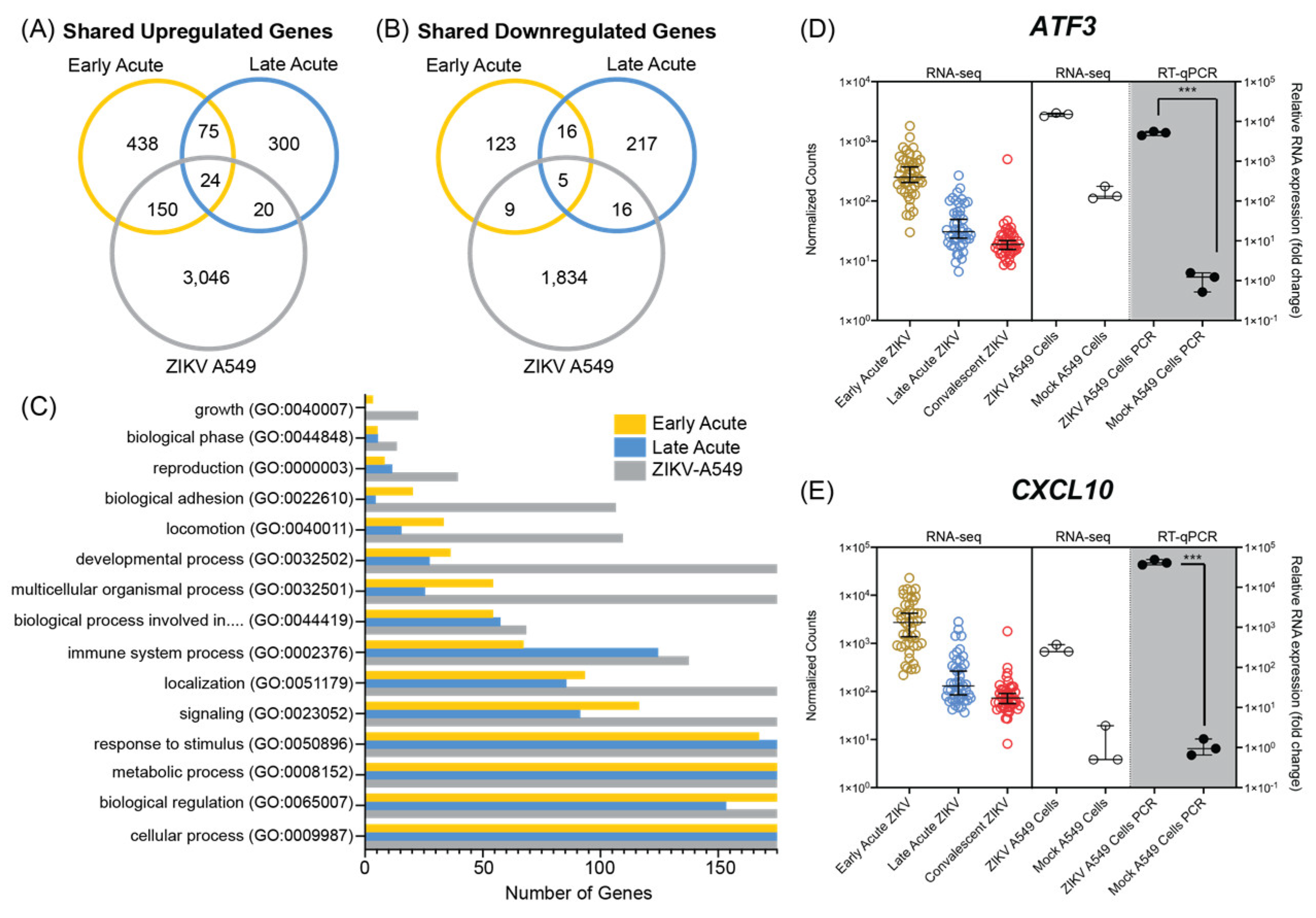
3.3. ZIKV Infection of A549 Cultured Cells Shares Transcriptional Profiles with Patients during Early and Late Acute Stages of ZIKV Infection
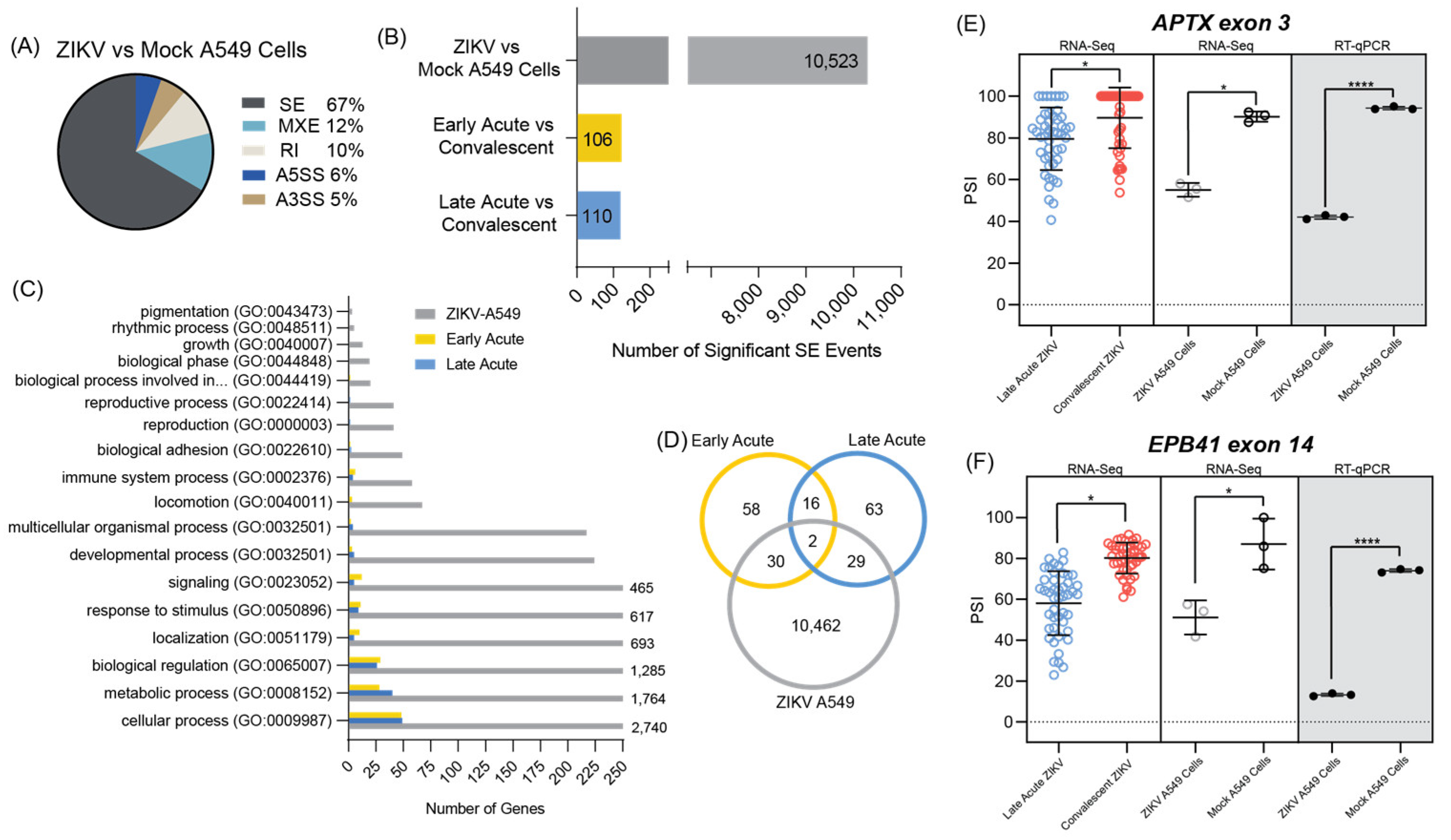
3.4. ZIKV Infected A549 Cell Line Models Alternative Splicing Changes Found in Infected Patient Data
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AS | Alternative Splicing |
| A3SS | Alternative to 3′ splice site |
| A5SS | Alternative to 5′ splice site |
| ΔPSI | Change in percent spliced in |
| FC | Fold change |
| FDR | False discovery rate |
| GO | Gene ontology |
| moi | Multiplicity of infection |
| MXE | Mutually exclusive exons |
| PBMC | Peripheral blood mononuclear cells |
| PCA | Principal component analysis |
| PRVABC56 | Puerto Rican Zika virus isolate |
| SE | Skipped exon |
| RI | Retained intron |
| ZIKV | Zika virus |
References
- Lazear, H.M.; Diamond, M.S. Zika Virus: New Clinical Syndromes and Its Emergence in the Western Hemisphere. J. Virol. 2016, 90, 4864–4875. [Google Scholar] [CrossRef] [PubMed]
- Coyne, C.B.; Lazear, H.M. Zika Virus — Reigniting the TORCH. Nat. Rev. Microbiol. 2016, 14, 707–715. [Google Scholar] [CrossRef] [PubMed]
- Fagbami, A.H. Zika Virus Infections in Nigeria: Virological and Seroepidemiological Investigations in Oyo State. Epidemiol. Infect. 1979, 83, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Robin, Y.; Mouchet, J. Serological and Entomological Study on Yellow Fever in Sierra Leone. Bull. Soc. Pathol. Exot. Filiales 1975, 68, 249–258. [Google Scholar] [PubMed]
- Jan, C.; Languillat, G.; Renaudet, J.; Robin, Y. A Serological Survey of Arboviruses in Gabon. Bull. Soc. Pathol. Exot. Filiales 1978, 71, 140–146. [Google Scholar] [PubMed]
- McCrae, A.W.R.; Kirya, B.G. Yellow Fever and Zika Virus Epizootics and Enzootics in Uganda. Trans. R. Soc. Trop. Med. Hyg. 1982, 76, 552–562. [Google Scholar] [CrossRef]
- Saluzzo, J.F.; Gonzalez, J.P.; Herve, J.P.; Georges, A.J. Serological Survey for the Prevalence of Certain Arboviruses in the Human Population of the South-East Area of Central African Republic (Author’s Transl). Bull. Soc. Pathol. Exot. Filiales 1981, 74, 490–499. [Google Scholar]
- Darwish, M.A.; Hoogstraal, H.; Roberts, T.J.; Ahmed, I.P.; Omar, F. A Sero-Epidemiological Survey for Certain Arboviruses (Togaviridae) in Pakistan. Trans. R. Soc. Trop. Med. Hyg. 1983, 77, 442–445. [Google Scholar] [CrossRef]
- Olson, J.; Ksiazek, T.G. Zika Virus, a Cause of Fever in Central Java, Indonesia. Trans. R. Soc.Trop. Med. Hyg. 1981, 75, 389–393. [Google Scholar] [CrossRef]
- Akoua-Koffi, C.; Diarrassouba, S.; Benie, V.B.; Ngbichi, J.M.; Bozoua, T.; Bosson, A.; Akran, V.; Carnevale, P.; Ehouman, A. Investigation Surrounding a Fatal Case of Yellow Fever in Cote d’Ivoire in 1999. Bull. Soc. Pathol. Exot. 2001, 94, 227–230. [Google Scholar]
- Duffy, M.R.; Chen, T.H.; Hancock, W.T.; Powers, A.M.; Kool, J.L.; Lanciotti, R.S.; Pretrick, M.; Marfel, M.; Holzbauer, S.; Dubray, C.; et al. Zika Virus Outbreak on Yap Island, Federated States of Micronesia. N. Engl. J. Med. 2009, 360, 2536–2543. [Google Scholar] [CrossRef] [PubMed]
- Musso, D.; Nilles, E.J.; Cao-Lormeau, V.M. Rapid Spread of Emerging Zika Virus in the Pacific Area. Clin. Microbiol. Infect. 2014, 20, O595–O596. [Google Scholar] [CrossRef] [PubMed]
- Haddow, A.D.; Schuh, A.J.; Yasuda, C.Y.; Kasper, M.R.; Heang, V.; Huy, R.; Guzman, H.; Tesh, R.B.; Weaver, S.C. Genetic Characterization of Zika Virus Strains: Geographic Expansion of the Asian Lineage. PLoS Negl. Trop. Dis. 2012, 6, e1477. [Google Scholar] [CrossRef] [PubMed]
- 1Cao-Lormeau, V.-M.; Roche, C.; Teissier, A.; Robin, E.; Berry, A.-L.; Mallet, H.-P.; Sall, A.A.; Musso, D. Zika Virus, French Polynesia, South Pacific, 2013. Emerg. Infect. Dis. 2014, 20, 1085–1086. [Google Scholar] [CrossRef]
- de Oliveira, W.K.; de França, G.V.A.; Carmo, E.H.; Duncan, B.B.; de Souza Kuchenbecker, R.; Schmidt, M.I. Infection-Related Microcephaly after the 2015 and 2016 Zika Virus Outbreaks in Brazil: A Surveillance-Based Analysis. The Lancet 2017, 390, 861–870. [Google Scholar] [CrossRef] [PubMed]
- Cao-Lormeau, V.M.; Blake, A.; Mons, S.; Lastère, S.; Roche, C.; Vanhomwegen, J.; Dub, T.; Baudouin, L.; Teissier, A.; Larre, P.; et al. Guillain-Barré Syndrome Outbreak Associated with Zika Virus Infection in French Polynesia: A Case-Control Study. Lancet 2016, 387, 1531–1539. [Google Scholar] [CrossRef] [PubMed]
- Salinas, J.L.; Walteros, D.M.; Styczynski, A.; Garzón, F.; Quijada, H.; Bravo, E.; Chaparro, P.; Madero, J.; Acosta-Reyes, J.; Ledermann, J.; et al. Zika Virus Disease-Associated Guillain-Barré Syndrome—Barranquilla, Colombia 2015–2016. J. Neurol. Sci. 2017, 381, 272–277. [Google Scholar] [CrossRef] [PubMed]
- Oehler, E.; Watrin, L.; Larre, P.; Leparc-Goffart, I.; Lastere, S.; Valour, F.; Baudouin, L.; Mallet, H.; Musso, D.; Ghawche, F. Zika Virus Infection Complicated by Guillain-Barre Syndrome–Case Report, French Polynesia, December 2013. Eurosurveillance 2014, 19, 20720. [Google Scholar] [CrossRef] [PubMed]
- Styczynski, A.R.; Malta, J.M.A.S.; Krow-Lucal, E.R.; Percio, J.; Nóbrega, M.E.; Vargas, A.; Lanzieri, T.M.; Leite, P.L.; Staples, J.E.; Fischer, M.X.; et al. Increased Rates of Guillain-Barré Syndrome Associated with Zika Virus Outbreak in the Salvador Metropolitan Area, Brazil. PLoS Negl. Trop. Dis. 2017, 11, e0005869. [Google Scholar] [CrossRef]
- Nascimento, O.J.M.; da Silva, I.R.F. Guillain–Barré Syndrome and Zika Virus Outbreaks. Curr. Opin. Neurol. 2017, 30, 500–507. [Google Scholar] [CrossRef]
- Parra, B.; Lizarazo, J.; Jiménez-Arango, J.A.; Zea-Vera, A.F.; González-Manrique, G.; Vargas, J.; Angarita, J.A.; Zuñiga, G.; Lopez-Gonzalez, R.; Beltran, C.L.; et al. Guillain–Barré Syndrome Associated with Zika Virus Infection in Colombia. N. Engl. J. Med. 2016, 375, 1513–1523. [Google Scholar] [CrossRef] [PubMed]
- Melo, A.S.d.O.; Aguiar, R.S.; Amorim, M.M.R.; Arruda, M.B.; Melo, F.d.O.; Ribeiro, S.T.C.; Batista, A.G.M.; Ferreira, T.; dos Santos, M.P.; Sampaio, V.V.; et al. Congenital Zika Virus Infection. JAMA Neurol. 2016, 73, 1407–1416. [Google Scholar] [CrossRef] [PubMed]
- Hughes, B.W.; Addanki, K.C.; Sriskanda, A.N.; McLean, E.; Bagasra, O. Infectivity of Immature Neurons to Zika Virus: A Link to Congenital Zika Syndrome. EBioMedicine 2016, 10, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Adams Waldorf, K.M.; Nelson, B.R.; Stencel-Baerenwald, J.E.; Studholme, C.; Kapur, R.P.; Armistead, B.; Walker, C.L.; Merillat, S.; Vornhagen, J.; Tisoncik-Go, J.; et al. Congenital Zika Virus Infection as a Silent Pathology with Loss of Neurogenic Output in the Fetal Brain. Nat. Med. 2018, 24, 368–374. [Google Scholar] [CrossRef] [PubMed]
- França, G.V.A.; Schuler-Faccini, L.; Oliveira, W.K.; Henriques, C.M.P.; Carmo, E.H.; Pedi, V.D.; Nunes, M.L.; Castro, M.C.; Serruya, S.; Silveira, M.F.; et al. Congenital Zika Virus Syndrome in Brazil: A Case Series of the First 1501 Livebirths with Complete Investigation. Lancet 2016, 388, 891–897. [Google Scholar] [CrossRef] [PubMed]
- van der Linden, V.; Pessoa, A.; Dobyns, W.; Barkovich, A.J.; Júnior, H.v.d.L.; Filho, E.L.R.; Ribeiro, E.M.; Leal, M.d.C.; Coimbra, P.P.d.A.; Aragão, M.d.F.V.V.; et al. Description of 13 Infants Born During October 2015–January 2016 With Congenital Zika Virus Infection Without Microcephaly at Birth — Brazil. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 1343–1348. [Google Scholar] [CrossRef] [PubMed]
- D’Ortenzio, E.; Matheron, S.; de Lamballerie, X.; Hubert, B.; Piorkowski, G.; Maquart, M.; Descamps, D.; Damond, F.; Yazdanpanah, Y.; Leparc-Goffart, I. Evidence of Sexual Transmission of Zika Virus. N. Engl. J. Med. 2016, 374, 2195–2198. [Google Scholar] [CrossRef] [PubMed]
- Moreira, J.; Lamas, C.C.; Siqueira, A. Sexual Transmission of Zika Virus: Implications for Clinical Care and Public Health Policy. Clin. Infect. Dis. 2016, 63, 141–142. [Google Scholar] [CrossRef]
- Musso, D.; Roche, C.; Robin, E.; Nhan, T.; Teissier Anita, C.-L.V.-M. Potential Sexual Transmission of Zika Virus. MCN Am. J. Matern. /Child Nurs. 2015, 21, 358. [Google Scholar] [CrossRef]
- Ryan, S.J.; Carlson, C.J.; Mordecai, E.A.; Johnson, L.R. Global Expansion and Redistribution of Aedes-Borne Virus Transmission Risk with Climate Change. PLoS Negl. Trop. Dis. 2019, 13, e0007213. [Google Scholar] [CrossRef] [PubMed]
- Hamel, R.; Dejarnac, O.; Wichit, S.; Ekchariyawat, P.; Neyret, A.; Luplertlop, N.; Perera-Lecoin, M.; Surasombatpattana, P.; Talignani, L.; Thomas, F.; et al. Biology of Zika Virus Infection in Human Skin Cells. J. Virol. 2015, 89, 8880–8896. [Google Scholar] [CrossRef] [PubMed]
- Grove, J.; Marsh, M. The Cell Biology of Receptor-Mediated Virus Entry. J. Cell Biol. 2011, 195, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- Hou, W.; Armstrong, N.; Obwolo, L.A.; Thomas, M.; Pang, X.; Jones, K.S.; Tang, Q. Determination of the Cell Permissiveness Spectrum, Mode of RNA Replication, and RNA-Protein Interaction of Zika Virus. BMC Infect. Dis. 2017, 17, 239. [Google Scholar] [CrossRef] [PubMed]
- Quicke, K.M.; Bowen, J.R.; Johnson, E.L.; McDonald, C.E.; Ma, H.; O’Neal, J.T.; Rajakumar, A.; Wrammert, J.; Rimawi, B.H.; Pulendran, B.; et al. Zika Virus Infects Human Placental Macrophages. Cell Host Microbe 2016, 20, 83–90. [Google Scholar] [CrossRef]
- Miner, J.J.; Sene, A.; Richner, J.M.; Smith, A.M.; Santeford, A.; Ban, N.; Weger-Lucarelli, J.; Manzella, F.; Rückert, C.; Govero, J.; et al. Zika Virus Infection in Mice Causes Panuveitis with Shedding of Virus in Tears. Cell Rep. 2016, 16, 3208–3218. [Google Scholar] [CrossRef] [PubMed]
- Govero, J.; Esakky, P.; Scheaffer, S.M.; Fernandez, E.; Drury, A.; Platt, D.J.; Gorman, M.J.; Richner, J.M.; Caine, E.A.; Salazar, V.; et al. Zika Virus Infection Damages the Testes in Mice. Nature 2016, 540, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Wu, D.; Zhong, H.; Guan, D.; Zhang, H.; Tan, Q.; Ke, C. Presence of Zika Virus in Conjunctival Fluid. JAMA Ophthalmol. 2016, 134, 1330–1332. [Google Scholar] [CrossRef]
- Prisant, N.; Bujan, L.; Benichou, H.; Hayot, P.H.; Pavili, L.; Lurel, S.; Herrmann, C.; Janky, E.; Joguet, G. Zika Virus in the Female Genital Tract. Lancet Infect. Dis. 2016, 16, 1000–1001. [Google Scholar] [CrossRef]
- Murray, K.O.; Gorchakov, R.; Carlson, A.R.; Berry, R.; Lai, L.; Natrajan, M.; Garcia, M.N.; Correa, A.; Patel, S.M.; Aagaard, K.; et al. Prolonged Detection of Zika Virus in Vaginal Secretions and Whole Blood. Emerg. Infect. Dis. 2017, 23, 99–101. [Google Scholar] [CrossRef]
- Miner, J.J.; Diamond, M.S. Zika Virus Pathogenesis and Tissue Tropism. Cell Host Microbe 2017, 21, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Barzon, L.; Pacenti, M.; Franchin, E.; Lavezzo, E.; Trevisan, M.; Sgarabotto, D. Palù G Infection Dynamics in a Traveller with Persistent Shedding of Zika Virus RNA in Semen for Six Months after Returning from Haiti to Italy. Eurosurveillance 2016, 21, 30316. [Google Scholar] [CrossRef] [PubMed]
- Vue, D.; Tang, Q. Zika Virus Overview: Transmission, Origin, Pathogenesis, Animal Model and Diagnosis. Zoonoses 2021, 1, 10.15212/zoonoses-2021-0017. [Google Scholar] [CrossRef] [PubMed]
- Silva, I.B.B.; Silva, A.S.d.; Cunha, M.S.; Cabral, A.D.; Oliveira, K.C.A.d.; Gaspari, E.D.; Prudencio, C.R. Zika Virus Serological Diagnosis: Commercial Tests and Monoclonal Antibodies as Tools. J. Venom. Anim. Toxins Incl. Trop. Dis. 2020, 26, e20200019. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Costa-Ramos, P.; Pham, J.; Tian, Y.; Rosales, S.L.; Seumois, G.; Sidney, J.; de Silva, A.D.; Premkumar, L.; Collins, M.H.; et al. Cutting Edge: Transcriptional Profiling Reveals Multifunctional and Cytotoxic Antiviral Responses of Zika Virus–Specific CD8+ T Cells. J. Immunol. 2018, 201, 3487–3491. [Google Scholar] [CrossRef] [PubMed]
- Michlmayr, D.; Kim, E.Y.; Rahman, A.H.; Raghunathan, R.; Kim-Schulze, S.; Che, Y.; Kalayci, S.; Gümüş, Z.H.; Kuan, G.; Balmaseda, A.; et al. Comprehensive Immunoprofiling of Pediatric Zika Reveals Key Role for Monocytes in the Acute Phase and No Effect of Prior Dengue Virus Infection. Cell Rep. 2020, 31, 107569. [Google Scholar] [CrossRef] [PubMed]
- Bonenfant, G.; Meng, R.; Shotwell, C.; Badu, P.; Payne, A.F.; Ciota, A.T.; Sammons, M.A.; Berglund, J.A.; Pager, C.T. Asian Zika Virus Isolate Significantly Changes the Transcriptional Profile and Alternative RNA Splicing Events in a Neuroblastoma Cell Line. Viruses 2020, 12, 510. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Hammack, C.; Ogden, S.C.; Cheng, Y.; Lee, E.M.; Wen, Z.; Qian, X.; Nguyen, H.N.; Li, Y.; Yao, B.; et al. Molecular Signatures Associated with ZIKV Exposure in Human Cortical Neural Progenitors. Nucleic Acids Res. 2016, 44, 8610–8620. [Google Scholar] [CrossRef]
- Tang, H.; Hammack, C.; Ogden, S.C.; Wen, Z.; Qian, X.; Li, Y.; Yao, B.; Shin, J.; Zhang, F.; Lee, E.M.; et al. Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell 2016, 18, 587–590. [Google Scholar] [CrossRef]
- Tiwari, S.K.; Dang, J.; Qin, Y.; Lichinchi, G.; Bansal, V.; Rana, T.M. Zika Virus Infection Reprograms Global Transcription of Host Cells to Allow Sustained Infection. Emerg. Microbes Infect. 2017, 6, e24. [Google Scholar] [CrossRef]
- Dang, J.; Tiwari, S.K.; Lichinchi, G.; Qin, Y.; Patil, V.S.; Eroshkin, A.M.; Rana, T.M. Zika Virus Depletes Neural Progenitors in Human Cerebral Organoids through Activation of the Innate Immune Receptor TLR3. Cell Stem Cell 2016, 19, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Huo, Y.; Yang, L.; Chen, G.; Luo, M.; Yang, J.; Zhou, J. ZIKV Infection Effects Changes in Gene Splicing, Isoform Composition and LncRNA Expression in Human Neural Progenitor Cells. Virol. J. 2017, 14, 217. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Hua, S.; Gao, C.; Blackmer, J.E.; Ouyang, Z.; Ard, K.; Ciaranello, A.; Yawetz, S.; Sax, P.E.; Rosenberg, E.S.; et al. Immune-Profiling of ZIKV-Infected Patients Identifies a Distinct Function of Plasmacytoid Dendritic Cells for Immune Cross-Regulation. Nat. Commun. 2020, 11, 2421. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Khatri, I.; Jha, A.; Pretto, C.D.; Spindler, K.R.; Arumugaswami, V.; Giri, S.; Kumar, A.; Bhasin, M.K. Determination of System Level Alterations in Host Transcriptome Due to Zika Virus (ZIKV) Infection in Retinal Pigment Epithelium. Sci. Rep. 2018, 8, 11209. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.C.; de Mendonça, L.R.; Rezende, A.M.; Carrera, R.M.; Aníbal-Silva, C.E.; Demers, M.; D’Aiuto, L.; Wood, J.; Chowdari, K.V.; Griffiths, M.; et al. The Transcriptional and Protein Profile from Human Infected Neuroprogenitor Cells Is Strongly Correlated to Zika Virus Microcephaly Cytokines Phenotype Evidencing a Persistent Inflammation in the CNS. Front. Immunol. 2019, 10, 01928. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.L.; Sharp, T.M.; Torres, J.; Armstrong, P.A.; Munoz-Jordan, J.; Ryff, K.R.; Martinez-Quiñones, A.; Arias-Berríos, J.; Mayshack, M.; Garayalde, G.J.; et al. Local Transmission of Zika Virus — Puerto Rico, November 23, 2015–January 28, 2016. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 63–67. [Google Scholar] [CrossRef]
- Wang, G.S.; Cooper, T.A. Splicing in Disease: Disruption of the Splicing Code and the Decoding Machinery. Nat. Rev. Genet. 2007, 8, 749–761. [Google Scholar] [CrossRef] [PubMed]
- Nojima, T.; Oshiro-Ideue, T.; Nakanoya, H.; Kawamura, H.; Morimoto, T.; Kawaguchi, Y.; Kataoka, N.; Hagiwara, M. Herpesvirus Protein ICP27 Switches PML Isoform by Altering MRNA Splicing. Nucleic Acids Res. 2009, 37, 6515–6527. [Google Scholar] [CrossRef]
- Schwartz, S.; Felber, B.K.; Fenyo, E.-M.; Pavlakisl, G.N. Env and Vpu Proteins of Human Immunodeficiency Virus Type 1 Are Produced from Multiple Bicistronic MRNAs. J. Virol. 1990, 64, 5448–5456. [Google Scholar] [CrossRef]
- Hu, B.; Li, X.; Huo, Y.; Yu, Y.; Zhang, Q.; Chen, G.; Zhang, Y.; Fraser, N.W.; Wu, D.; Zhou, J. Cellular Responses to HSV-1 Infection Are Linked to Specific Types of Alterations in the Host Transcriptome. Sci. Rep. 2016, 6, 28075. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, X.; Zhao, Z.; Wang, L.; Li, B.; Li, G.; Dean, M.; Yu, Q.; Wang, Y.; Lin, X.; et al. Full-Length Single-Cell RNA-Seq Applied to a Viral Human Cancer: Applications to HPV Expression and Splicing Analysis in HeLa S3 Cells. Gigascience 2015, 4, s13742-015-0091-4. [Google Scholar] [CrossRef]
- De Maio, F.A.; Risso, G.; Iglesias, N.G.; Shah, P.; Pozzi, B.; Gebhard, L.G.; Mammi, P.; Mancini, E.; Yanovsky, M.J.; Andino, R.; et al. The Dengue Virus NS5 Protein Intrudes in the Cellular Spliceosome and Modulates Splicing. PLoS Pathog. 2016, 12, e1005841. [Google Scholar] [CrossRef]
- Rolfe, A.J.; Bosco, D.B.; Wang, J.; Nowakowski, R.S.; Fan, J.; Ren, Y. Bioinformatic Analysis Reveals the Expression of Unique Transcriptomic Signatures in Zika Virus Infected Human Neural Stem Cells. Cell Biosci. 2016, 6, 42. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Saucedo-Cuevas, L.; Regla-Nava, J.A.A.; Chai, G.; Sheets, N.; Tang, W.; Terskikh, A.V.V.; Shresta, S.; Gleeson, J.G.G. Zika Virus Infects Neural Progenitors in the Adult Mouse Brain and Alters Proliferation. Cell Stem Cell 2016, 19, 593–598. [Google Scholar] [CrossRef]
- Sun, X.; Hua, S.; Chen, H.R.; Ouyang, Z.; Einkauf, K.; Tse, S.; Ard, K.; Ciaranello, A.; Yawetz, S.; Sax, P.; et al. Transcriptional Changes during Naturally Acquired Zika Virus Infection Render Dendritic Cells Highly Conducive to Viral Replication. Cell Rep. 2017, 21, 3471–3482. [Google Scholar] [CrossRef]
- Brand, C.; Deschamps-Francoeur, G.; Bullard-Feibelman, K.M.; Scott, M.S.; Geiss, B.J.; Bisaillon, M. Kunjin Virus, Zika Virus, and Yellow Fever Virus Infections Have Distinct Effects on the Coding Transcriptome and Proteome of Brain-Derived U87 Cells. Viruses 2023, 15, 1419. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huo, Z.; Lin, Q.; Lin, Y.; Chen, C.; Huang, Y.; Huang, C.; Zhang, J.; He, J.; Liu, C.; et al. Positive Feedback Loop of Long Noncoding RNA OASL-IT1 and Innate Immune Response Restricts the Replication of Zika Virus in Epithelial A549 Cells. J. Innate. Immun. 2021, 13, 179–193. [Google Scholar] [CrossRef] [PubMed]
- Frumence, E.; Roche, M.; Krejbich-Trotot, P.; El-Kalamouni, C.; Nativel, B.; Rondeau, P.; Missé, D.; Gadea, G.; Viranaicken, W.; Desprès, P. The South Pacific Epidemic Strain of Zika Virus Replicates Efficiently in Human Epithelial A549 Cells Leading to IFN-β Production and Apoptosis Induction. Virology 2016, 493, 217–226. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. PANTHER: A Library of Protein Families and Subfamilies Indexed by Function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef]
- Shen, S.; Park, J.W.; Lu, Z.X.; Lin, L.; Henry, M.D.; Wu, Y.N.; Zhou, Q.; Xing, Y. RMATS: Robust and Flexible Detection of Differential Alternative Splicing from Replicate RNA-Seq Data. Proc. Natl. Acad. Sci. USA 2014, 111, E5593–E5601. [Google Scholar] [CrossRef]
- Venny 2.1.0. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 31 March 2024).
- Kumar, S.; Barouch-Bentov, R.; Xiao, F.; Schor, S.; Pu, S.; Biquand, E.; Lu, A.; Lindenbach, B.D.; Jacob, Y.; Demeret, C.; et al. MARCH8 Ubiquitinates the Hepatitis C Virus Nonstructural 2 Protein and Mediates Viral Envelopment. Cell Rep. 2019, 26, 1800–1814.e5. [Google Scholar] [CrossRef]
- Yu, C.; Li, S.; Zhang, X.; Khan, I.; Ahmad, I.; Zhou, Y.; Li, S.; Shi, J.; Wang, Y.; Zheng, Y.H. MARCH8 Inhibits Ebola Virus Glycoprotein, Human Immunodeficiency Virus Type 1 Envelope Glycoprotein, and Avian Influenza Virus H5N1 Hemagglutinin Maturation. mBio 2020, 11, 1–15. [Google Scholar] [CrossRef]
- Bartee, E.; Mansouri, M.; Hovey Nerenberg, B.T.; Gouveia, K.; Früh, K. Downregulation of Major Histocompatibility Complex Class I by Human Ubiquitin Ligases Related to Viral Immune Evasion Proteins. J. Virol. 2004, 78, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Shi, C.; Li, H.; Shen, S.; Su, C.; Yin, H. MARCH8 Attenuates CGAS-Mediated Innate Immune Responses through Ubiquitylation. Sci. Signal 2022, 15, eabk3067. [Google Scholar] [CrossRef] [PubMed]
- Yuki, K.E.; Marei, H.; Fiskin, E.; Eva, M.M.; Gopal, A.A.; Schwartzentruber, J.A.; Majewski, J.; Cellier, M.; Mandl, J.N.; Vidal, S.M.; et al. CYRI/FAM49B Negatively Regulates RAC1-Driven Cytoskeletal Remodelling and Protects against Bacterial Infection. Nat. Microbiol. 2019, 4, 1516–1531. [Google Scholar] [CrossRef] [PubMed]
- Nislow, C.; Lombillo, V.A.; Kuriyama, R.; McIntosh, J.R. A Plus-End-Directed Motor Enzyme That Moves Antiparallel Microtubules in Vitro Localizes to the Interzone of Mitotic Spindles. Nature 1992, 359, 543–547. [Google Scholar] [CrossRef]
- Badu, P.; Baniulyte, G.; Sammons, M.A.; Pager, C.T. Activation of ATF3 via the Integrated Stress Response Pathway Regulates Innate Immune Response to Restrict Zika Virus. bioRxiv 2024. [Google Scholar] [CrossRef]
- Aragão, M.D.F.V.V.; Van Der Linden, V.; Petribu, N.C.; Valenca, M.M.; Parizel, P.M.; De Mello, R.J.V. Congenital Zika Syndrome: The Main Cause of Death and Correspondence Between Brain CT and Postmortem Histological Section Findings From the Same Individuals. Top. Magn. Reson. Imaging 2019, 28, 29–33. [Google Scholar] [CrossRef]
- Chen, Z.L.; Yin, Z.J.; Qiu, T.Y.; Chen, J.; Liu, J.; Zhang, X.Y.; Xu, J. qing Revealing the Characteristics of ZIKV Infection through Tissue-Specific Transcriptome Sequencing Analysis. BMC Genomics 2022, 23, 1–16. [Google Scholar] [CrossRef]
- Michlmayr, D.; Andrade, P.; Gonzalez, K.; Balmaseda, A.; Harris, E. CD14+CD16+ Monocytes Are the Main Target of Zika Virus Infection in Peripheral Blood Mononuclear Cells in a Paediatric Study in Nicaragua. Nat. Microbiol. 2017, 2, 1462–1470. [Google Scholar] [CrossRef] [PubMed]
- Chu, L.; Qian, L.; Chen, Y.; Duan, S.; Ding, M.; Sun, W.; Meng, W.; Zhu, J.; Wang, Q.; Hao, H.; et al. HERC5-Catalyzed ISGylation Potentiates CGAS-Mediated Innate Immunity. Cell Rep. 2024, 43, 113870. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Perez, J.M.; Faull, P.A.; Chan, C.; Munting, F.W.; Canadeo, L.A.; Cenik, C.; Huibregtse, J.M. Cellular Targets and Lysine Selectivity of the HERC5 ISG15 Ligase. iScience 2024, 27, 108820. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, N.A.; Paparisto, E.; Barr, S.D.; Spratt, D.E. HERC5 and the ISGylation Pathway: Critical Modulators of the Antiviral Immune Response. Viruses 2021, 13, 1102. [Google Scholar] [CrossRef] [PubMed]
- Mladinich, M.C.; Schwedes, J.; Mackow, E.R. Zika Virus Persistently Infects and Is Basolaterally Released from Primary Human Brain Microvascular Endothelial Cells. mBio 2017, 8, e00952-17. [Google Scholar] [CrossRef] [PubMed]
- Tran, P.-T.-H.; Chiramel, A.I.; Johansson, M.; Melik, W. Roles of ESCRT Proteins ALIX and CHMP4A and Their Interplay with Interferon-Stimulated Gene 15 during Tick-Borne Flavivirus Infection. J. Virol. 2022, 96. [Google Scholar] [CrossRef]
- Gizzi, A.S.; Grove, T.L.; Arnold, J.J.; Jose, J.; Jangra, R.K.; Garforth, S.J.; Du, Q.; Cahill, S.M.; Dulyaninova, N.G.; Love, J.D.; et al. A Naturally Occurring Antiviral Ribonucleotide Encoded by the Human Genome. Nature 2018, 558, 610–614. [Google Scholar] [CrossRef]
- Pawlak, J.B.; Hsu, J.C.C.; Xia, H.; Han, P.; Suh, H.W.; Grove, T.L.; Morrison, J.; Shi, P.Y.; Cresswell, P.; Laurent-Rolle, M. CMPK2 Restricts Zika Virus Replication by Inhibiting Viral Translation. PLoS Pathog. 2023, 19, e1011286. [Google Scholar] [CrossRef]
- Zhu, Y.; Tan, Q.; Shi, Y.; Li, Q.; Li, S.; Wen, W.; Xie, H.; Li, B.; Duan, X.; Chen, L. UMP-CMP Kinase 2 Inhibits ZIKV Replication through Activation of Type I IFN Signaling Pathway. J. Med. Virol. 2024, 96, e29533. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Li, Z.; Lan, S.; Hao, H.; Baz, A.A.; Yan, X.; Gao, P.; Chen, S.; Chu, Y. The Dual Roles of Activating Transcription Factor 3 (ATF3) in Inflammation, Apoptosis, Ferroptosis, and Pathogen Infection Responses. Int. J. Mol. Sci. 2024, 25, 824. [Google Scholar] [CrossRef] [PubMed]
- Hai, T.; Wolford, C.C.; Chang, Y.S. ATF3, a Hub of the Cellular Adaptive-Response Network, in the Pathogenesis of Diseases: Is Modulation of Inflammation a Unifying Component? Gene Expr. 2010, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Gladwyn-Ng, I.; Cordón-Barris, L.; Alfano, C.; Creppe, C.; Couderc, T.; Morelli, G.; Thelen, N.; America, M.; Bessières, B.; Encha-Razavi, F.; et al. Stress-Induced Unfolded Protein Response Contributes to Zika Virus-Associated Microcephaly. Nat. Neurosci. 2018, 21, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Horibata, S.; Teramoto, T.; Vijayarangan, N.; Kuhn, S.; Padmanabhan, R.; Vasudevan, S.; Gottesman, M.; Padmanabhan, R. Host Gene Expression Modulated by Zika Virus Infection of Human-293 Cells. Virology 2021, 552, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Martinez, M.G.; Lin, J.; Gregory, J.; Nguyen, T.U.; Abdelaal, R.; Kang, K.; Brennand, K.; Grünweller, A.; Ouyang, Z.; et al. Transcriptional and Translational Dynamics of Zika and Dengue Virus Infection. Viruses 2022, 14, 1418. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.-B.; Yang, W.; Chang, J.-T.; Fan, D.-Y.; Wu, Y.-H.; Wang, P.-G.; An, J. Zika Virus Infection Leads to Hormone Deficiencies of the Hypothalamic-Pituitary-Gonadal Axis and Diminished Fertility in Mice. J. Virol. 2023, 97, e0100623. [Google Scholar] [CrossRef] [PubMed]
- Hammack, C.; Ogden, S.C.; Madden, J.C.; Medina, A.; Xu, C.; Phillips, E.; Son, Y.; Cone, A.; Giovinazzi, S.; Didier, R.A.; et al. Zika Virus Infection Induces DNA Damage Response in Human Neural Progenitors That Enhances Viral Replication. J. Virol. 2019, 93, e00638-19. [Google Scholar] [CrossRef] [PubMed]
- Rychlowska, M.; Agyapong, A.; Weinfeld, M.; Schang, L.M. Zika Virus Induces Mitotic Catastrophe in Human Neural Progenitors by Triggering Unscheduled Mitotic Entry in the Presence of DNA Damage While Functionally Depleting Nuclear PNKP. J. Virol. 2022, 96, e0033322. [Google Scholar] [CrossRef]
- Su, S.; Liu, X.; Tian, R.-R.; Qiao, K.-X.; Zheng, C.-B.; Gao, W.-C.; Yang, L.-M.; Kang, Q.-Z.; Zheng, Y.-T. Cell Membrane Skeletal Protein 4.1R Participates in Entry of Zika Virus into Cells. Virus Res. 2021, 306, 198593. [Google Scholar] [CrossRef]
- McIntyre, W.; Netzband, R.; Bonenfant, G.; Biegel, J.M.; Miller, C.; Fuchs, G.; Henderson, E.; Arra, M.; Canki, M.; Fabris, D.; et al. Positive-Sense RNA Viruses Reveal the Complexity and Dynamics of the Cellular and Viral Epitranscriptomes during Infection. Nucleic Acids Res. 2018, 46, 5776–5791. [Google Scholar] [CrossRef] [PubMed]
- Gokhale, N.S.; McIntyre, A.B.R.; McFadden, M.J.; Roder, A.E.; Kennedy, E.M.; Gandara, J.A.; Hopcraft, S.E.; Quicke, K.M.; Vazquez, C.; Willer, J.; et al. N6-Methyladenosine in Flaviviridae Viral RNA Genomes Regulates Infection. Cell Host Microbe 2016, 20, 654–665. [Google Scholar] [CrossRef] [PubMed]
- Lichinchi, G.; Zhao, B.S.; Wu, Y.; Lu, Z.; Qin, Y.; He, C.; Rana, T.M. Dynamics of Human and Viral RNA Methylation during Zika Virus Infection. Cell Host Microbe 2016, 20, 666–673. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, L.A.; Edgar, A.J.; Ehley, J.; Gottesfeld, J.M. Cyclin L Is an RS Domain Protein Involved in Pre-MRNA Splicing. J. Biol. Chem. 2002, 277, 25465–25473. [Google Scholar] [CrossRef]
- Gupta, S.; Jani, J.; Vijayasurya; Mochi, J.; Tabasum, S.; Sabarwal, A.; Pappachan, A. Aminoacyl-TRNA Synthetase – a Molecular Multitasker. FASEB J. 2023, 37, e23219. [Google Scholar] [CrossRef] [PubMed]
- Ahn, Y.H.; Park, S.; Choi, J.J.; Park, B.-K.; Rhee, K.H.; Kang, E.; Ahn, S.; Lee, C.-H.; Lee, J.S.; Inn, K.-S.; et al. Secreted Tryptophanyl-TRNA Synthetase as a Primary Defence System against Infection. Nat. Microbiol. 2016, 2, 16191. [Google Scholar] [CrossRef]
- Lee, H.-C.; Lee, E.-S.; Uddin, M.B.; Kim, T.-H.; Kim, J.-H.; Chathuranga, K.; Chathuranga, W.A.G.; Jin, M.; Kim, S.; Kim, C.-J.; et al. Released Tryptophanyl-TRNA Synthetase Stimulates Innate Immune Responses against Viral Infection. J. Virol. 2019, 93, e01291-18. [Google Scholar] [CrossRef]
- Yeung, M.L.; Jia, L.; Yip, C.C.Y.; Chan, J.F.W.; Teng, J.L.L.; Chan, K.H.; Cai, J.P.; Zhang, C.; Zhang, A.J.; Wong, W.M.; et al. Human Tryptophanyl-TRNA Synthetase Is an IFN-γ-Inducible Entry Factor for Enterovirus. J. Clin. Investig. 2018, 128, 5163–5177. [Google Scholar] [CrossRef]
- Karlebach, G.; Aronow, B.; Baylin, S.B.; Butler, D.; Foox, J.; Levy, S.; Meydan, C.; Mozsary, C.; Saravia-Butler, A.M.; Taylor, D.M.; et al. Betacoronavirus-Specific Alternate Splicing. Genomics 2022, 114, 110270. [Google Scholar] [CrossRef]
- Selinger, M.; Věchtová, P.; Tykalová, H.; Ošlejšková, P.; Rumlová, M.; Štěrba, J.; Grubhoffer, L. Integrative RNA Profiling of TBEV-Infected Neurons and Astrocytes Reveals Potential Pathogenic Effectors. Comput. Struct. Biotechnol. J. 2022, 20, 2759–2777. [Google Scholar] [CrossRef]
- Feng, M.; Zhang, H. Aminoacyl-TRNA Synthetase: A Non-Negligible Molecule in RNA Viral Infection. Viruses 2022, 14, 613. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, N.; Miya, F.; Tsunoda, T.; Kanemura, Y.; Saitoh, S.; Kato, M.; Yanagi, K.; Kaname, T.; Kosaki, K. Four Pedigrees with Aminoacyl-TRNA Synthetase Abnormalities. Neurol. Sci. 2022, 43, 2765–2774. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.J.; Vona, B.; Porter, H.M.; Izadi, M.; Huang, K.; Lacassie, Y.; Rosenfeld, J.A.; Khan, S.; Petree, C.; Ali, T.A.; et al. Biallelic Variants in WARS1 Cause a Highly Variable Neurodevelopmental Syndrome and Implicate a Critical Exon for Normal Auditory Function. Hum. Mutat. 2022, 43, 1472–1489. [Google Scholar] [CrossRef] [PubMed]
- Bögershausen, N.; Krawczyk, H.E.; Jamra, R.A.; Lin, S.J.; Yigit, G.; Hüning, I.; Polo, A.M.; Vona, B.; Huang, K.; Schmidt, J.; et al. WARS1 and SARS1: Two TRNA Synthetases Implicated in Autosomal Recessive Microcephaly. Hum. Mutat. 2022, 43, 1454–1471. [Google Scholar] [CrossRef] [PubMed]
- Mlakar, J.; Korva, M.; Tul, N.; Popović, M.; Poljšak-Prijatelj, M.; Mraz, J.; Kolenc, M.; Resman Rus, K.; Vesnaver Vipotnik, T.; Fabjan Vodušek, V.; et al. Zika Virus Associated with Microcephaly. N. Engl. J. Med. 2016, 374, 951–958. [Google Scholar] [CrossRef] [PubMed]
- Schuler-Faccini, L.; Ribeiro, E.M.; Feitosa, I.M.L.; Horovitz, D.D.G.; Cavalcanti, D.P.; Pessoa, A.; Doriqui, M.J.R.; Neri, J.I.; Neto, J.M.d.P.; Wanderley, H.Y.C.; et al. Possible Association Between Zika Virus Infection and Microcephaly—Brazil, 2015. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 59–62. [Google Scholar] [CrossRef]
- Pozzi, B.; Bragado, L.; Mammi, P.; Torti, M.F.; Gaioli, N.; Gebhard, L.G.; García Solá, M.E.; Vaz-Drago, R.; Iglesias, N.G.; García, C.C.; et al. Dengue Virus Targets RBM10 Deregulating Host Cell Splicing and Innate Immune Response. Nucleic Acids Res. 2020, 48, 6824–6838. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berglund, G.; Lennon, C.D.; Badu, P.; Berglund, J.A.; Pager, C.T. Transcriptomic Signatures of Zika Virus Infection in Patients and a Cell Culture Model. Microorganisms 2024, 12, 1499. https://doi.org/10.3390/microorganisms12071499
Berglund G, Lennon CD, Badu P, Berglund JA, Pager CT. Transcriptomic Signatures of Zika Virus Infection in Patients and a Cell Culture Model. Microorganisms. 2024; 12(7):1499. https://doi.org/10.3390/microorganisms12071499
Chicago/Turabian StyleBerglund, Gillian, Claudia D. Lennon, Pheonah Badu, John Andrew Berglund, and Cara T. Pager. 2024. "Transcriptomic Signatures of Zika Virus Infection in Patients and a Cell Culture Model" Microorganisms 12, no. 7: 1499. https://doi.org/10.3390/microorganisms12071499
APA StyleBerglund, G., Lennon, C. D., Badu, P., Berglund, J. A., & Pager, C. T. (2024). Transcriptomic Signatures of Zika Virus Infection in Patients and a Cell Culture Model. Microorganisms, 12(7), 1499. https://doi.org/10.3390/microorganisms12071499





