Abstract
Since nitrogen limitation is known to be an important trigger of triacylglycerol (TAG) accumulation in most microorganisms, we first assessed the global lipid content of 21 strains derived from Streptomyces coelicolor M145 deleted for genes involved in nitrogen metabolism. Seven of these strains deleted for genes encoding proteins involved in polyamine (GlnA2/SCO2241, GlnA3/SCO6962, GlnA4/SCO1613), or protein (Pup/SCO1646) degradation, in the regulation of nitrogen metabolism (GlnE/SCO2234 and GlnK/SCO5584), or the global regulator DasR/SCO5231 that controls negatively the degradation of N-acetylglucosamine, a constituent of peptidoglycan, had a higher TAG content than the original strain, whereas five of these strains (except the glnA2 and pup mutants) had a lower cardiolipin (CL) content. The production of the blue polyketide actinorhodin (ACT) was totally abolished in the dasR mutant in both Pi conditions, whereas the deletion of pup, glnA2, glnA3, and glnA4 was correlated with a significant increase in total ACT production, but mainly in Pi limitation. Unexpectedly, ACT production was strongly reduced in the glnA3 mutant in Pi proficiency. Altogether, our data suggest that high TAG and ACT biosynthesis and low CL biosynthesis might all contribute to the lowering of oxidative stress resulting from nitrogen limitation or from other causes.
1. Introduction
Nitrogen (N) and phosphate (P) are constitutive elements of all living cells since, with carbon (C), oxygen (O), hydrogen (H), and sulfur (S), these elements are necessary for the biosynthesis of building blocks (nucleotides and amino acids) used for the biosynthesis of cellular components such as nucleic acids, proteins, membranous phospholipids, cell wall peptidoglycan, etc. However, the inorganic forms of N (ammonium, nitrates, nitrites, etc.) and P are often scarce in the environment of most microorganisms, and these elements are thus usually present in biological molecules such as nucleic acids, proteins, polyamines, lipids, and the cell wall resulting from plant or animal death. These macromolecules have to be extracellularly degraded into smaller compounds to be transported and assimilated by microorganisms. Alternatively, if external inorganic or organic N and P sources are scarce, the bacteria activates the degradation of its own biological molecules in order to re-cycle N and P present in the latter’s.
In this study, we assessed the content of lipids/triacylglycerol (TAG) and the level of production of the specialized metabolite actinorhodin (ACT) of various strains derived from S. coelicolor (SC) M145 and grown in the classical medium R2YE. These strains were deleted for genes encoding proteins involved in the degradation of proteins (Pup/SCO1646) [1,2] or polyamines (GlnA2/SCO2241, GlnA3/SCO6962, and GlnA4/SCO1613) [3,4,5,6,7] as well as others genes involved in nitrogen metabolism (glnA/sco2198, glnII/sco2210, and nnar/sco2958, gdhA/sco4683) or its regulation (glnR/sco4159, glnRII/sco2213, glnK/sco5584, glnE/sco2234, and glnD/sco5585 and the double mutants afsQ1/sco4907&afsQ2/sco4906 and amtB/sco5583&glnK/sco5584) and of strains derived from S. coelicolor M600 and deleted for genes encoding the global regulator DasR/SCO5231 [8] that controls negatively the expression of proteins involved in the up-take of N-acetylglucosamine (NAG) [9], a component of peptidoglycan, by a phosphotransferase system including the permease NagE2/SCO2907 [10] as well as proteins involved in the intracellular degradation of NAG (NagB/SCO5236, NagK/SCO4285, and NagA/SCO4284) [11].
This study revealed that only 7 deleted strains for the genes pup, glnA2, glnA3, glnA4, dasR, glnK, and glnE, to a lesser extent, had a significant positive impact on the total lipid/TAG content. These 7 strains were grown on R2YE either limited (1 mM) or proficient (5 mM) in phosphate (Pi), and their TAG content and level of ACT production were compared to those of the original strain, S. coelicolor M145 (SC). This study revealed that it was the deletion of the pup, glnA2, and dasR genes that had the strongest positive impact on TAG content in both Pi conditions but more so in Pi proficiency. Unexpectedly, the cardiolipin content of most of these strains (except that of glnA2 and pup mutant strains) was lower than that of the native strain, and that of the dasR mutant strain was the lowest, being threefold lower than that of the original strain. Interestingly, ACT production was totally abolished in the dasR mutant in both Pi conditions, whereas the deletion of pup, glnA2, glnA3, and glnA4 was correlated with a significant increase in total ACT production in Pi limitation, when this production was only slightly enhanced in the pup, glnA2, and glnA4 mutants and was strongly reduced in the glnA3 mutant as well as in the glnE and glnK mutants in Pi proficiency.
Our study confirmed that N limitation is an important trigger of TAG biosynthesis in SC, as in most other microorganisms studied [12,13,14,15]. Surprisingly, the high TAG content of 5 of the 7 mutant strains studied (with the exception of glnA2 and pup mutant strains) was correlated with a low cardiolipin content, suggesting that TAG biosynthesis might occur at the expense of that of CL. N limitation also triggers ACT biosynthesis, but the intensity of this trigger was limited by high Pi availability, whereas high Pi availability stimulated TAG biosynthesis. At last, our data indicated that the biosynthesis of TAG and that of ACT were not mutually exclusive, even if many reports in the literature mentioned that a high TAG content was usually correlated with low specialized metabolite production and conversely [16,17,18,19,20]. In conclusion, we propose that the biosynthesis of ACT, which was shown to have anti-oxidant properties [21] and is likely to be induced by oxidative stress (OxS) [22], together with the enhanced or reduced biosynthesis of TAG and CL, respectively, might contribute to the reduction of OxS resulting from nitrogen limitation or from other causes.
2. Materials and Methods
2.1. Strains and Culture Conditions
The strains used in this study were derived from S. coelicolor (SC) M145 or M600 [23]. These were generous gifts of Wolfgang Wholleben for strains deleted for genes involved in N metabolism (glnA/sco2198, glnII/sco2210, nnar/sco2958,gdhA/sco4683) or its regulation (glnR/sco4159, glnRII/sco2213, glnK/sco5584, glnE/sco2234, glnD/sco5585 and the double mutants afsQ1/sco4907&afsQ2/sco4906 and amtB/sco5583&glnK/sco5584) as well as in polyamine degradation (GlnA2/SCO2241, GlnA3/SCO6962, GlnA4/SCO1613) [3,4,5,6,7]; of Jean-Luc Pernodet and Hasna Boubakri for the strain deleted for pup/sco1646 involved in protein degradation [1,2] and of Gilles Van Wezel and Sébastien Rigali for strains derived from S. coelicolor M600 including strains deleted for the genes encoding the regulator DasR/SCO5231 [8], the permease NagE2/SCO2907 [10] as well as proteins involved in the intracellular degradation of NAG (NagB/SCO5236, NagK/SCO4285, NagA/SCO4284) [11].
These strains were grown in triplicates on agar plates of the modified R2YE medium [24] devoid of sucrose and with (5 mM final, condition of phosphate proficiency) or without (1 mM final, condition of phosphate limitation) addition of K2HPO4. This medium is thought to be limited both in nitrogen and phosphate. Its main nitrogen sources are proline (3 g·L−1), a mixture of casamino acids Difco (0.1 g·L−1) and yeast extract (5 g·L−1), whereas free Pi (1 mM as determined by the Pi blue test from Gentaur, France) originates mainly from yeast extract. Approximately 106 spores of the strains were plated on the surface of cellophane disks (Focus Packaging & Design Ltd., Louth, UK) laid down on the surface of solid R2YE plates. The plates were incubated for 48 h or 72 h at 28 °C, in obscurity.
2.2. Analysis of the Lipid Content
Approximately 106 spores of the 21 mutant strains and of the native strain of SC were plated on cellophane disks deposited on the surface of plates of solid modified R2YE medium proficient in Pi (5 mM). Plates were incubated for 72 h at 28 °C, and mycelial lawns were scraped off the cellophane disks with a spatula and used to determine the global lipid content by Fourier transform infrared spectroscopy (FTIRS, Pike ZnSe ATR-system and FTIR spectrophotometer Vertex 70 from Bruker, Bruker Optics, Germany) as described previously [25]. The accurate determination of the lipid classes present in 7 of these strains (glnA2/sco2241, glnA3/sco6962, glnA4/sco1613, glnE/sco2234, glnK/sco5584, pup/sco1646, and dasR/sco5231 mutant strains) was achieved by LC/MS2 as described previously [26,27].
2.3. Assay of Actinorhodin Production
The 106 spores of the 6 mutant strains and of the native strain of SC were plated on cellophane disks deposited on the surface of plates of solid modified R2YE medium, either proficient (5 mM) or limited (1 mM) in Pi. Plates were incubated for 48 h at 28 °C. The mycelium of the strains was scraped off the cellophane disks with a spatula and used to determine the intracellular ACT production, whereas the agar medium present under the cellophane disk was used to determine the extracellular ACT production as described previously [20,24].
3. Results
3.1. Impact of the Deletion of Genes Encoding Proteins Playing a Role in Nitrogen Metabolism on the Lipid Content of Streptomyces Coelicolor M145
The lipid content of SC (M145 and M600) (Figure S3B) and of the 21 deletion mutants of genes involved in nitrogen metabolism mentioned in the introduction was first assessed using the Fourier transform infrared spectroscopy (FTIRS) method as described previously [25]. This method gives an indication of the total lipid content of the strains, and results are shown in Supplementary Figures S1–S3. Six deletion mutants showing a significant variation in their total lipid content in comparison with the original strain, as well as one (glnE) that did not, were chosen to determine more accurately their content in different lipid species using LC/MS2 [26], when grown on the solid modified R2YE medium either limited (1 mM) or proficient (5 mM) in phosphate (Pi) at 28 °C for 72 h. Results are shown in Figure 1, Figure 2 and Figure 3.
3.1.1. glnA2, glnA3, and glnA4 Deletion Mutants
Streptomyces coelicolor M145 was shown to be able to grow in the presence of high concentrations of polyamines, such as putrescine, cadaverine, spermidine, or spermine, as a sole nitrogen source [3,4,5]. However, these polyamines can also be synthesized intracellularly by bacteria, constituting nitrogen storage molecules that can be degraded in conditions of low external nitrogen availability [7,28]. Genes encoding glutamine synthetase-like proteins were characterized in SC and shown to be involved in the degradation of polyamines. These enzymes, GlnA2 (SCO2241), GlnA3 (SCO6962), and GlnA4 (SCO1613), catalyze the glutamylation of their favorite polyamine substrates, which constitutes the first step of their catabolism. GlnA2 possesses the highest specificity towards short-chain polyamines (putrescine and cadaverine) [3], while GlnA3 (SCO6962) prefers long-chain polyamines (spermidine and spermine) [4] and GlnA4 (SCO1613) accepts only monoamines such as ethanolamine [5]. These enzymes thus contribute to the survival of SC in conditions of high and potentially toxic external polyamine concentrations [28] or in conditions of low external N availability via the degradation of intracellular polyamines, leading to the re-cycling of N present in these biological molecules.
Analysis of the lipid content of S. coelicolor M145 and of these 3 mutant strains grown on solid R2YE medium either limited (1 mM) or proficient (5 mM) in Pi (Figure 1A–C) indicated that (1) the TAG content of S. coelicolor M145 did not vary with Pi availability, as reported previously [27]. In contrast, the TAG content of the glnA2 mutant strain was 2.22 and 2.82, and that of the gln4 mutant was 1.22- and 1.64-fold higher than that of the original strain in Pi limitation and proficiency, respectively. In these conditions, the TAG content of the glnA4 mutant was approximately 40% lower than that of the glnA2 mutant. In contrast, the TAG content of the glnA3 mutant was similar to that of the original strain in Pi limitation and 1.6-fold higher than that of the latter in Pi proficiency, and its TAG content was also approximately 40% lower than that of the glnA2 mutant. (2) The phosphatidyl ethanolamine (PE) content of the glnA2 and glnA3 mutants was similar to that of the original strain in both Pi conditions, whereas that of the glnA4 mutant was slightly lower. This was unexpected since GlnA4 was proposed to be the first enzyme of the ethanolamine degradative pathway; its absence was predicted to be correlated with higher ethanolamine and thus PE levels. The absence of this degradative pathway might thus stimulate the expression of alternative ones [29]. (3) Ornithine lipids (OL) were unfortunately co-eluting with PA and Ac-PIM2, however, since OL are phosphate-free lipids known to replace phospholipids (PL) in conditions of Pi limitation [27,30,31,32], their content is likely to be higher in Pi limitation than in Pi proficiency in the original strain. The putative OL content of the original strain and of the glnA2 mutant were similar, whereas that of the glnA3 and glnA4 mutants was over twofold lower than that of the original strain in Pi limitation. However, the co-elution of OL with PA and Ac-PIM2 did not allow rigorous interpretation of these data. (4) The phosphatidylinositol (PI) content was at a similar level in the original strain and in the glnA2 and glnA4 mutants but was slightly higher in the glnA3 mutant in both Pi conditions. In the 3 mutant strains, the PI content was higher in Pi proficiency than in Pi limitation. (5) The cardiolipin (CL) content of the original strain and of the glnA2 mutant was similar, whereas the CL content of the glnA3 and glnA4 mutants was approximately 30 to 40% lower than that of the original strain in both Pi conditions.
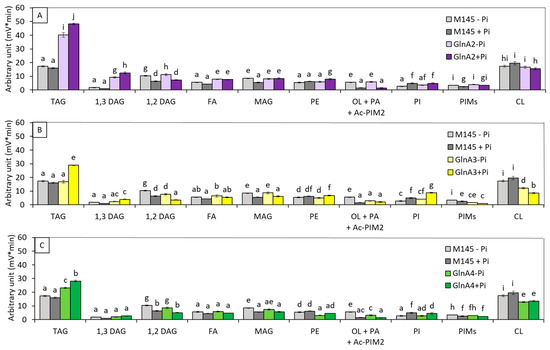
Figure 1.
LC/Corona-CAD analysis of the total lipid content of the original strain of S. coelicolor M145 (grey histograms) and of derivatives of this strain deleted for glnA2/sco2241 (purple histograms) (A), glnA3/sco6962 (yellow histograms) (B) or gnlA4/sco1613 (green histograms) (C). The strains were grown on modified solid R2YE medium either limited (1 mM, light color histograms) or proficient (5 mM, dark color histograms) in phosphate at 28 °C for 72 h. TAG, triacylglycerol; DAG, diacylglycerol (1,2 or 1,3); FA, fatty acids; MAG, monoacylglycerol; PE, phosphatidylethanolamine; OL, ornithine lipids; PI, phosphatidylinositol; Ac-PIM2, acetylated phosphatidylinositol mannoside 2; CL, cardiolipid. Mean values are shown as histograms with error bars representing standard error. Means sharing a letter are not significantly different (p > 0.05; Tukey-adjusted comparisons).
Our results indicate that a default in internal polyamine degradation leads to nitrogen deficiency that triggers TAG biosynthesis in SC, as in most microorganisms studied [12,13,14,15]. Interestingly, external Pi availability did not have any impact on the TAG content of S. coelicolor M145 nor on that of S. lividans TK24, as reported previously [27], whereas TAG and PI content were higher in Pi proficiency in the glnA3 mutant strain as well as in the glnA2 and glnA4 mutant strains, to a lesser extent. This effect is not clearly understood, but it might be due to the higher expression of these genes in Pi proficiency than in Pi limitation; in consequence, their deletion would have more drastic negative consequences on N availability and thus a positive one on TAG biosynthesis in Pi proficiency than in Pi limitation.
3.1.2. Pup Mutant
Pup/SCO1646 (72 AA) is a prokaryotic ubiquitin-like protein that plays an important role in the turnover and degradation of proteins [1,2]. The covalent attachment of this small protein to specific proteins targets the latter to the 20S proteasome, which achieves their degradation. In the pup mutant, protein degradation and thus N turnover is expected to be less efficient than in the original strain leading to reduced N availability that would have a positive impact on TAG biosynthesis.
Analysis of the lipid content of the pup mutant grown in Pi limitation or proficiency (Figure 2) revealed that (1) the TAG content of the pup was 1.6- and 2.25-fold higher than that of the original strain in Pi limitation and proficiency, respectively. (2) The phosphatidyl ethanolamine (PE) content of the original strain and of the pup mutant were rather similar in both Pi conditions. (3) The putative ornithine lipid (OL) content of the pup mutant strain was similar to that of the original strain and, as expected, higher in Pi limitation than in Pi proficiency. (4) The phosphatidylinositol (PI) content of the original strain and of the pup mutant strains was similar and higher in Pi proficiency than in Pi limitation. (5) The cardiolipin (CL) contents of the original strain and of the pup mutant were similar in both Pi conditions.
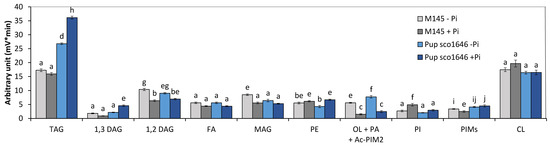
Figure 2.
LC/Corona-CAD analysis of the total lipid content of the original strain of S. coelicolor M145 (grey histograms) and of derivatives of this strain deleted for pup/sco1646 (marine blue histograms). The strains were grown on modified solid R2YE medium either limited (1 mM, light color histograms) or proficient (5 mM, dark color histograms) in phosphate at 28 °C for 72 h. TAG, triacylglycerol; DAG, diacylglycerol (1,2 or 1,3); FA, fatty acids; MAG, monoacylglycerol; PE, phosphatidylethanolamine; OL, ornithine lipids; PI, phosphatidylinositol; Ac-PIM2, acetylated phosphatidylinositol mannoside 2; CL, cardiolipid. Mean values are shown as histograms with error bars representing standard error. Means sharing a letter are not significantly different (p > 0.05; Tukey-adjusted comparisons).
3.1.3. dasR Mutant
The transcriptional regulator DasR was shown to negatively control the uptake of N-acetylglucosamine (NAG), a molecule resulting from the autolytic degradation of cell wall peptidoglycan [8,11], by a phosphotransferase transport system including the permease NagE2/SCO2907 [10]. DasR also negatively controls the intracellular catabolism of NAG by specific enzymes encoded by the genes nagB/sco5236, nagK/sco4285, and nagA/sco4284 [11]. However, DasR is a pleiotropic regulator with multiple regulatory targets. It also negatively controls the expression of acetyl-CoA synthetase [33] as well as citrate synthases [34,35], encoding genes that are directly involved in lipid metabolism, as well as that of the regulator dmdR1, which negatively controls the expression of genes involved in siderophore biosynthesis and uptake [36,37,38,39]. In consequence, in a DasR mutant, genes involved in the catabolism of NAG and those directly involved in lipid metabolism should be up-regulated, whereas those involved in siderophore-mediated iron uptake should be down-regulated.
Analysis of the total lipid content of the nagB/sco5236, nagK/sco4285, nagA/sco4284, and nagE2/sco2907 mutant strains by FTIRS (Figure S3) revealed that these strains had a slightly lower lipid content than the original strain, whereas the dasR mutant had a twofold higher lipid content than the original strain. Analysis of the lipid content of the dasR mutant strain grown in Pi limitation or proficiency in LC/MS2 (Figure 3) revealed that (1) the TAG content of the dasR mutant was 1.82- and 1.68-fold higher than that of the original strain in Pi limitation and proficiency, respectively. (2) The phosphatidyl ethanolamine (PE) content of the original strain and of the dasR mutant were similar in Pi limitation but the PE content of the dasR mutant was 1.5-fold higher than that of the original strain in Pi proficiency. (3) The putative ornithine lipid (OL) contents of the original strain and of the dasR mutant were similar and, as expected, higher in Pi limitation than in Pi proficiency. (4) The phosphatidylinositol (PI) content of the original strain and of the dasR mutant strain was similar and higher in Pi proficiency than in Pi limitation. (5) The cardiolipin (CL) content of the dasR mutant was unexpectedly approximately threefold lower than that of the original strain in both Pi conditions.
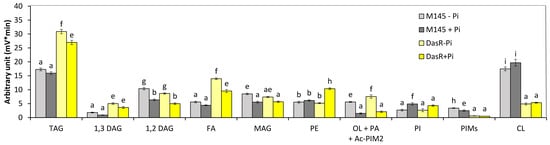
Figure 3.
LC/Corona-CAD analysis of the total lipid content of the original strain of S. coelicolor M145 (grey histograms) and of derivatives of this strain deleted for dasR/sco5231 (brown histograms). The strains were grown on modified solid R2YE medium either limited (1 mM, light color histograms) or proficient (5 mM, dark color histograms) in phosphate, at 28 °C for 72 h. TAG, triacylglycerol; DAG, diacylglycerol (1,2 or 1,3); FA, fatty acids; MAG, monoacylglycerol; PE, phosphatidylethanolamine; OL, ornithine lipids; PI, phosphatidylinositol; Ac-PIM2, acetylated phosphatidylinositol mannoside 2; CL, cardiolipid. Mean values are shown as histograms with error bars representing standard error. Means sharing a letter are not significantly different (p > 0.05; Tukey-adjusted comparisons).
3.1.4. glnK and glnE Deletion Mutants
The glnK/sco5584 gene of S. coelicolor M145 encodes a P-II-like nitrogen regulatory protein. Regulatory PII proteins are thought to sense and integrate the intracellular levels of carbon (2-oxogluratate) and nitrogen (glutamine), as well as the energetic status (ATP/ADP ratio), and control the activities of various transcription factors, transport systems, or enzymes by direct interaction with the latter [40,41,42,43].
The glnE/sco2234 gene of S. coelicolor M145 encodes an adenylyltransferase that adenylates glutamine synthase I/GlnA/SCO2198 [44]. GlnA plays a crucial role in nitrogen assimilation since it catalyzes the condensation of glutamate and ammonium to yield glutamine. The adenylylation of GlnA by GlnE downregulates GlnA activity after an ammonium shock in order to maintain glutamate/glutamine homeostasis [45]. In contrast to the E. coli situation, in Streptomyces, neither GlnK nor GlnD are required for the regulation of GlnE in response to nitrogen availability [46].
Analysis of the lipid content of these 2 mutant strains grown in Pi limitation or proficiency (Figure 4A,B) revealed that (1) the TAG content of the glnE mutant was rather similar to that of the original strain (1.1- and 1.2-fold higher in Pi limitation and Pi proficiency, respectively), whereas that of the glnK mutant was 1.23- and 2-fold higher than that of the original strain in Pi limitation and Pi proficiency, respectively. (2) The phosphatidyl ethanolamine (PE) content of the original strain and of the glnK mutant were similar in both Pi conditions, whereas that of the glnE mutant was approximately twofold lower than that of the original strain in Pi limitation. (3) The putative ornithine lipid (OL) contents of the original strain and of the glnE mutant were similar in both Pi conditions, whereas that of the glnK mutant strains was lower than that of the original strain in both Pi conditions. (4) The phosphatidylinositol (PI) content of the original strain and of the glnK mutant were similar in both Pi conditions, whereas that of the glnE mutant was approximately 1.5- to 2-fold higher than that of the original strain in Pi limitation and proficiency, respectively. (5) The cardiolipin (CL) content of the glnK mutant and the glnE mutant, to a lesser extent, was lower than that of the original strain in both Pi conditions and were lower in Pi proficiency than in Pi limitation.
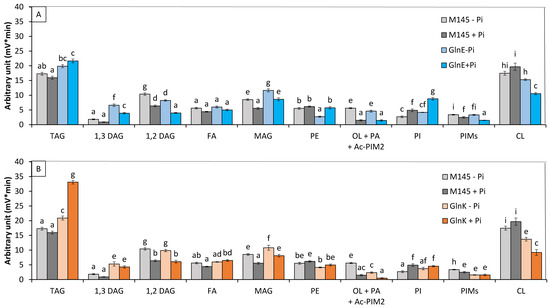
Figure 4.
LC/Corona-CAD analysis of the total lipid content of the original strain of S. coelicolor M145 (grey histograms) and of derivatives of this strain deleted for glnE/sco2234 (blue histograms) (A) and glnK/sco5584 (orange histograms) (B). The strains were grown on modified solid R2YE medium either limited (1 mM, light color histograms) or proficient (5 mM, dark color histograms) in phosphate, at 28 °C for 72 h. TAG, triacylglycerol; DAG, diacylglycerol (1,2 or 1,3); FA, fatty acids; MAG, monoacylglycerol; PE, phosphatidylethanolamine; OL, ornithine lipids; PI, phosphatidylinositol; Ac-PIM2, acetylated phosphatidylinositol mannoside 2; CL, cardiolipid. Mean values are shown as histograms with error bars representing standard error. Means sharing a letter are not significantly different (p > 0.05; Tukey-adjusted comparisons).
The deletion of glnE led only to limited TAG accumulation in both Pi conditions, whereas the deletion of glnK led to twofold increase in TAG content compared to the original strain, but mainly in Pi proficiency. The deletion of genes encoding PII proteins was also reported to enhance fatty acid biosynthesis in E. coli [47]. This might be related to the negative impact that these proteins exert on the activity of acetylCoA-carboxylase as reported in Synechococcus [48]. Furthermore, the activity of GlnK is stimulated when the ATP/ADP ratio is high [40], as in Pi proficiency; the deletion of glnK might have more drastic consequences in Pi proficiency than in Pi limitation. At last, interestingly, the concomitant deletion of amtB/sco5583 and glnK/sco5584 (Figure S1) did not lead to TAG accumulation, suggesting that the ammonium transporter AmtB might be involved in the efflux of intracellular ammonium [49] resulting from the usual degradation of proteins, amino acids, and polyamines. Its deletion would thus lead to an increase in intracellular N availability that would impair the triggering of TAG biosynthesis.
3.2. Impact of the Deletion of Genes Encoding Proteins Playing a Role in the Degradation of Nitrogen-Containing Biological Molecules on the Production of Actinorhodin in Streptomyces Coelicolor M145
Since N as well as P limitations are known to trigger specialized metabolite biosynthesis in Streptomyces species [50,51], we assessed intra- and extracellular production of the blue polyketide actinorhodin (ACT) in our mutants. This study revealed that ACT production was totally abolished in the dasR mutant in both Pi conditions and thus could not be assayed. The biosynthesis of other specialized metabolites was also reported to be abolished in dasR mutants of other Streptomyces species [52,53], as well as in Saccharopolyspora erythraea [54]. Interestingly, the original strain and the 6 mutant strains excreted the most ACT produced when grown in conditions of Pi limitation (Figure 5A), whereas ACT remained mainly intracellular when the strains were grown in conditions of Pi proficiency (Figure 5B).
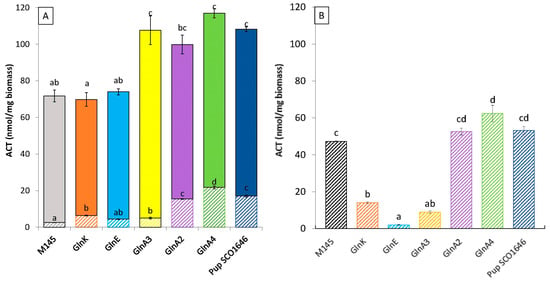
Figure 5.
Quantification of total cellular actinorhodin produced by the original strain of S. coelicolor M145 (grey histograms) and derivatives of this strain deleted for the genes glnK/sco5584 (orange histograms) and glnE/sco2234 (blue histograms), glnA3/sco6962 (yellow histograms), glnA2/sco2241 (purple histograms), gnlA4/sco1613 (green histograms) and pup/sco1646 (marine blue histograms) on modified solid R2YE medium either limited (1 mM) (A) or proficient (5 mM) in phosphate (B) grown at 28 °C for 72 h. Plain and hatched parts of the histograms represent extracellular and intracellular ACT production, respectively. Mean values are shown as histograms with error bars representing standard error. Means sharing a letter are not significantly different (p > 0.05; Tukey-adjusted comparisons).
In Pi limitation, the level of total ACT production of the glnA2, glnA3, and glnA4 mutant strains, as well as the pup mutant, was 1.4- to 1.6-fold higher than that of the original strain. In these conditions, the glnA2, pup, and gln4 mutants had a 2.21-, 1.46-, and 1.19-fold higher TAG content than that of the wild-type strain, respectively, whereas GlnA3 had a TAG content similar to that of the original strain. The level of ACT production of the glnK and glnE mutants was also similar to that of the original strain, and their TAG was only slightly higher than that of the original strain.
In Pi proficiency, the glnA2, glnA4 and pup mutant strains are producing 1.1- to 1.3-fold more intracellular ACT than the original strain. In these conditions, the glnA2, glnA3, glnA4, and pup had a 3-, 1.8-, 1.75-, and 2.25-fold higher TAG content than the original strain, respectively. In contrast, the glnE, glnA3, and glnK mutants produces 12-, 6-, and 3.4-fold less ACT than the original strain. In these conditions, the glnE, glnA3, and glnK mutants had a 1.3-, 1.81-, and 2.1-fold higher TAG content than the original strain.
4. Discussion
In this issue, we demonstrated that mutant strains deleted for the genes pup and glnA2 as well as for glnA4 and glnA3, to a lesser extent, that encode proteins involved in the degradation of N-containing biological molecules, proteins, and polyamines, accumulate a higher level of TAG than the original strain. Since TAG biosynthesis is known to be triggered in conditions of N deprivation in most microorganisms [12,13,14,15], the higher TAG content of the pup and glnA2 mutant strains, compared to that of glnA3 and glnA4 mutant strains, suggested that the degradation of the Pup-targeted proteins by the proteasome and of short-chain polyamines (putrescine and cadaverine) by GlnA2 play a more important role in the internal N supply via the recycling of N present in these biological molecules than GlnA3 and GlnA4 involved in the degradation of long-chain polyamines (spermidine and spermine) and ethanolamine, respectively. This might be due to the higher intracellular abundance of the GlnA2 substrates than of the GlnA3 and GlnA4 substrates. As a result, the deletion of pup and glnA2 led to more severe nitrogen limitation, resulting in higher TAG content than that of glnA3 and glnA4.
The deletion of dasR also had a strong positive impact on TAG biosynthesis, whereas the deletion of genes belonging to the DasR regulon involved in NAG uptake and degradation did not (Figure S3). Since DasR is a pleiotropic regulator with numerous regulatory targets [55], its high TAG content is likely to result from multiple causes. TAG accumulation in the DasR mutant cannot be due to the negative control it exerts on the expression of enzymes involved in NAG uptake and degradation, since in the dasR mutant, the expression of these enzymes is higher than in the original strain [8,11] and should result in an enhanced N availability that has a negative impact on TAG biosynthesis. The high TAG content of the dasR mutant might thus be due to the negative regulation that DasR exerts on the expression of genes encoding citrate synthases, enzymes catalyzing the conversion of citrate into acetylCoA [34,35] and acetyl-CoA synthetase [33]. These regulatory features were demonstrated in Saccharopolyspora erythraea, but not in SC yet. However, if these enzymes are over-expressed in the dasR mutant of SC, as they are in the dasR mutant S. erythraea, an excess acetylCoA might be generated and used for the biosynthesis of fatty acids present in TAG. Furthermore, DasR was shown to repress the expression of the regulator dmdR1, which negatively controls the expression of genes involved in siderophore biosynthesis and uptake in SC [36,37,38,39]. In consequence, in a DasR mutant, these genes are not expressed, and siderophore-mediated iron uptake does not occur resulting into iron deprivation. Interestingly, iron deprivation was shown to promote TAG accumulation, at least in Chlamydomonas species [56,57]. Since iron is a necessary co-factor of numerous enzymes, including enzymes of the TCA cycle (aconitase, citrate synthase, isocitrate dehydrogenase, and succinate dehydrogenase) and of the respiratory chain, reduced iron availability ought to result in low TCA activity, whereas TCA activity is crucial for N assimilation. Iron deprivation might thus indirectly result in low N assimilation, triggering TAG biosynthesis. Interestingly, the lower TAG content of the dasR mutant in Pi proficiency than in Pi limitation might be due to the fact that Pi could be transported as an iron chelate [58] and thus co –Pi/iron transport might reduce the severity of iron shortage and thus TAG accumulation.
At last, we noticed that the dasR mutant had the lowest cardiolipin (CL) content of all strains. Its CL content was approximately threefold lower than that of the original strain in both Pi conditions. The very low CL content of the dasR mutant was unexpected and is not understood, but an anti-correlation between CL and TAG content was previously reported in Saccharomyces cerevisiae [59], suggesting that these two lipid species could be inter-converted. DasR might positively control CL biosynthesis as it does for iron uptake. CL and iron are both known to play an important role in the good functioning of enzymes in the respiratory chain [60,61,62], so the co-regulation of these two processes by DasR could make sense. However, sco1389, encoding a putative eukaryotic-like cardiolipin synthase [30,63], was not found among the DasR target genes [55]. In contrast, sco5773, encoding putative phosphatidylglycerol phosphate synthase [30,63], could possibly be one of the DasR target genes since it is located downstream, transcribed in the same direction, and perhaps co-transcribed with sco5751, which is listed as a potential DasR target gene in Table S1 of [55]. If, as in other organisms, CL plays an important role in the good functioning of enzymes of the respiratory chain [60,61,62] and thus of respiration, the low CL content of the dasR mutant might lead to a reduced respiration and thus a lower generation of OxS, which was proposed to be an important trigger of ACT biosynthesis [22]. Indeed, the deletion of dasR totally abolished ACT production in both Pi conditions, whereas the deletion of glnA2, glnA3, glnA4, and pup led to an increase in ACT production (1.5-fold in average), compared to the original strain, in Pi limitation. As expected, in Pi proficiency, this increase was more moderate (1.2 in average) in the glnA2, glnA4, and pup mutant, since ACT biosynthesis is known to be repressed in this condition [50,64]. In contrast and unexpectedly, ACT production was sixfold lower in the glnA3 mutant than in the original strain in Pi proficiency. The deletion of glnE and glnK also led to a strong reduction of ACT production (12- and 3.4-fold, respectively), but only in Pi proficiency. Interestingly, in all strains, ACT was mainly excreted in Pi limitation, whereas in Pi proficiency, ACT remained intracellular. Since we have previously demonstrated that ACT bears an anti-oxidant function [21] and is thus likely to be triggered by oxidative stress [65,66,67], the strains producing high levels of ACT are likely to be those experiencing high levels of oxidative stress, and conversely. The glnA2, glnA3, glnA4, and pup deletion mutants that are suffering from N limitation are likely to suffer from oxidative stress. Indeed, some reports in the literature mention that N limitation induces oxidative stress (OxS) [68] and that OxS plays a positive role in the triggering of lipid/TAG biosynthesis in various organisms [48,69,70]. The total absence of ACT biosynthesis in the dasR mutant might be attributed to a low level of OxS due to the important storage of acetylCoA as TAG that limits the activation of the oxidative metabolism, the generator of OxS, as well as to iron deficiency that limits the generation of OxS by the Fenton reaction [71]. The very low ACT production of the glnE, glnK, and glnA3 mutant strains in Pi proficiency suggested that oxidative stress was lower in these mutants than in the original strain. GlnE is known to inhibit GlnA activity [44], so in its absence, N assimilation might be stimulated, resulting in N proficiency that is correlated with a lower level of OxS and thus lower ACT biosynthesis. The lower OxS of the glnK mutant might also result from better N assimilation. At last, the low level of OxS of the GlnA3 mutant, which is correlated with reduced ACT biosynthesis, might be due to its high spermine and spermidine content. Indeed, several reports in the literature mention that these molecules protect the cell against OxS [72,73].
In conclusion, our study confirmed that N limitation is an important trigger of TAG biosynthesis in SC, as in most microorganisms [12,13,14,15]. Surprisingly, the high TAG content of 5 of the 7 mutant strains studied (with the exception of glnA2 and pup mutant strains) was correlated with a low cardiolipin content, suggesting that TAG biosynthesis might occur at the expense of that of CL. N limitation also triggers specialized metabolite biosynthesis, but the intensity of this trigger was limited by high Pi availability, whereas high Pi availability stimulated TAG biosynthesis. At last, our data indicated that the biosynthesis of TAG and that of ACT were not mutually exclusive, even if many reports in the literature mentioned that a high TAG content was usually correlated with low specialized metabolite production and conversely [16,17,18,19,20]. Interestingly, the enhanced and reduced TAG and CL content, respectively, and the triggering of ACT biosynthesis might all contribute to the lowering of OxS. Indeed, the storage of acetylCoA as TAG limits the feeding of the TCA cycle and thus the activation of the oxidative metabolism that generates OxS, whereas low CL content might contribute to a lower respiratory activity that would lead to a reduction of OxS. At last, ACT, via its anti-oxidant activity [21], would also limit OxS resulting from nitrogen limitation or from other causes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms12081560/s1, Figure S1: Total lipid content, determined with FTIRS, of S. coelicolor M145 and derivatives of this strain deleted for the genes glnR/sco4159, glnRII/sco2213, glnA/sco2198, glnII/sco2210, glnE/sco2234, nnar/sco2958,gdhA/sco4683, glnD/sco5585, glnK/sco5585, glnA2/sco2241, glnA3/sco6962, glnA4/sco1613 as well as the double mutants afsQ1/sco4907&afsQ2/sco4906 and amtB/sco5583 & glnK/sco5584). The strains were grown, in triplicates, on modified solid R2YE proficient in phosphate (5mM) at 28 °C for 72 h. Mean values of the three replicates are shown as histograms with error bars representing standard error (p > 0.05; Tukey-adjusted comparisons); Figure S2: Total lipid content of S. coelicolor M145 (grey histograms), determined with FTIRS, and of a derivative of this strain deleted for the gene pup/sco1646 (white histograms) and this mutant strain complemented by the gene pup/sco1646 cloned into the vector pSET152 (white histograms) (1). The three strains were grown, in triplicates, in solid R2YE medium limited in phosphate (1mM), for 48 h, 72h and 96h. Mean values of the three replicates are shown as histograms with error bars representing standard error (p > 0.05; Tukey-adjusted comparisons); Figure S3: Total lipid content, determined with FTIRS of S. coelicolor M600 and derivatives of this strain deleted for dasR/sco5231 and for genes of the DasR regulon involved in N-acetylglucosamine (NAG) degradation and up-take, including the genes nagA/sco2758 (NAG-6-phosphate deacetylase), nagB/sco5236 (glucosamine-6-phosphate deaminase/isomerase), nagK/sco4285 (NAG kinase) and nagE2/sco2907 (high-affinity NAG permease) (A) as well as of S. coelicolor M145 and S. coelicolor M600 (B). These strains were grown, in triplicates, on modified solid R2YE medium limited in phosphate (1mM) at 28 °C for 72 h. Mean values of the three replicates are shown as histograms with error bars representing standard error (p > 0.05; Tukey-adjusted comparisons).
Author Contributions
Conceptualization: M.-J.V.; Methodology: M.D. (analysis of global lipid content using FTIRS); S.A. (preparation and analysis of lipid using LC/MS2); C.L. (antibiotic assays and formatting and representation of data); Original draft preparation: M.-J.V.; Reviewing and editing original draft: S.A., P.C. and C.L.; Supervision: M.-J.V. and P.C.; Project administration: M.-J.V.; Funding acquisition: M.-J.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by public institutions (CNRS and University Paris-Saclay) as well as by the program “Investment for the Future: Biotechnologies et Bioressources” n◦11-BTBR-0003 (PROBIO3), the ANR BioSound-IR (ANR-15-CE09-0002) and the ANR-17-ASTR-0018INNOVANTIBIO from the “Direction Générale des Armées” (DGA) and “l’Agence de l’Innovationde Défense” (AID).
Data Availability Statement
The data supporting the findings of this study are available from the corresponding author.
Acknowledgments
We wish to thank Gilles Van Wezel, Sébastien Rigali, Wolfgang Wholleben, Jean-Luc Pernodet and Hasna Boubakri for generous gift of strains as well as Christian Sohlenkamp for stimulating discussions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Boubakri, H.; Seghezzi, N.; Duchateau, M.; Gominet, M.; Kofronova, O.; Benada, O.; Mazodier, P.; Pernodet, J.L. The Absence of Pupylation (Prokaryotic Ubiquitin-Like Protein Modification) Affects Morphological and Physiological Differentiation in Streptomyces coelicolor. J. Bacteriol. 2015, 197, 3388–3399. [Google Scholar] [CrossRef] [PubMed]
- Compton, C.L.; Fernandopulle, M.S.; Nagari, R.T.; Sello, J.K. Genetic and Proteomic Analyses of Pupylation in Streptomyces coelicolor. J. Bacteriol. 2015, 197, 2747–2753. [Google Scholar] [CrossRef] [PubMed]
- Krysenko, S.; Okoniewski, N.; Nentwich, M.; Matthews, A.; Bauerle, M.; Zinser, A.; Busche, T.; Kulik, A.; Gursch, S.; Kemeny, A.; et al. A Second Gamma-Glutamylpolyamine Synthetase, GlnA2, Is Involved in Polyamine Catabolism in Streptomyces coelicolor. Int. J. Mol. Sci. 2022, 23, 3752. [Google Scholar] [CrossRef]
- Krysenko, S.; Okoniewski, N.; Kulik, A.; Matthews, A.; Grimpo, J.; Wohlleben, W.; Bera, A. Gamma-Glutamylpolyamine Synthetase GlnA3 Is Involved in the First Step of Polyamine Degradation Pathway in Streptomyces coelicolor M145. Front. Microbiol. 2017, 8, 726. [Google Scholar] [CrossRef] [PubMed]
- Krysenko, S.; Matthews, A.; Okoniewski, N.; Kulik, A.; Girbas, M.G.; Tsypik, O.; Meyners, C.S.; Hausch, F.; Wohlleben, W.; Bera, A. Initial Metabolic Step of a Novel Ethanolamine Utilization Pathway and Its Regulation in Streptomyces coelicolor M145. Mbio 2019, 10, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Gevrekci, A.O. The roles of polyamines in microorganisms. World J. Microbiol. Biotechnol. 2017, 33, 204. [Google Scholar] [CrossRef]
- Krysenko, S.; Wohlleben, W. Polyamine and Ethanolamine Metabolism in Bacteria as an Important Component of Nitrogen Assimilation for Survival and Pathogenicity. Med. Sci. 2022, 10, 40. [Google Scholar] [CrossRef] [PubMed]
- Rigali, S.; Titgemeyer, F.; Barends, S.; Mulder, S.; Thomae, A.W.; Hopwood, D.A.; van Wezel, G.P. Feast or famine: The global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008, 9, 670–675. [Google Scholar] [CrossRef] [PubMed]
- Rigali, S.; Nothaft, H.; Noens, E.E.; Schlicht, M.; Colson, S.; Muller, M.; Joris, B.; Koerten, H.K.; Hopwood, D.A.; Titgemeyer, F.; et al. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Mol. Microbiol. 2006, 61, 1237–1251. [Google Scholar] [CrossRef]
- Nothaft, H.; Rigali, S.; Boomsma, B.; Swiatek, M.; McDowall, K.J.; van Wezel, G.P.; Titgemeyer, F. The permease gene nagE2 is the key to N-acetylglucosamine sensing and utilization in Streptomyces coelicolor and is subject to multi-level control. Mol. Microbiol. 2010, 75, 1133–1144. [Google Scholar] [CrossRef]
- Swiatek, M.A.; Tenconi, E.; Rigali, S.; van Wezel, G.P. Functional analysis of the N-acetylglucosamine metabolic genes of Streptomyces coelicolor and role in control of development and antibiotic production. J. Bacteriol. 2012, 194, 1136–1144. [Google Scholar] [CrossRef] [PubMed]
- Santucci, P.; Johansen, M.D.; Point, V.; Poncin, I.; Viljoen, A.; Cavalier, J.F.; Kremer, L.; Canaan, S. Nitrogen deprivation induces triacylglycerol accumulation, drug tolerance and hypervirulence in mycobacteria. Sci. Rep. 2019, 9, 8667. [Google Scholar] [CrossRef] [PubMed]
- Boyle, N.R.; Page, M.D.; Liu, B.; Blaby, I.K.; Casero, D.; Kropat, J.; Cokus, S.J.; Hong-Hermesdorf, A.; Shaw, J.; Karpowicz, S.J.; et al. Three acyltransferases and nitrogen-responsive regulator are implicated in nitrogen starvation-induced triacylglycerol accumulation in Chlamydomonas. J. Biol. Chem. 2012, 287, 15811–15825. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cen, K.; Lu, Y.; Zhang, S.; Shang, Y.; Wang, C. Nitrogen-starvation triggers cellular accumulation of triacylglycerol in Metarhizium robertsii. Fungal. Biol. 2018, 122, 410–419. [Google Scholar] [CrossRef]
- Hernandez, M.A.; Lara, J.; Gago, G.; Gramajo, H.; Alvarez, H.M. The pleiotropic transcriptional regulator NlpR contributes to the modulation of nitrogen metabolism, lipogenesis and triacylglycerol accumulation in oleaginous rhodococci. Mol. Microbiol. 2017, 103, 366–385. [Google Scholar] [CrossRef]
- Banchio, C.; Gramajo, H. A stationary-phase acyl-coenzyme A synthetase of Streptomyces coelicolor A3(2) is necessary for the normal onset of antibiotic production. Appl. Environ. Microbiol. 2002, 68, 4240–4246. [Google Scholar] [CrossRef] [PubMed]
- Craney, A.; Ozimok, C.; Pimentel-Elardo, S.M.; Capretta, A.; Nodwell, J.R. Chemical perturbation of secondary metabolism demonstrates important links to primary metabolism. Chem. Biol. 2012, 19, 1020–1027. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, S.; Li, Z.; Zhang, J.; Fan, K.; Tan, G.; Ai, G.; Lam, S.M.; Shui, G.; Yang, Z.; et al. Harnessing the intracellular triacylglycerols for titer improvement of polyketides in Streptomyces. Nat. Biotechnol. 2020, 38, 76–83. [Google Scholar] [CrossRef] [PubMed]
- David, M.; Lejeune, C.; Abreu, S.; Thibessard, A.; Leblond, P.; Chaminade, P.; Virolle, M.J. Negative Correlation between Lipid Content and Antibiotic Activity in Streptomyces: General Rule and Exceptions. Antibiotics 2020, 9, 280. [Google Scholar] [CrossRef]
- Dulermo, T.; Lejeune, C.; Aybeke, E.; Abreu, S.; Bleton, J.; David, M.; Deniset-Besseau, A.; Chaminade, P.; Thibessard, A.; Leblond, P.; et al. Genome Analysis of a Variant of Streptomyces coelicolor M145 with High Lipid Content and Poor Ability to Synthetize Antibiotics. Microorganisms 2023, 11, 1470. [Google Scholar] [CrossRef]
- Millan-Oropeza, A.; Henry, C.; Lejeune, C.; David, M.; Virolle, M.J. Expression of genes of the Pho regulon is altered in Streptomyces coelicolor. Sci. Rep. 2020, 10, 8492. [Google Scholar] [CrossRef] [PubMed]
- Virolle, M.J. A Challenging View: Antibiotics Play a Role in the Regulation of the Energetic Metabolism of the Producing Bacteria. Antibiotics 2020, 9, 83. [Google Scholar] [CrossRef] [PubMed]
- Bentley, S.D.; Chater, K.F.; Cerdeno-Tarraga, A.M.; Challis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Hopwood DA. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Millan-Oropeza, A.; Rebois, R.; David, M.; Moussa, F.; Dazzi, A.; Bleton, J.; Virolle, M.J.; Deniset-Besseau, A. Attenuated Total Reflection Fourier Transform Infrared (ATR FT-IR) for Rapid Determination of Microbial Cell Lipid Content: Correlation with Gas Chromatography-Mass Spectrometry (GC-MS). Appl. Spectrosc. 2017, 71, 2344–2352. [Google Scholar] [CrossRef] [PubMed]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; John Innes Foundation: Foulsham, UK, 2000. [Google Scholar]
- Abreu, S.; Solgadi, A.; Chaminade, P. Optimization of normal phase chromatographic conditions for lipid analysis and comparison of associated detection techniques. J. Chromatogr. A 2017, 1514, 54–71. [Google Scholar] [CrossRef] [PubMed]
- Lejeune, C.; Abreu, S.; Chaminade, P.; Dulermo, T.; David, M.; Werten, S.; Virolle, M.J. Impact of Phosphate Availability on Membrane Lipid Content of the Model Strains, Streptomyces lividans and Streptomyces coelicolor. Front. Microbiol. 2021, 12, 623919. [Google Scholar] [CrossRef]
- Krysenko, S.; Matthews, A.; Busche, T.; Bera, A.; Wohlleben, W. Poly-and Monoamine Metabolism in Streptomyces coelicolor: The New Role of Glutamine Synthetase-Like Enzymes in the Survival under Environmental Stress. Microb. Physiol. 2021, 31, 233–247. [Google Scholar] [CrossRef]
- Kaval, K.G.; Garsin, D.A. Ethanolamine Utilization in Bacteria. Mbio 2018, 9, 10–1128. [Google Scholar] [CrossRef]
- Sandoval-Calderon, M.; Guan, Z.; Sohlenkamp, C. Knowns and unknowns of membrane lipid synthesis in streptomycetes. Biochimie 2017, 141, 21–29. [Google Scholar] [CrossRef]
- Sandoval-Calderon, M.; Nguyen, D.D.; Kapono, C.A.; Herron, P.; Dorrestein, P.C.; Sohlenkamp, C. Plasticity of Streptomyces coelicolor Membrane Composition Under Different Growth Conditions and During Development. Front. Microbiol. 2015, 6, 1465. [Google Scholar] [CrossRef]
- Sohlenkamp, C.; Geiger, O. Bacterial membrane lipids: Diversity in structures and pathways. FEMS Microbiol. Rev. 2016, 40, 133–159. [Google Scholar] [CrossRef]
- You, D.; Zhang, B.Q.; Ye, B.C. GntR Family Regulator DasR Controls Acetate Assimilation by Directly Repressing the acsA Gene in Saccharopolyspora erythraea. J. Bacteriol. 2018, 200, 10–1128. [Google Scholar] [CrossRef]
- Liao, C.H.; Yao, L.L.; Ye, B.C. Three genes encoding citrate synthases in Saccharopolyspora erythraea are regulated by the global nutrient-sensing regulators, G.l.n.R, DasR and CRP. Mol. Microbiol. 2014, 94, 1065–1084. [Google Scholar] [CrossRef]
- Sato, R.; Ara, S.; Yamazaki, H.; Ishiya, K.; Aburatani, S.; Takaku, H. Citrate-Mediated Acyl-CoA Synthesis Is Required for the Promotion of Growth and Triacylglycerol Production in Oleaginous Yeast Lipomyces starkeyi. Microorganisms 2021, 9, 1693. [Google Scholar] [CrossRef]
- Craig, M.; Lambert, S.; Jourdan, S.; Tenconi, E.; Colson, S.; Maciejewska, M.; Ongena, M.; Martin, J.F.; van Wezel, G.; Rigali, S. Unsuspected control of siderophore production by N-acetylglucosamine in streptomycetes. Environ. Microbiol. Rep. 2012, 4, 512–521. [Google Scholar] [CrossRef]
- Flores, F.J.; Martin, J.F. Iron-regulatory proteins DmdR1 and DmdR2 of Streptomyces coelicolor form two different DNA-protein complexes with iron boxes. Biochem. J. 2004, 380, 497–503. [Google Scholar] [CrossRef]
- Flores, F.J.; Barreiro, C.; Coque, J.J.; Martin, J.F. Functional analysis of two divalent metal-dependent regulatory genes dmdR1 and dmdR2 in Streptomyces coelicolor and proteome changes in deletion mutants. FEBS J. 2005, 272, 725–735. [Google Scholar] [CrossRef]
- Bunet, R.; Brock, A.; Rexer, H.U.; Takano, E. Identification of genes involved in siderophore transport in Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 2006, 262, 57–64. [Google Scholar] [CrossRef][Green Version]
- Forchhammer, K.; Selim, K.A.; Huergo, L.F. New views on PII signaling: From nitrogen sensing to global metabolic control. Trends Microbiol. 2022, 30, 722–735. [Google Scholar] [CrossRef]
- Selim, K.A.; Alva, V. PII-like signaling proteins: A new paradigm in orchestrating cellular homeostasis. Curr. Opin. Microbiol. 2024, 79, 102453. [Google Scholar] [CrossRef]
- Jiang, P.; Ninfa, A.J. Escherichia coli PII signal transduction protein controlling nitrogen assimilation acts as a sensor of adenylate energy charge in Vitro. Biochemistry 2007, 46, 12979–12996. [Google Scholar] [CrossRef]
- Gosztolai, A.; Schumacher, J.; Behrends, V.; Bundy, J.G.; Heydenreich, F.; Bennett, M.H.; Buck, M.; Barahona, M. GlnK Facilitates the Dynamic Regulation of Bacterial Nitrogen Assimilation. Biophys. J. 2017, 112, 2219–2230. [Google Scholar] [CrossRef]
- Fink, D.; Falke, D.; Wohlleben, W.; Engels, A. Nitrogen metabolism in Streptomyces coelicolor A3(2): Modification of glutamine synthetase I by an adenylyltransferase. Microbiology 1999, 145 Pt 9, 2313–2322. [Google Scholar] [CrossRef]
- Reuther, J.; Wohlleben, W. Nitrogen metabolism in Streptomyces coelicolor: Transcriptional and post-translational regulation. J. Mol. Microbiol. Biotechnol. 2007, 12, 139–146. [Google Scholar] [CrossRef]
- Hesketh, A.; Fink, D.; Gust, B.; Rexer, H.U.; Scheel, B.; Chater, K.; Wohlleben, W.; Engels, A. The GlnD and GlnK homologues of Streptomyces coelicolor A3(2) are functionally dissimilar to their nitrogen regulatory system counterparts from enteric bacteria. Mol. Microbiol. 2002, 46, 319–330. [Google Scholar] [CrossRef]
- Rodrigues, T.E.; Sassaki, G.L.; Valdameri, G.; Pedrosa, F.O.; Souza, E.M.; Huergo, L.F. Fatty acid biosynthesis is enhanced in Escherichia coli strains with deletion in genes encoding the PII signaling proteins. Arch. Microbiol. 2019, 201, 209–214. [Google Scholar] [CrossRef]
- Verma, E.C.S.; Kharwar, S.; Tiwari, B.; Shila Singh, S.S.; Mishra, A.K. Nitrogen starvation–induced oxidative stress relieves PII-mediated inhibition of acetyl-CoA carboxylase (ACCase) activity and signals enhanced lipid synthesis in Synechococcus PCC 7942. J. Appl. Phycol. 2021, 33, 313–329. [Google Scholar] [CrossRef]
- Allen, W.J.; Collinson, I. Ammonium Transporters: A molecular dual carriageway. eLife 2020, 9, e61148. [Google Scholar] [CrossRef]
- Barreiro, C.; Martinez-Castro, M. Regulation of the phosphate metabolism in Streptomyces genus: Impact on the secondary metabolites. Appl. Microbiol. Biotechnol. 2019, 103, 1643–1658. [Google Scholar] [CrossRef]
- Krysenko, S. Impact of Nitrogen-Containing Compounds on Secondary Metabolism in Streptomyces spp.—A Source of Metabolic Engineering Strategies. Synbio 2023, 1, 204–225. [Google Scholar] [CrossRef]
- Pai, H.; Liu, Y.; Zhang, C.; Su, J.; Lu, W. Effects of the pleiotropic regulator DasR on lincomycin production in Streptomyces lincolnensis. Appl. Microbiol. Biotechnol. 2024, 108, 373. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lin, C.Y.; Li, X.M.; Tang, Z.K.; Qiao, J.; Zhao, G.R. DasR positively controls monensin production at two-level regulation in Streptomyces cinnamonensis. J. Ind. Microbiol. Biotechnol. 2016, 43, 1681–1692. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.H.; Xu, Y.; Rigali, S.; Ye, B.C. DasR is a pleiotropic regulator required for antibiotic production, pigment biosynthesis, and morphological development in Saccharopolyspora erythraea. Appl. Microbiol. Biotechnol. 2015, 99, 10215–10224. [Google Scholar] [CrossRef] [PubMed]
- Swiatek-Polatynska, M.A.; Bucca, G.; Laing, E.; Gubbens, J.; Titgemeyer, F.; Smith, C.P.; Rigali, S.; van Wezel, G.P. Genome-wide analysis of in Vivo binding of the master regulator DasR in Streptomyces coelicolor identifies novel non-canonical targets. PLoS ONE 2015, 10, e0122479. [Google Scholar] [CrossRef] [PubMed]
- Urzica, E.I.; Vieler, A.; Hong-Hermesdorf, A.; Page, M.D.; Casero, D.; Gallaher, S.D.; Kropat, J.; Pellegrini, M.; Benning, C.; Merchant, S.S. Remodeling of membrane lipids in iron-starved Chlamydomonas. J. Biol. Chem. 2013, 288, 30246–30258. [Google Scholar] [CrossRef]
- Devadasu, E.; Subramanyam, R. Enhanced Lipid Production in Chlamydomonas reinhardtii Caused by Severe Iron Deficiency. Front. Plant. Sci. 2021, 12, 615577. [Google Scholar] [CrossRef] [PubMed]
- van Veen, H.W.; Abee, T.; Kortstee, G.J.; Konings, W.N.; Zehnder, A.J. Translocation of metal phosphate via the phosphate inorganic transport system of Escherichia coli. Biochemistry 1994, 33, 1766–1770. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.K.; Rajasekharan, R. Cardiolipin deficiency causes triacylglycerol accumulation in Saccharomyces cerevisiae. Mol. Cell. Biochem. 2017, 434, 89–103. [Google Scholar] [CrossRef] [PubMed]
- Pfeiffer, K.; Gohil, V.; Stuart, R.A.; Hunte, C.; Brandt, U.; Greenberg, M.L.; Schagger, H. Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 2003, 278, 52873–52880. [Google Scholar] [CrossRef]
- Haines, T.H.; Dencher, N.A. Cardiolipin: A proton trap for oxidative phosphorylation. FEBS Lett. 2002, 528, 35–39. [Google Scholar] [CrossRef]
- Gohil, V.M.; Hayes, P.; Matsuyama, S.; Schagger, H.; Schlame, M.; Greenberg, M.L. Cardiolipin biosynthesis and mitochondrial respiratory chain function are interdependent. J. Biol. Chem. 2004, 279, 42612–42618. [Google Scholar] [CrossRef]
- Sandoval-Calderon, M.; Geiger, O.; Guan, Z.; Barona-Gomez, F.; Sohlenkamp, C. A eukaryote-like cardiolipin synthase is present in Streptomyces coelicolor and in most actinobacteria. J. Biol. Chem. 2009, 284, 17383–17390. [Google Scholar] [CrossRef]
- Martin, J.F.; Demain, A.L. Control of antibiotic biosynthesis. Microbiol. Rev. 1980, 44, 230–251. [Google Scholar] [CrossRef]
- Esnault, C.; Dulermo, T.; Smirnov, A.; Askora, A.; David, M.; Deniset-Besseau, A.; Holland, I.B.; Virolle, M.J. Strong antibiotic production is correlated with highly active oxidative metabolism in Streptomyces coelicolor M145. Sci. Rep. 2017, 7, 200. [Google Scholar] [CrossRef]
- Lejeune, C.; Sago, L.; Cornu, D.; Redeker, V.; Virolle, M.J. A Proteomic Analysis Indicates That Oxidative Stress Is the Common Feature Triggering Antibiotic Production in Streptomyces coelicolor and in the pptA Mutant of Streptomyces lividans. Front. Microbiol. 2022, 12, 813993. [Google Scholar] [CrossRef]
- Millan-Oropeza, A.; Henry, C.; Blein-Nicolas, M.; Aubert-Frambourg, A.; Moussa, F.; Bleton, J.; Virolle, M.J. Quantitative Proteomics Analysis Confirmed Oxidative Metabolism Predominates in Streptomyces coelicolor versus Glycolytic Metabolism in Streptomyces lividans. J. Proteome. Res. 2017, 16, 2597–2613. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Chen, H.; He, C.L.; Wang, Q. Nitrogen starvation induced oxidative stress in an oil-producing green alga Chlorella sorokiniana C3. PLoS ONE 2013, 8, e69225. [Google Scholar] [CrossRef]
- Yilancioglu, K.; Cokol, M.; Pastirmaci, I.; Erman, B.; Cetiner, S. Oxidative stress is a mediator for increased lipid accumulation in a newly isolated Dunaliella salina strain. PLoS ONE 2014, 9, e91957. [Google Scholar] [CrossRef]
- Liufu, W.D.M.; Yingying, P.; Liqiu, S.; Wenzhi, W. Nitrogen limitation and hydrogen peroxide act synergistically to enhance lipids accumulation via ROS/Ca2+ dependent mechanism in Chlorella sorokiniana. Algal. Research. 2023, 70, 102974. [Google Scholar] [CrossRef]
- Winterbourn, C.C. Toxicity of iron and hydrogen peroxide: The Fenton reaction. Toxicol Lett. 1995, 82–83, 969–974. [Google Scholar] [CrossRef]
- Chattopadhyay, M.K.; Tabor, C.W.; Tabor, H. Polyamines protect Escherichia coli cells from the toxic effect of oxygen. Proc. Natl. Acad. Sci. USA 2003, 100, 2261–2265. [Google Scholar] [CrossRef] [PubMed]
- Rider, J.E.; Hacker, A.; Mackintosh, C.A.; Pegg, A.E.; Woster, P.M.; Casero, R.A., Jr. Spermine and spermidine mediate protection against oxidative damage caused by hydrogen peroxide. Amino Acids 2007, 33, 231–240. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).