Effects of Deep Tillage on Wheat Regarding Soil Fertility and Rhizosphere Microbial Community
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site and Experimental Design
2.2. Sampling and Soil Fertility Measurement
2.3. DNA Extraction and PCR Amplification
2.4. Illumina MiSeq Sequencing
2.5. Data Processing
2.6. Statistical Analysis
3. Results
3.1. Changes in Edaphic Properties
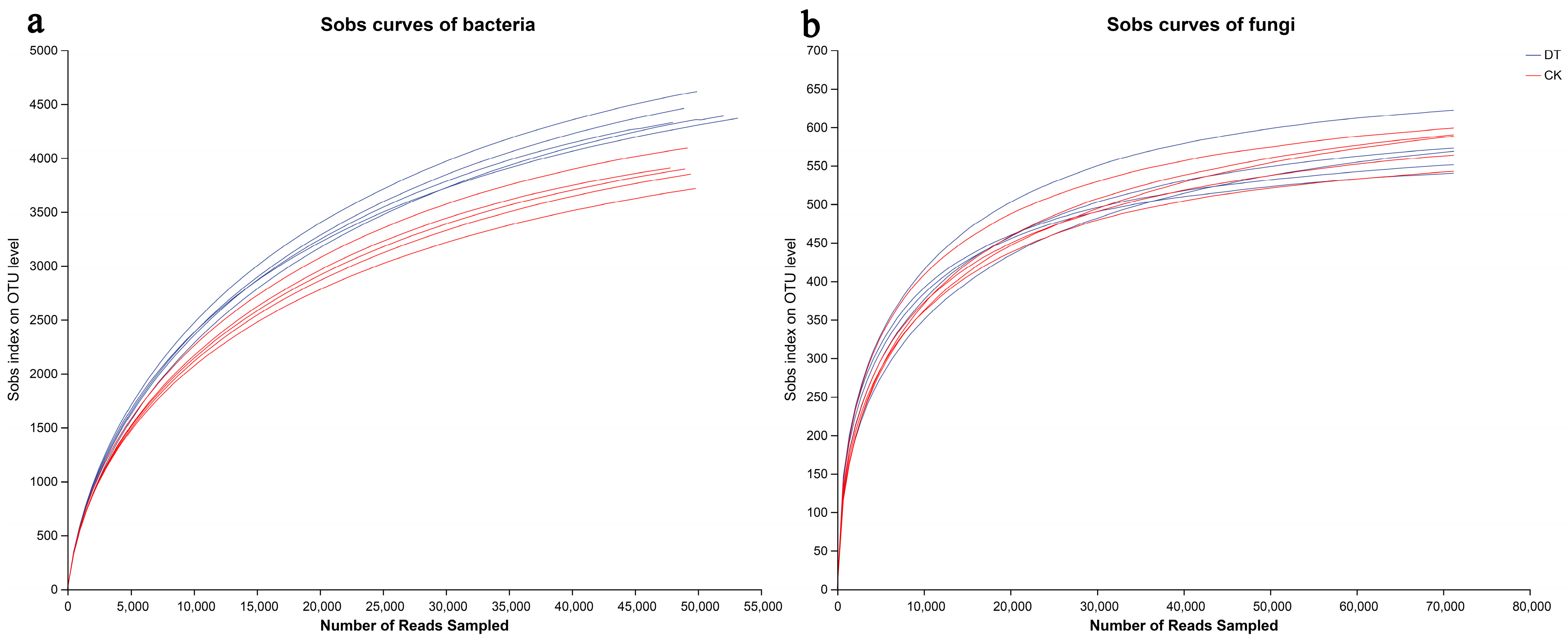
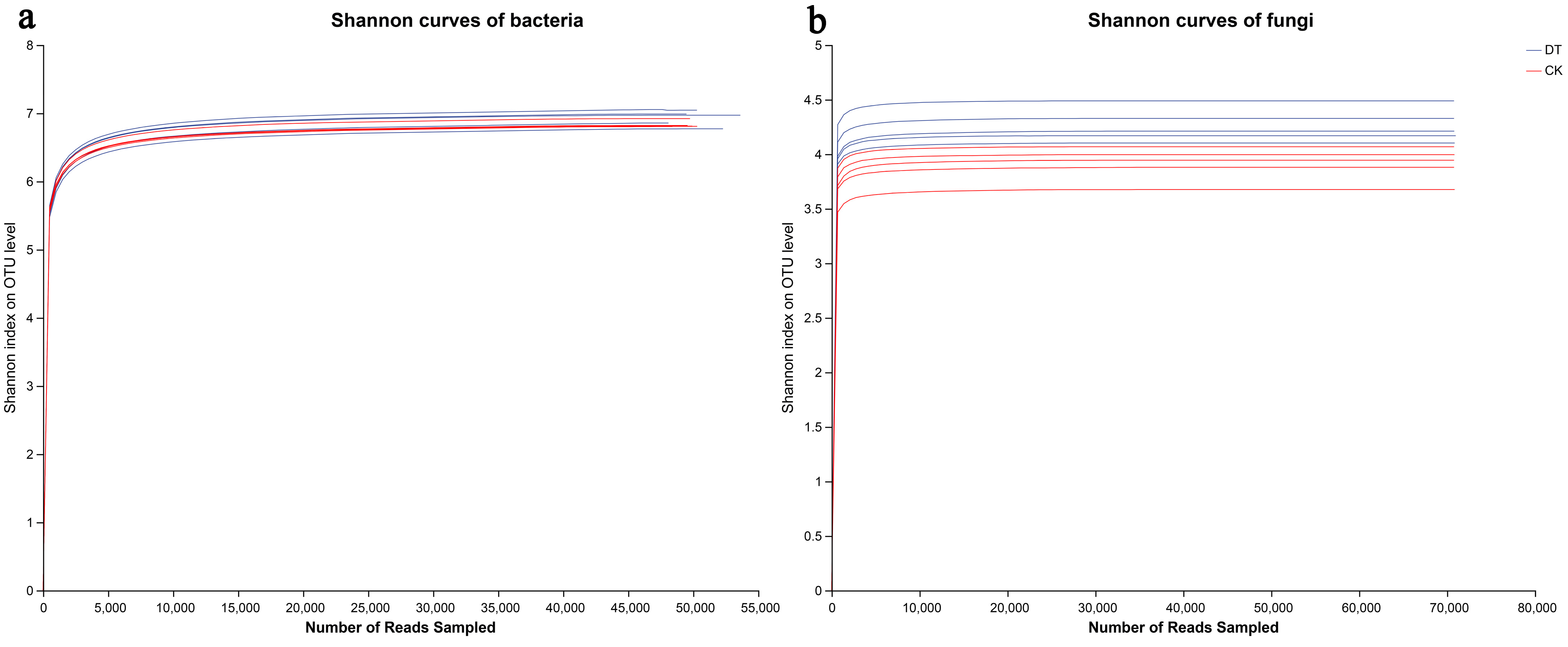
3.2. Sequencing Quality Evaluation
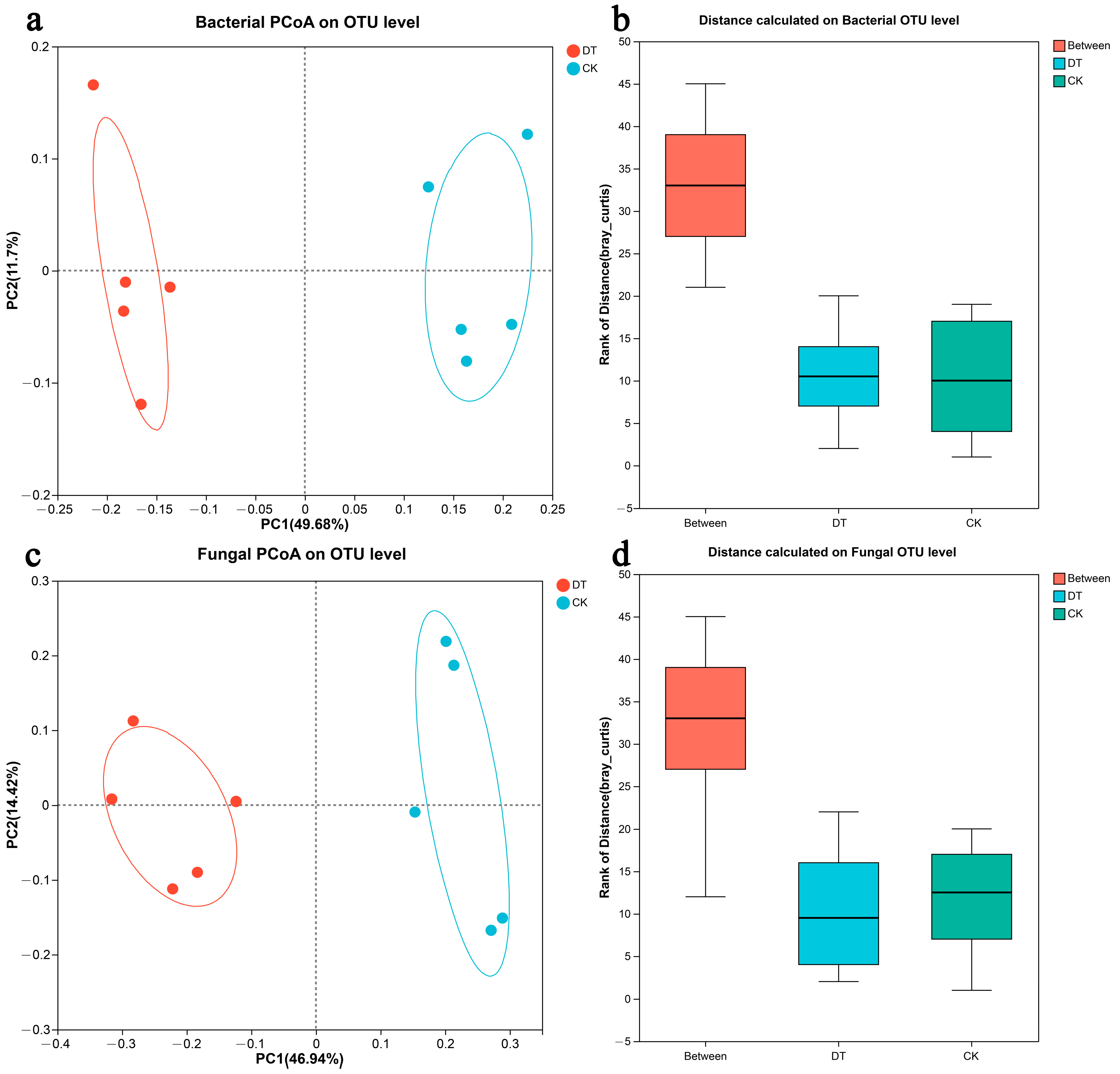
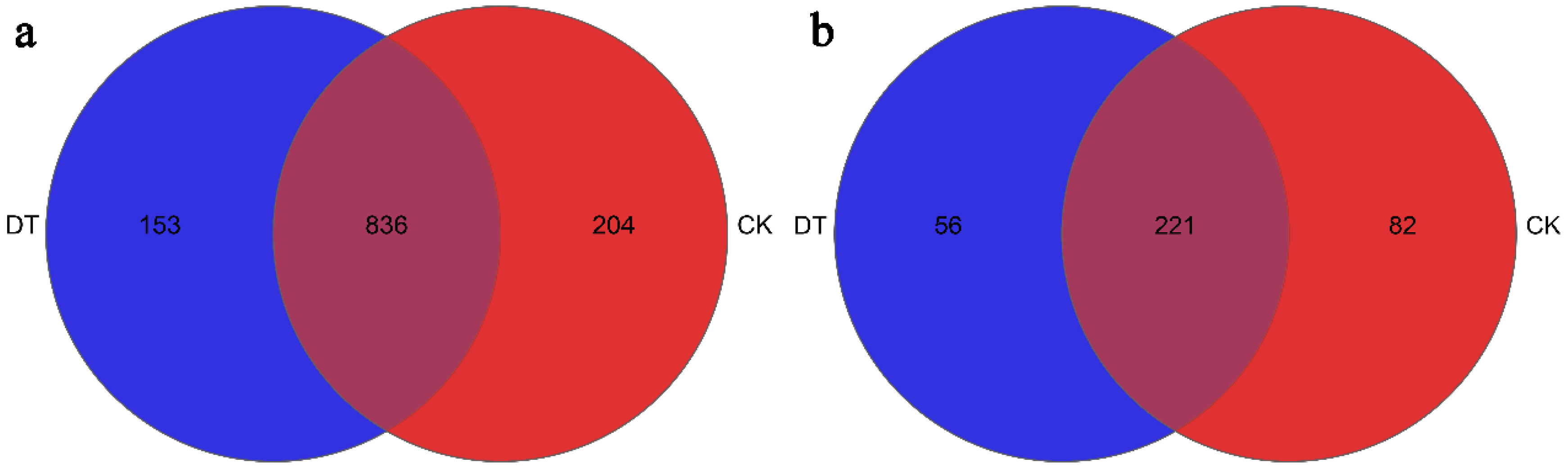
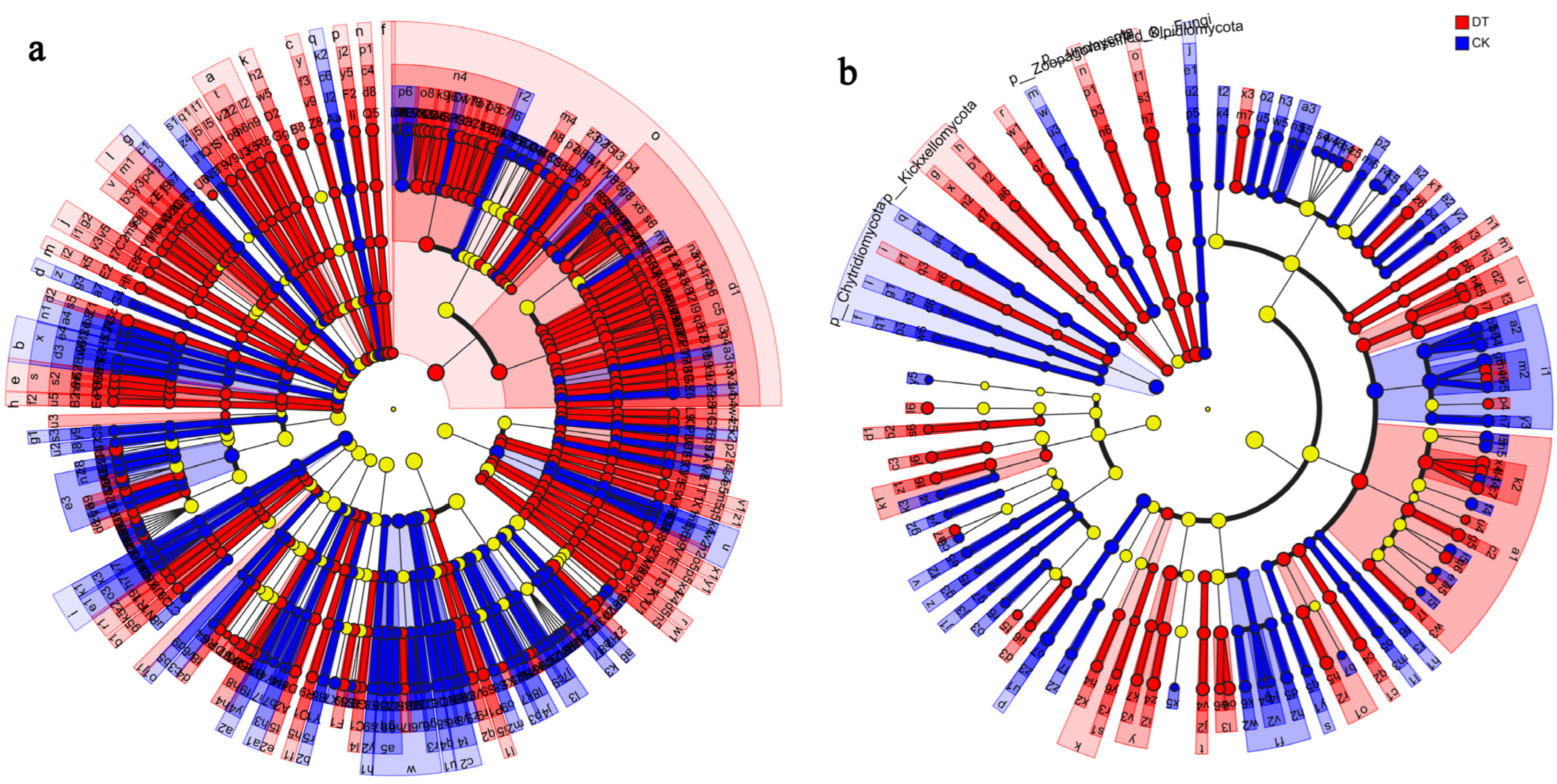
3.3. Microbial Community Composition and Structure
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, Y.; Shen, K.; Yin, C.; Xu, X.; Yu, X.; Ye, B.; Sun, Z.; Dong, J.; Bi, A.; Zhao, X.; et al. Genetic basis of geographical differentiation and breeding selection for wheat plant architecture traits. Genome Biol. 2023, 24, 114. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Li, C.; Fu, K.; Li, C.; Li, C. Phosphorus Alters Starch Morphology and Gene Expression Related to Starch Biosynthesis and Degradation in Wheat Grain. Front. Plant Sci. 2017, 8, 2252. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Li, L.; Zhao, L.; Liu, Z.; Li, X. Effects of Nitrogen Application in the Wheat Booting Stage on Glutenin Polymerization and Structural-Thermal Properties of Gluten with Variations in HMW-GS at the Glu-D1 Locus. Foods 2020, 9, 353. [Google Scholar] [CrossRef] [PubMed]
- Sajid Ansari, M.; Ahmad, G.; Khan, A.A.; Mohamed, H.I.; Elhakem, A. Coal fly ash and nitrogen application as eco-friendly approaches for modulating the growth, yield, and biochemical constituents of radish plants. Saudi J. Biol. Sci. 2022, 29, 103306. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; La, Y.; Dong, T.; Liu, M.; Sun, X. Soil physical properties and vertical distribution of root systems affected by tillage methods in black soil slope farmlands in Northeast China. Trans. Chin. Soc. Agric. Eng. 2021, 37, 125–132. [Google Scholar]
- Chen, H.; Zhao, S.; Zhao, J.; Zhang, K.; Jiang, J.; Guan, Z.; Chen, S.; Chen, F.; Fang, W. Deep tillage combined with biofertilizer following soil fumigation improved chrysanthemum growth by regulating the soil microbiome. Microbiol. Open 2020, 9, e1045. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Xu, P.; Zhang, Z.; Li, S.; Xie, R.; Zhai, L.; Wei, B. Effects of deep vertical rotary tillage on dry matter accumulation and grain yield of summer maize in the Huang-Huai-Hai Plain of China. Soil Tillage Res. 2017, 170, 167–174. [Google Scholar] [CrossRef]
- Guo, X.; Wang, H.; Yu, Q.; Wang, R.; Wang, X.; Li, J. Effects of Tillage on Soil Moisture and Yield of Wheat-Maize Rotation Field in Weibei Upland Plateau. Sci. Agric. Sin. 2021, 54, 2977–2990. [Google Scholar]
- Li, F.; Xu, D.; Wu, P.; Le, T.; Zhu, M.; Li, C.; Zhu, X.; Yang, S.; Ding, J.; Guo, W. Effects of Mechanical Tillage and sowing methods on photosynthetic production and yield of Rice stubble Wheat. Trans. Chin. Soc. Agric. Eng. 2021, 37, 41–49. [Google Scholar]
- Ramadhan, M.N. Yield and yield components of maize and soil physical properties as affected by tillage practices and organic mulching. Saudi J. Biol. Sci. 2021, 28, 7152–7159. [Google Scholar] [CrossRef] [PubMed]
- Alcántara, V.; Don, A.; Vesterdal, L.; Well, R.; Nieder, R. Stability of buried carbon in deep-ploughed forest and cropland soils—Implications for carbon stocks. Sci. Rep. 2017, 7, 5511. [Google Scholar] [CrossRef] [PubMed]
- Shen, F.; Zhu, C.; Jiang, G.; Yang, J.; Zhu, X.; Wang, S.; Wang, R.; Liu, F.; Jie, X.; Liu, S. Differentiation in Nitrogen Transformations and Crop Yield as Affected by Tillage Modes in a Fluvo-Aquic Soil. Plants 2023, 12, 783. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Guan, X.; Liu, C.; Wang, Y.; Wu, P.; Gao, C.; Li, Y.; Qiao, Y.; Zhao, Z.; Shao, J.; et al. Appropriate tillage regime for the improvement of winter wheat yield, soil water, and nutrients condition in Huanghuaihai Plain of China. Trans. Chin. Soc. Agric. Eng. 2023, 39, 82–91. [Google Scholar]
- Meena, K.K.; Sorty, A.M.; Bitla, U.M.; Choudhary, K.; Gupta, P.; Pareek, A.; Singh, D.P.; Prabha, R.; Sahu, P.K.; Gupta, V.K.; et al. Abiotic Stress Responses and Microbe-Mediated Mitigation in Plants: The Omics Strategies. Front. Plant Sci. 2017, 8, 172. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Kang, D.; Wu, J.p.; Zhao, F.; Yang, G.; Han, X.; Feng, Y.; Ren, G. Temporal variation in soil enzyme activities after afforestation in the Loess Plateau, China. Geoderma 2016, 282, 103–111. [Google Scholar] [CrossRef]
- Escudero-Martinez, C.; Coulter, M.; Alegria Terrazas, R.; Foito, A.; Kapadia, R.; Pietrangelo, L.; Maver, M.; Sharma, R.; Aprile, A.; Morris, J.; et al. Identifying plant genes shaping microbiota composition in the barley rhizosphere. Nat. Commun. 2022, 13, 3443. [Google Scholar] [CrossRef] [PubMed]
- Li, E.; de Jonge, R.; Liu, C.; Jiang, H.; Friman, V.P.; Pieterse, C.M.J.; Bakker, P.; Jousset, A. Rapid evolution of bacterial mutualism in the plant rhizosphere. Nat. Commun. 2021, 12, 3829. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Shao, C.; Jin, Q.; Li, M.; Zhang, Z.; Liang, H.; Lei, H.; Qian, J.; Zhang, Y. Effects of cadmium contamination on bacterial and fungal communities in Panax ginseng-growing soil. BMC Microbiol. 2022, 22, 77. [Google Scholar] [CrossRef] [PubMed]
- Ji, B.; Hu, H.; Zhao, Y.; Mu, X.; Liu, K.; Li, C. Effects of deep tillage and straw returning on soil microorganism and enzyme activities. Sci. World J. 2014, 2014, 451493. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Zheng, B.; Liu, Y.; Peng, S.; Gong, J.; Li, J.; Qin, T.; Liang, J.; Xiong, K.; Shao, L.; et al. Deep tillage enhances the spatial homogenization of bacterial communities by reducing deep soil compaction. Soil Tillage Res. 2024, 239, 106062. [Google Scholar] [CrossRef]
- Yao, R.; Gao, Q.; Liu, Y.; Li, H.; Yang, J.; Bai, Y.; Zhu, H.; Wang, X.; Xie, W.; Zhang, X. Deep vertical rotary tillage mitigates salinization hazards and shifts microbial community structure in salt-affected anthropogenic-alluvial soil. Soil Tillage Res. 2023, 227, 105627. [Google Scholar] [CrossRef]
- Gu, Y.; Liu, Y.; Li, J.; Cao, M.; Wang, Z.; Li, J.; Meng, D.; Cao, P.; Duan, S.; Zhang, M.; et al. Mechanism of Intermittent Deep Tillage and Different Depths Improving Crop Growth From the Perspective of Rhizosphere Soil Nutrients, Root System Architectures, Bacterial Communities, and Functional Profiles. Front. Microbiol. 2022, 12, 759374. [Google Scholar] [CrossRef]
- Zhong, R.; Wang, P.R.; Sun, P.J.; Lin, W.; Ren, A.X.; Ren, Y.K.; Sun, M.; Gao, Z.Q. Effects of Long-term Tillage on Soil Bacterial Community Structure and Physicochemical Properties of Dryland Wheat Fields in Northern China. Huan Jing Ke Xue 2023, 44, 5800–5812. [Google Scholar] [PubMed]
- Guan, Y.; Xu, B.; Zhang, X.; Yang, W. Tillage Practices and Residue Management Manipulate Soil Bacterial and Fungal Communities and Networks in Maize Agroecosystems. Microorganisms 2022, 10, 1056. [Google Scholar] [CrossRef] [PubMed]
- Belghit, I.; Lock, E.J.; Fumière, O.; Lecrenier, M.C.; Renard, P.; Dieu, M.; Berntssen, M.H.G.; Palmblad, M.; Rasinger, J.D. Species-Specific Discrimination of Insect Meals for Aquafeeds by Direct Comparison of Tandem Mass Spectra. Animals 2019, 9, 222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Cui, M.; Duan, J.; Zhuang, X.; Zhuang, G.; Ma, A. Abundance, rather than composition, of methane-cycling microbes mainly affects methane emissions from different vegetation soils in the Zoige alpine wetland. Microbiol. Open 2019, 8, e00699. [Google Scholar] [CrossRef] [PubMed]
- Afify, A.; El Baroty, G.S.; El Baz, F.K.; Abd El Baky, H.H.; Murad, S.A. Scenedesmus obliquus: Antioxidant and antiviral activity of proteins hydrolyzed by three enzymes. J. Genet. Eng. Biotechnol. 2018, 16, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Bai, Z.; Jia, A.; Bai, Z.; Qu, S.; Zhang, M.; Kong, L.; Sun, R.; Wang, M. Photovoltaic panels have altered grassland plant biodiversity and soil microbial diversity. Front. Microbiol. 2022, 13, 1065899. [Google Scholar] [CrossRef] [PubMed]
- Hao, Q.; Shang, W.; Zhang, C.; Chen, H.; Chen, L.; Yuan, S.; Chen, S.; Zhang, X.; Zhou, X. Identification and Comparative Analysis of CBS Domain-Containing Proteins in Soybean (Glycine max) and the Primary Function of GmCBS21 in Enhanced Tolerance to Low Nitrogen Stress. Int. J. Mol. Sci. 2016, 17, 620. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.Z.; Zhang, W.J.; Zhao, Z.F.; Qiu, X.C. Determinants and Assembly Mechanism of Bacterial Community Structure in Ningxia Section of the Yellow River. Microorganisms 2023, 11, 496. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, J.; Shen, W.; Xu, H.; Li, Y.; Luo, T. The Structure and Species Co-Occurrence Networks of Soil Denitrifying Bacterial Communities Differ Between A Coniferous and A Broadleaved Forests. Microorganisms 2019, 7, 361. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhao, D.; Ma, W.; Guo, Y.; Lee, D.J. Denitrifying sulfide removal process on high-salinity wastewaters in the presence of Halomonas sp. Appl. Microbiol. Biotechnol. 2016, 100, 1421–1426. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wei, Y.; Yin, Y.; Zhu, W.; Bai, X.; Zhou, Y. Characteristics of Soil Physicochemical Properties and Microbial Community of Mulberry (Morus alba L.) and Alfalfa (Medicago sativa L.) Intercropping System in Northwest Liaoning. Microorganisms 2023, 11, 114. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Mago, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. Taxonomic Note: A Place for DNA-DNA Reassociation and 16S rRNA Sequence Analysis in the Present Species Definition in Bacteriology. Int. J. Syst. Evol. Microbiol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Niu, R.; Zhu, C.; Jiang, G.; Yang, J.; Zhu, X.; Li, L.; Shen, F.; Jie, X.; Liu, S. Variations in Soil Nitrogen Availability and Crop Yields under a Three-Year Annual Wheat and Maize Rotation in a Fluvo-Aquic Soil. Plants 2023, 12, 808. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wu, L.; Zhu, S.; Sun, H.; Xu, C.; Fu, J.; Ning, T. Sensitivities of Physical and Chemical Attributes of Soil Quality to Different Tillage Management. Agronomy 2022, 12, 1153. [Google Scholar] [CrossRef]
- Zhu, C.-w.; Long, Q.; Dong, S.-g.; Shi, K.; Jiang, G.-y.; Li, X.-l.; Zhang, C.-y.; Liu, F.; Shen, F.-m.; Liu, S.-l. Effects of rotary and deep tillage modes on soil microbial biomass carbon and nitrogen and enzyme activities in fluvo-aquic soil under wheat–maize rotation system. J. Plant Nutr. Fertil. 2020, 26, 51–63. [Google Scholar]
- Ghorbani, M.; Konvalina, P.; Neugschwandtner, R.W.; Kopecký, M.; Amirahmadi, E.; Bucur, D.; Walkiewicz, A. Interaction of Biochar with Chemical, Green and Biological Nitrogen Fertilizers on Nitrogen Use Efficiency Indices. Agronomy 2022, 12, 2106. [Google Scholar] [CrossRef]
- Liu, M.; Linna, C.; Ma, S.; Ma, Q.; Song, W.; Shen, M.; Song, L.; Cui, K.; Zhou, Y.; Wang, L. Biochar combined with organic and inorganic fertilizers promoted the rapeseed nutrient uptake and improved the purple soil quality. Front. Nutr. 2022, 9, 997151. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Luo, Y.; Gao, Y.; Gao, X.; Gao, J.; Wang, P.; Feng, B. Soil properties, bacterial and fungal community compositions and the key factors after 5-year continuous monocropping of three minor crops. PLoS ONE 2020, 15, e0237164. [Google Scholar] [CrossRef] [PubMed]
- Yao, D.; Yu, Q.; Xu, L.; Su, T.; Ma, L.; Wang, X.; Wu, M.; Li, Z.; Zhang, D.; Wang, C. Wheat supplement with buckwheat affect gut microbiome composition and circulate short-chain fatty acids. Front. Nutr. 2022, 9, 952738. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liu, Y.; Zhao, L.; Zhang, W.; Liu, L. Change of soil microbial community under long-term fertilization in a reclaimed sandy agricultural ecosystem. PeerJ 2019, 7, e6497. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Dou, X.; Li, J.; Li, F. Impervious Surfaces Alter Soil Bacterial Communities in Urban Areas: A Case Study in Beijing, China. Front. Microbiol. 2018, 9, 226. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.F.; Abdullah, H.; Hassan, M.N.; Jamaludin, M.I.; Sivam, A.; Komatsu, K.; Sapian, I.S.; Alias, H.; Mat Isa, M.N.; Kuwahara, V.S.; et al. Topographically Distinguished Microbiome Taxonomy and Stress-Response Genes of Royal Belum Rainforest and Raja Muda Musa Peat Swamp Revealed through Metagenomic Inquisition. Int. J. Mol. Sci. 2023, 24, 872. [Google Scholar] [CrossRef] [PubMed]
- Ashworth, A.J.; DeBruyn, J.M.; Allen, F.L.; Radosevich, M.; Owens, P.R. Microbial community structure is affected by cropping sequences and poultry litter under long-term no-tillage. Soil. Biol. Biochem. 2017, 114, 210–219. [Google Scholar] [CrossRef]
- Zhou, Z.; Zhang, B.; Wang, Q.; Meng, X.; Wu, Q.; Zheng, T.; Huhe, T. Designing Multi-Stage 2 A/O-MBR Processes for a Higher Removal Rate of Pollution in Wastewater. Membranes 2022, 12, 377. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Wang, R.; Kang, F.; Yan, X.; Sun, L.; Wang, N.; Gong, Y.; Gao, X.; Huang, L. Microbial diversity composition of apple tree roots and resistance of apple Valsa canker with different grafting rootstock types. BMC Microbiol. 2022, 22, 148. [Google Scholar] [CrossRef] [PubMed]
- Aguirre de Cárcer, D. A conceptual framework for the phylogenetically constrained assembly of microbial communities. Microbiome 2019, 7, 142. [Google Scholar] [CrossRef]
- Yu-Te, L.; Whitman, W.B.; Coleman, D.C.; Chih-Yu, C. Effects of reforestation on the structure and diversity of bacterial communities in subtropical low mountain forest soils. Front. Microbiol. 2018, 9, 1968. [Google Scholar]
- Odeberg, G.; Bläckberg, A.; Sunnerhagen, T. Infection or Contamination with Rothia, Kocuria, Arthrobacter and Pseudoglutamicibacter-a Retrospective Observational Study of Non-Micrococcus Micrococcaceae in the Clinic. J. Clin. Microbiol. 2023, 61, e01484-22. [Google Scholar] [CrossRef] [PubMed]
- Huq, M.A.; Akter, S. Characterization and Genome Analysis of Arthrobacter bangladeshi sp. nov., Applied for the Green Synthesis of Silver Nanoparticles and Their Antibacterial Efficacy against Drug-Resistant Human Pathogens. Pharmaceutics 2021, 13, 1691. [Google Scholar] [CrossRef] [PubMed]
- Thomas, F.; Corre, E.; Cébron, A. Stable isotope probing and metagenomics highlight the effect of plants on uncultured phenanthrene-degrading bacterial consortium in polluted soil. ISME J. 2019, 13, 1814–1830. [Google Scholar] [CrossRef] [PubMed]
- Pylak, M.; Oszust, K.; Frąc, M. Searching for New Beneficial Bacterial Isolates of Wild Raspberries for Biocontrol of Phytopathogens-Antagonistic Properties and Functional Characterization. Int. J. Mol. Sci. 2020, 21, 9361. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lee, S.Y.; Akter, S.; Huq, M.A. Probiotic-Mediated Biosynthesis of Silver Nanoparticles and Their Antibacterial Applications against Pathogenic Strains of Escherichia coli O157:H7. Polymers 2022, 14, 1834. [Google Scholar] [CrossRef] [PubMed]
- Mazhar, S.; Khokhlova, E.; Colom, J.; Simon, A.; Deaton, J.; Rea, K. In vitro and in silico assessment of probiotic and functional properties of Bacillus subtilis DE111(®). Front. Microbiol. 2022, 13, 1101144. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Rufty, T.; Shi, W. Predominant Microbial Colonizers in the Root Endosphere and Rhizosphere of Turfgrass Systems: Pseudomonas veronii, Janthinobacterium lividum, and Pseudogymnoascus spp. Front. Microbiol. 2021, 12, 643904. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, V.N.; Lirio, J.M.; Coria, S.H.; Lopes, F.A.C.; Convey, P.; de Oliveira, F.S.; Carvalho-Silva, M.; Câmara, P.; Rosa, L.H. Soil Fungal Diversity and Ecology Assessed Using DNA Metabarcoding along a Deglaciated Chronosequence at Clearwater Mesa, James Ross Island, Antarctic Peninsula. Biology 2023, 12, 275. [Google Scholar] [CrossRef] [PubMed]
- Sui, X.; Zeng, X.; Li, M.; Weng, X.; Frey, B.; Yang, L.; Li, M. Influence of Different Vegetation Types on Soil Physicochemical Parameters and Fungal Communities. Microorganisms 2022, 10, 829. [Google Scholar] [CrossRef] [PubMed]
- Somenahally, A.; DuPont, J.I.; Brady, J.; McLawrence, J.; Northup, B.; Gowda, P. Microbial communities in soil profile are more responsive to legacy effects of wheat-cover crop rotations than tillage systems. Soil Biol. Biochem. 2018, 123, 126–135. [Google Scholar] [CrossRef]
- Wang, Y.; Chang, Y.; Ortañez, J.; Peña, J.F.; Carter-House, D.; Reynolds, N.K.; Smith, M.E.; Benny, G.; Mondo, S.J.; Salamov, A.; et al. Divergent Evolution of Early Terrestrial Fungi Reveals the Evolution of Mucormycosis Pathogenicity Factors. Genome Biol. Evol. 2023, 15, evad046. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Wang, D.; Mao, Z.; Pan, L.; Liao, J.; Cai, Z. Infection of Plasmodiophora brassicae changes the fungal endophyte community of tumourous stem mustard roots as revealed by high-throughput sequencing and culture-dependent methods. PLoS ONE 2019, 14, e0214975. [Google Scholar] [CrossRef] [PubMed]
- Patkowska, E. Biostimulants Managed Fungal Phytopathogens and Enhanced Activity of Beneficial Microorganisms in Rhizosphere of Scorzonera (Scorzonera hispanica L.). Agriculture 2021, 11, 347. [Google Scholar] [CrossRef]
- Besaury, L.; Martinet, L.; Mühle, E.; Clermont, D.; Rémond, C. Streptomyces silvae sp. nov., isolated from forest soil. Int. J. Syst. Evol. Microbiol. 2021, 71, 005147. [Google Scholar] [CrossRef] [PubMed]
- Likar, M.; Grašič, M.; Stres, B.; Regvar, M.; Gaberščik, A. Original Leaf Colonisers Shape Fungal Decomposer Communities of Phragmites australis in Intermittent Habitats. J. Fungi 2022, 8, 284. [Google Scholar] [CrossRef] [PubMed]
- Halo, B.A.; Al-Yahyai, R.A.; Maharachchikumbura, S.S.N.; Al-Sadi, A.M. Talaromyces variabilis interferes with Pythium aphanidermatum growth and suppresses Pythium-induced damping-off of cucumbers and tomatoes. Sci. Rep. 2019, 9, 11255. [Google Scholar] [CrossRef] [PubMed]



| TN g/kg | TP g/kg | TK g/kg | AP mg/kg | AK mg/kg | NN mg/kg | AN mg/kg | OC g/kg | MBC mg/kg | |
|---|---|---|---|---|---|---|---|---|---|
| DT | 0.78 ± 0.10 b | 0.75 ± 0.15 b | 19.34 ± 0.55 b | 11.81 ± 2.06 b | 94.89 ± 11.92 b | 70.77 ± 6.22 a | 10.43 ± 1.06 a | 11.62 ± 1.65 b | 215.94 ± 13.42 b |
| CK | 1.41 ± 0.09 a | 1.21 ± 0.10 a | 21.28 ± 0.72 a | 24.11 ± 7.82 a | 180.97 ± 40.57 a | 49.30 ± 5.97 b | 13.98 ± 3.68 a | 23.79 ± 3.18 a | 571.51 ± 143.23 a |
| Sample | ACE | Chao | Sobs | Simpson | Shannon | Coverage | |
|---|---|---|---|---|---|---|---|
| Bacterial | DT | 5228.87 ± 182.36 a | 4981.95 ± 164.23 a | 4378.20 ± 127.97 a | 0.0048 ± 0.0013 a | 6.92 ± 0.12 a | 0.9775 |
| CK | 4552.78 ± 119.48 b | 4328.56 ± 107.71 b | 3866.40 ± 144.60 b | 0.0036 ± 0.0004 a | 6.83 ± 0.05 a | 0.9815 | |
| Fungal | DT | 599.85 ± 34.66 a | 600.35 ± 29.80 a | 570.80 ± 31.52 a | 0.035 ± 0.008 b | 4.27 ± 0.15 a | 0.9992 |
| CK | 614.75 ± 31.74 a | 610.31 ± 30.82 a | 576.60 ± 23.05 a | 0.062 ± 0.018 a | 3.92 ± 0.15 b | 0.9991 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sui, J.; Wang, C.; Ren, C.; Hou, F.; Zhang, Y.; Shang, X.; Zhao, Q.; Hua, X.; Liu, X.; Zhang, H. Effects of Deep Tillage on Wheat Regarding Soil Fertility and Rhizosphere Microbial Community. Microorganisms 2024, 12, 1638. https://doi.org/10.3390/microorganisms12081638
Sui J, Wang C, Ren C, Hou F, Zhang Y, Shang X, Zhao Q, Hua X, Liu X, Zhang H. Effects of Deep Tillage on Wheat Regarding Soil Fertility and Rhizosphere Microbial Community. Microorganisms. 2024; 12(8):1638. https://doi.org/10.3390/microorganisms12081638
Chicago/Turabian StyleSui, Junkang, Chenyu Wang, Changqing Ren, Feifan Hou, Yuxuan Zhang, Xueting Shang, Qiqi Zhao, Xuewen Hua, Xunli Liu, and Hengjia Zhang. 2024. "Effects of Deep Tillage on Wheat Regarding Soil Fertility and Rhizosphere Microbial Community" Microorganisms 12, no. 8: 1638. https://doi.org/10.3390/microorganisms12081638
APA StyleSui, J., Wang, C., Ren, C., Hou, F., Zhang, Y., Shang, X., Zhao, Q., Hua, X., Liu, X., & Zhang, H. (2024). Effects of Deep Tillage on Wheat Regarding Soil Fertility and Rhizosphere Microbial Community. Microorganisms, 12(8), 1638. https://doi.org/10.3390/microorganisms12081638






