The Diatom Diversity and Ecological Status of a Tufa-Depositing River through eDNA Metabarcoding vs. a Morphological Approach—A Case Study of the Una River (Bosnia and Herzegovina)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Sampling
2.3. Microscopical and Molecular Analysis
2.4. Bioinformatic Processing
2.5. Statistical Analysis
3. Results
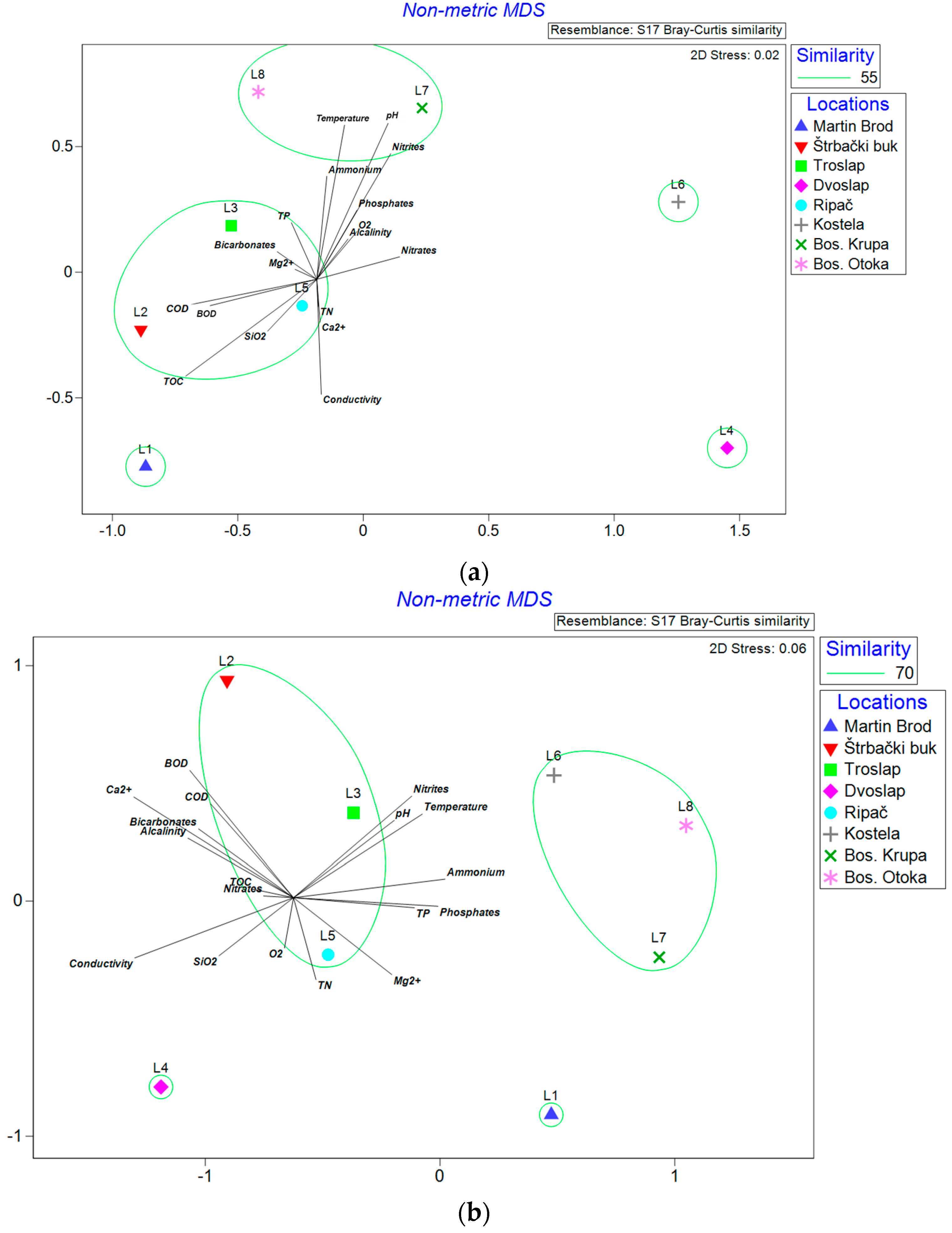
3.1. Physical and Chemical Properties
3.2. Morphological Identification
3.3. Molecular Approach
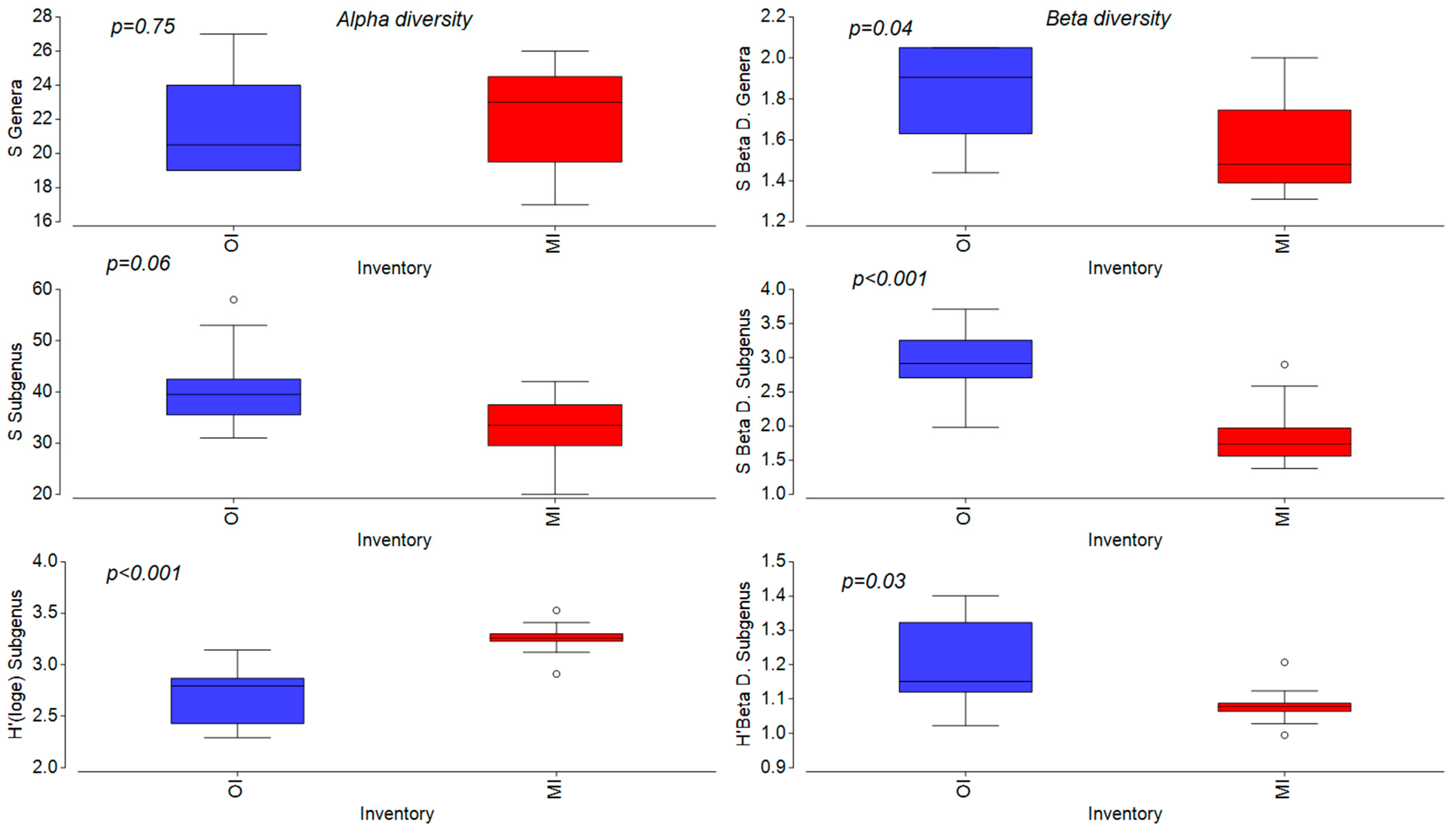
3.4. Comparison of Two Approaches
3.5. Comparison of Methods for Evaluation of Ecological Status
4. Discussion
4.1. The Diatom Communities of Tufa Deposits
4.2. Differences in Methods
4.3. Diversity of Diatoms
4.4. Communities in Different Environmental Conditions
4.5. Assessing the Ecological Status of the Sampling Sites
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ford, T.D.; Pedley, H.M. A Review of Tufa and Travertine Deposits of the World. Earth Sci. Rev. 1996, 41, 117–175. [Google Scholar] [CrossRef]
- Matoničkin, I.; Pavletić, Z. Životni uvjeti na sedrenim slapovima krških voda u Jugoslaviji. Acta Bot. Croat. 1962, 20, 175–198. (In Croatian) [Google Scholar]
- Primc-Habdija, B.; Habdija, I.; Plenković-Moraj, A.E. Tufa deposition and periphyton overgrowth as factors affecting the ciliate community on travertine barriers in different current velocity conditions. Hydrobiologia 2001, 457, 87–96. [Google Scholar] [CrossRef]
- Pavlović, G.; Zupanić, J.; Prohić, E.; Tibljaš, D. Impressions of the biota associated with waterfalls and cascades from a Holocene tufa in the Zrmanja River Canyon, Croatia. Geol. Croat. 2002, 55, 25–37. [Google Scholar] [CrossRef]
- Pedley, H.M.; Rogerson, M. Introduction to tufas and speleothems. Geol. Soc. Lond. Spec. Publ. 2010, 336, 1–5. [Google Scholar] [CrossRef]
- Golubić, S.; Violante, C.; Plenković-Moraj, A.; Grgasović, T. Travertines and Calcareous Tufa Deposits: An Insight into Diagenesis. Geol. Croat. 2008, 61, 363–378. [Google Scholar] [CrossRef]
- Reichardt, E. Zur Diatomeenflora (Bacillariophyceae) tuffabscheidender Quellen und Bäche im Südlichen Frankenjura. Berichte Der Bayer. Bot. Ges. 1994, 64, 119–133. [Google Scholar]
- Winsborough, B.M.; Golubić, S. The role of diatoms in stromatolite growth: Two examples from modern freshwater settings. J. Phycol. 1987, 43, 195–201. [Google Scholar] [CrossRef]
- Plenković-Moraj, A.; Horvatinčić, N.; Primc-Habdija, B. Periphyton and its role in tufa deposition at karstic waters (Plitvice Lakes, Croatia). Biologia 2002, 57, 423–431. [Google Scholar]
- Žutinić, P.; Kulaš, A.; Levkov, Z.; Šušnjara, M.; Orlić, S.; Kukić, S.; Goreta, G.; Valić, D.; Udovič, M.G. Ecological status assessment using periphytic diatom communities—Case study Krka River. Maced. J. Ecol. Environ. 2020, 22, 29–44. [Google Scholar]
- Arp, G.; Bissett, A.; Brinkmann, N.; Cousin, S.; De Beer, D.; Friedl, T.K.I.M.; Neu, T.R.; Reimer, A.; Shiraishi, F.; Stackebrandt, E.; et al. Tufa-Forming Biofilms of German Karstwater Streams: Microorganisms, Exopolymers, Hydrochemistry and Calcification. Geol. Soc. Lond. Spec. Publ. 2010, 336, 83–118. [Google Scholar] [CrossRef]
- Smol, J.P.; Stoermer, E.F. (Eds.) The Diatoms: Applications for the Environmental and Earth Sciences, 2nd ed.; Cambridge University Press: Cambridge, UK, 2010; p. 686. ISBN 9781107564961. [Google Scholar]
- WFD. Directive 2000/60/EC of the European Parliament and of the Council 22.12.2000. Off. J. Eur. Communities 2000, L327, 1–72. [Google Scholar]
- Pawlowski, J.; Kelly-Quinn, M.; Altermatt, F.; Apothéloz-Perret-Gentil, L.; Beja, P.; Boggero, A.; Borja, A.; Bouchez, A.; Cordier, T.; Domaizon, I.; et al. The future of biotic indices in the ecogenomic era: Integrating (e)DNA metabarcoding in biological assessment of aquatic ecosystems. Sci. Total Environ. 2018, 637–638, 1295–1310. [Google Scholar] [CrossRef] [PubMed]
- Kermarrec, L.; Franc, A.; Rimet, F.; Chaumeil, P.; Humbert, J.F.; Bouchez, A. Next-Generation Sequencing to Inventory Taxonomic Diversity in Eukaryotic Communities: A Test for Freshwater Diatoms. Mol. Ecol. Resour. 2013, 13, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Stein, E.D.; Martinez, M.C.; Stiles, S.; Miller, P.E.; Zakharov, E.V. Is DNA barcoding actually cheaper and faster than traditional morphological methods: Results from a survey of freshwater bioassessment efforts in the United States? PLoS ONE 2014, 9, e95525. [Google Scholar] [CrossRef]
- Zimmermann, J.; Glöckner, G.; Jahn, R.; Enke, N.; Gemeinholzer, B. Metabarcoding vs. morphological identification to assess diatom diversity in environmental studies. Mol. Ecol. Resour. 2015, 15, 526–542. [Google Scholar] [CrossRef] [PubMed]
- Keck, F.; Vasselon, V.; Tapolczai, K.; Rimet, F.; Bouchez, A. Freshwater Biomonitoring in the Information Age. Front. Ecol. Environ. 2017, 15, 266–274. [Google Scholar] [CrossRef]
- Vasselon, V.; Rimet, F.; Domaizon, I.; Monnier, O.; Reyjol, Y.; Bouchez, A. Assessing pollution of aquatic environments with diatoms’ DNA metabarcoding: Experience and developments from France water framework directive networks. Metabarcoding Metagenom. 2019, 3, 101–115. [Google Scholar] [CrossRef]
- Kelly, M.; Boonham, N.; Juggins, S.; Kille, P.; Mann, D.; Pass, D.; Sapp, M.; Sato, S.; Glover, R. A DNA Based Diatom Metabarcoding Approach for Water Framework Directive Classification of Rivers; Environment Agency: Bristol, UK, 2018; ISBN 978-1-84911-406-6.
- Rivera, S.F.; Vasselon, V.; Jacquet, S.; Bouchez, A.; Ariztegui, D.; Rimet, F. Metabarcoding of lake benthic diatoms: From structure assemblages to ecological assessment. Hydrobiologia 2018, 807, 37–51. [Google Scholar] [CrossRef]
- Vasselon, V.; Rimet, F.; Tapolczai, K.; Bouchez, A. Assessing ecological status with diatoms DNA metabarcoding: Scaling-up on a WFD monitoring network (Mayotte island, France). Ecol. Indic. 2017, 82, 1–12. [Google Scholar] [CrossRef]
- Leese, F.; Altermatt, F.; Bouchez, A.; Ekrem, T.; Hering, D.; Meissner, K.; Mergen, P.; Pawlowski, J.; Piggott, J.; Rimet, F.; et al. DNAqua-Net: Developing New Genetic Tools for Bioassessment and Monitoring of Aquatic Ecosystems in Europe. Res. Ideas Outcomes 2016, 2, e11321. [Google Scholar] [CrossRef]
- Vasselon, V.; Domaizon, I.; Rimet, F.; Kahlert, M.; Bouchez, A. Application of high-throughput sequencing (HTS) metabarcoding to diatom biomonitoring: Do DNA extraction methods matter? Freshw. Sci. 2017, 36, 162–177. [Google Scholar] [CrossRef]
- Elbrecht, V.; Leese, F. Can DNA-Based Ecosystem Assessments Quantify Species Abundance? Testing Primer Bias and Biomass-Sequence Relationships with an Innovative Metabarcoding Protocol. PLoS ONE 2015, 10, e0130324. [Google Scholar] [CrossRef] [PubMed]
- Bailet, B.; Apothéloz-Perret-Gentil, L.; Baričević, A.; Chonova, T.; Franc, A.; Frigerio, J.-M.; Kelly, M.; Mora, D.; Pfannkuchen, M.; Proft, S.; et al. Diatom DNA Metabarcoding for Ecological Assessment: Comparison Among Bioinformatics Pipelines Used in Six European Countries Reveals the Need for Standardization. Sci. Total Environ. 2020, 745, 140948. [Google Scholar] [CrossRef]
- Tapolczai, K.; Vasselon, V.; Bouchez, A.; Stenger-Kovács, C.; Padisák, J.; Rimet, F. The impact of OTU sequence similarity threshold on diatom-based bioassessment: A case study of the rivers of Mayotte (France, Indian Ocean). Ecol. Evol. 2019, 9, 166–179. [Google Scholar] [CrossRef] [PubMed]
- Tapolczai, K.; Selmeczy, G.B.; Szabó, B.; B-Béres, V.; Keck, F.; Bouchez, A.; Rimet, F.; Padisák, J. The potential of exact sequence variants (ESVs) to interpret and assess the impact of agricultural pressure on stream diatom assemblages revealed by DNA metabarcoding. Ecol. Indic. 2021, 122, 107322. [Google Scholar] [CrossRef]
- Rimet, F.; Vasselon, V.; A-Keszte, B.; Bouchez, A. Do we similarly assess diversity with microscopy and high-throughput sequencing? Case of microalgae in lakes. Org. Divers. Evol. 2018, 18, 51–62. [Google Scholar] [CrossRef]
- Kulaš, A.; Udovič, M.G.; Tapolczai, K.; Žutinić, P.; Orlić, S.; Levkov, Z. Diatom eDNA Metabarcoding and Morphological Methods for Bioassessment of Karstic River. Sci. Total Environ. 2022, 829, 154536. [Google Scholar] [CrossRef]
- Matoničkin, I.; Pavletić, Z. Sedrene naslage u rijeci Uni i njihova biološka uvjetovanost. Hrvat. Geogr. Glas. 1963, 25, 105–112. (In Croatian) [Google Scholar]
- Matoničkin, I.; Pavletić, Z. Životne zajednice na sedrenim slapovima rijeke Une i u brzacima pritoke Unca. Acta Musei. Mac. Sci. Nat. 1959, 6, 76–99. (In Croatian) [Google Scholar]
- Pavletić, Z.; Matoničkin, I. Biološka klasifikacija gornjih tijekova krških rijeka. Acta Bot. Croat. 1965, 24, 151–162. (In Croatian) [Google Scholar]
- Hafner, D. Floristička istraživanja mikrofita rijeke Une (Floristic Research Microphytes on the River Una). Bilt. Društva Ekol. Bosne Hercegovine 1991, 6, 177–185. (In Croatian) [Google Scholar]
- Hafner, D.; Mirković, G. Dijatomeje sedrenih barijera rijeke Une. In Proceedings of the International Conference Protected Areas in Function of Sustainable Development, Bihać, Bosnia and Herzegovina, 6–8 November 2008; Federal Ministry of Environment and Tourism: Sarajevo, Bosnia and Herzegovina, 2008; pp. 533–543. (In Croatian). [Google Scholar]
- Redžić, A. Uticaj onečišćenja na distribuciju fitobentosa na rijeci Uni. Bilt. Društva Ekol. Bosne I Hercegovine 1991, 6, 187–190. (In Bosnian) [Google Scholar]
- Bakrač, A.; Rimceska, B.; Bilbija, B.; Atanacković, A.; Džaferović, A.; Nikolić, V.; Marković, V. Aquatic Macroinvertebrates Diversity in the Upper Stretch of Una River (Una National Park, SW Bosnia and Herzegovina). Ecol. Balk. 2021, 13, 131–141. [Google Scholar]
- Žganec, K.; Gottstein, S.; Đurić, P. Distribution of native and alien gammarids (Crustacea: Amphipoda) along the course of the Una River. Nat. Croat. 2010, 19, 141–150. [Google Scholar]
- Žganec, K.; Lunko, P.; Stroj, A.; Mamos, T.; Grabowski, M. Distribution, ecology and conservation status of two endemic amphipods, Echinogammarus acarinatus and Fontogammarus dalmatinus, from the Dinaric karst rivers, Balkan Peninsula. Ann. De Limnol. Int. J. Limnol. 2016, 52, 13–26. [Google Scholar] [CrossRef]
- Illies, J. (Ed.) Limnofauna Europaea: Eine Zusammenstellung aller die Europäischen Binnengewässer Bewohnenden Mehrzelligen Tierarten mit Angaben über ihre Verbreitung und Ökologie; Gustav Fischer Verlag: Stuttgart, Germany, 1978. [Google Scholar]
- Spahić, M.; Korjenic, A.; Hrelja, E. Problems of genesis, evolution and protection of the Una tuff in Una National Park. Manag. Policy Issues Eco. Mont. 2014, 6, 47–53. [Google Scholar] [CrossRef]
- Spahić, M. The Una river—Potamologic studies. Bilt. Društva Ekol. Bosne i Hercegovine 1991, 6, 161–168. [Google Scholar]
- Ministry of Civil Affairs of Bosnia and Herzegovina, Complex of Travertine Waterfalls in Martin Brod—Una National Park, World Heritage Convention Unesco Home Page. Available online: https://whc.unesco.org/en/tentativelists/6400/ (accessed on 5 June 2024).
- Službene novine Federacije Bosne i Hercegovine, 44/2008. Zakon o Nacionalnom parku “Una”. JP NIO Službeni List Bosne I Hercegovine Home Page. Available online: http://www.sluzbenilist.ba/page/i/zGlzgUW4R5Q= (accessed on 20 February 2024). (In Bosnian).
- Službene novine Federacije Bosne i Hercegovine 01/2014, Odluka o Karakterizaciji Površinskih i Podzemnih Voda, Referentnim Uslovima i Parametrima za Ocjenu Stanja Voda i Monitoringu Voda. JP NIO Službeni List Bosne I Hercegovine Home Page. Available online: https://www.fbihvlada.gov.ba/bosanski/zakoni/2014/odluke%20registar%20glavna2014.htm (accessed on 15 February 2024). (In Bosnian)
- Narodne Novine, Službeni List Republike Hrvatske, 96/2019, Uredba o Standardu Kakvoće Voda. Narodne Novine Home Page. Available online: https://narodne-novine.nn.hr/clanci/sluzbeni/2019_10_96_1879.html (accessed on 15 February 2024). (In Croatian).
- American Public Health Association; American Water Works Association; Water Environment Federation. Standard Methods for the Examination of Water and Wastewater, 23rd ed.; American Public Health Association: Washington, DC, USA, 2017. [Google Scholar]
- Hustedt, F. Die Süßwasserflora Mitteleuropas. Heft 10: Bacillariophyta (Diatomeae), 2nd ed.; Gustav Fischer: Jena, Germany, 1930. [Google Scholar]
- Krammer, K.; Lange-Bertalot, H. Bacillariophyceae. 2. Teil: Bacillariaceae, Epithemiaceae, Surirellaceae. In Süsswasserflora von Mitteleuropa; Ettl, H., Gerloff, J., Heynig, H., Mollenhauer, D., Eds.; Gustav Fischer: Jena, Germany, 1988; Volume 2/2, pp. 1–596. [Google Scholar]
- Krammer, K.; Lange-Bertalot, H. Bacillariophyceae. 1. Teil: Naviculaceae. In Süsswasserflora von Mitteleuropa; Ettl, H., Gerloff, J., Heynig, H., Mollenhauer, D., Eds.; Gustav Fischer: Stuttgart, Germany, 1986; Volume 2/1, pp. 1–876. [Google Scholar]
- Krammer, K.; Lange-Bertalot, H. Bacillariophyceae. 3. Teil: Centrales, Fragilariaceae, Eunotiaceae. In Süsswasserflora von Mitteleuropa; Ettl, H., Gerloff, J., Heynig, H., Mollenhauer, D., Eds.; Gustav Fisher Verlag: Stuttgart, Germany, 1991; Volume 2/3, pp. 1–598. [Google Scholar]
- Krammer, K.; Lange-Bertalot, H. Bacillariophyceae 4. Teil: Achnanthaceae. Kritische Ergänzungen zu Navicula (Lineolatae) und Gomphonema Gesamtliteraturverzeichnis Teil 1–4. In Süßwasserflora von Mitteleuropa; Ettl, H., Gärtner, G., Gerloff, J., Heynig, H., Mollenhauer, D., Eds.; Spektrum Akademischer: Heidelberg/Berlin, Germany, 2004; Volume 2/4, pp. 1–468. [Google Scholar]
- Krammer, K. The Genus Pinnularia. In Diatoms of Europe, Volume 1; Lange-Bertalot, H., Ed.; A.R.G. Gantner Verlag K.G.: Ruggell, Liechtenstein, 2000; pp. 1–703. [Google Scholar]
- Krammer, K. Cymbella. In Diatoms of Europe; Lange-Bertalot, H., Ed.; A.R.G. Gantner Verlag K.G.: Ruggell, Liechtenstein, 2002; Volume 3, pp. 1–584. [Google Scholar]
- Hofmann, G.; Werum, M.; Lange-Bertalot, H. Diatomeen im Süßwasser—Benthos von Mitteleuropa: Bestimmungsflora Kieselalgen für die ökologische Praxis; Ganter: Rugell, Liechtenstein, 2011; pp. 1–1000. [Google Scholar]
- Lange-Bertalot, H.; Hofmann, G.; Werum, M.; Cantonati, M. Freshwater Benthic Diatoms of Central Europe: Over 800 Common Species Used in Ecological Assessment; Cantonati, M., Kelly, M.G., Lange-Bertalot, H., Eds.; Koeltz Botanical Books: Oberreifenberg, Germany, 2017; pp. 1–942. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Keck, F. DADA2 Pipeline for rbcL Diatoms. GitHub Repository. Available online: https://github.com/fkeck/DADA2_diatoms_pipeline (accessed on 10 January 2024).
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Keck, F.; Rimet, F.; Vasselon, V.; Bouchez, A. A Ready-to-Use Database for DADA2: Diat.barcode_rbcL_312bp_DADA2. 2019. Recherche Data Gouv, V2. Available online: https://entrepot.recherche.data.gouv.fr/dataset.xhtml?persistentId=doi:10.15454/HNI1EK (accessed on 24 January 2024). [CrossRef]
- Rimet, F.; Gusev, E.; Kahlert, M.; Kelly, M.G.; Kulikovskiy, M.; Maltsev, Y.; Mann, D.G.; Pfannkuchen, M.; Trobajo, R.; Vasselon, V.; et al. Diat. Barcode, an open-access curated barcode library for diatoms. Sci. Rep. 2019, 9, 15116. [Google Scholar] [CrossRef]
- ESRI, ArcGIS Online. Available online: https://www.arcgis.com/index.html (accessed on 13 February 2023).
- Clarke, K.R.; Gorley, R.N. PRIMER v7: User Manual/Tutorial; PRIMER-E: Plymouth, UK, 2015. [Google Scholar]
- Hammer, Ø.; Harper, D.A.T.; Ryan, P.D. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 9. Available online: http://palaeo-electronica.org/2001_1/past/issue1_01.htm (accessed on 23 April 2024).
- Gloor, G.B.; Macklaim, J.M.; Pawlowsky-Glahn, V.; Egozcue, J.J. Microbiome Datasets Are Compositional: And This Is Not Optional. Front. Microbiol. 2017, 8, 2224. [Google Scholar] [CrossRef]
- Mantel, N. The Detection of Disease Clustering and a Generalized Regression Approach. Cancer Res. 1967, 27, 209–220. [Google Scholar]
- Vasselon, V.; Bouchez, A.; Rimet, F.; Jacquet, S.; Trobajo, R.; Corniquel, M.; Tapolczai, K.; Domaizon, I. Avoiding quantification bias in metabarcoding: Application of a cell biovolume correction factor in diatom molecular biomonitoring. Methods Ecol. Evol. 2018, 9, 1060–1069. [Google Scholar] [CrossRef]
- Metodologija Uzorkovanja, Laboratorijskih Analiza i Određivanja Omjera Ekološke Kakvoće Bioloških Elemenata Kakvoće, Hrvatske Vode 2016. Hrvatske Vode Home Page. Available online: https://voda.hr/hr/node/1293 (accessed on 15 February 2024). (In Croatian).
- Rott, E.; Pfister, P.; van Dam, H.; Pipp, E.; Pall, K.; Binder, N.; Ortler, K. Indikationslisten für Aufwuchsalgen. Teil 2: Trophieindikation sowie geochemische. Präferenz, taxonomische und toxikologische Anmerkungen; Bundesministerium für Land- und Forstwirtschaft: Wien, Austria, 1999; pp. 1–248.
- Pantle, R.; Buck, H. Die Biologische Uberwachung der Gewässer und Die Darstellung der Ergebnisse. Gas Wasserfach Wasser Abwasser 1955, 96, 604. [Google Scholar]
- Kelly, M.G.; Adams, C.; Graves, A.C.; Jamieson, J.; Krokowski, J.; Lycett, E.B.; Wilkins, C. The Trophic Diatom Index: A User’s Manual Revised Edition R&D Technical Report E2/TR2; Environment Agency: Bristol, UK, 2001.
- Coste, M. Étude des Méthodes Biologiques d’Appréciation Quantitative de la Qualité des Eaux; Rapport d’Étude Cemagref; Agence de l’Eau: Corse, France, 1982; p. 218. [Google Scholar]
- Water Management Plan for the Water Area of the Sava River in the Federation of Bosnia and Herzegovina, 2022–2027. In Bosnian: Plan Upravljanja Vodama za Vodno Područje Rijeke Save u Federaciji Bosne i Hercegovine (2022–2027). Prateći dokument br.1. Karakterizacijski izvještaj_Dio 1. Available online: https://www.voda.ba/plan-upravljanja-2022-2027 (accessed on 25 January 2024).
- Lecointe, C.; Coste, M.; Prygiel, J. “Omnidia”: Software for Taxonomy, Calculation of Diatom Indices and Inventories Management. Hydrobiologia 1993, 269, 509–513. [Google Scholar] [CrossRef]
- Wegl, R. Index für Limnosaprobität. Wasser Abwasser 1983, 26, 1–175. [Google Scholar]
- Zabell, S.L. On Student’s 1908 article “The probable error of a mean”. J. Am. Stat. Assoc. 2008, 103, 1–7. [Google Scholar] [CrossRef]
- Bauer, D.F. Constructing Confidence Sets Using Rank Statistics. J. Am. Stat. Assoc. 1972, 67, 687–690. [Google Scholar] [CrossRef]
- Clarke, K.R. Non-Parametric Multivariate Analyses of Changes in Community Structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Bailet, B.; Bouchez, A.; Franc, A.; Frigerio, J.-M.; Keck, F.; Karjalainen, S.M.; Rimet, F.; Schneider, S.; Kahlert, M. Molecular versus Morphological Data for Benthic Diatoms Biomonitoring in Northern Europe Freshwater and Consequences for Ecological Status. Metabarcoding Metagenomics 2019, 3, 21–35. [Google Scholar] [CrossRef]
- Golubić, S. The Relationship Between Blue-Green Algae and Carbonate Deposits. In The Biology of Blue–Green Algae; Blackwell: Oxford, UK, 1973; pp. 434–472. [Google Scholar]
- Pentecost, A. Association of cyanobacteria with tufa deposits: Identity, enumeration, and nature of the sheath material revealed by histochemistry. Geomicrobiol. J. 1985, 4, 285–298. [Google Scholar] [CrossRef]
- Cantonati, M.; Angeli, N.; Bertuzzi, E.; Spitale, D.; Lange-Bertalot, H. Diatoms in Springs of the Alps: Spring Types, Environmental Determinants, and Substratum. Freshwater Sci. 2012, 31, 499–524. [Google Scholar] [CrossRef]
- Nistal-García, A.; García-García, P.; García-Girón, J.; Borrego-Ramos, M.; Blanco, S.; Bécares, E. DNA Metabarcoding and Morphological Methods Show Complementary Patterns in the Metacommunity Organization of Lentic Epiphytic Diatoms. Sci. Total Environ. 2021, 786, 147423. [Google Scholar] [CrossRef]
- Annunziata, R.; Balestra, C.; Marotta, P.; Ruggiero, A.; Manfellotto, F.; Benvenuto, G.; Biffali, E.; Ferrante, M.I. An Optimized Method for Intact Nuclei Isolation from Diatoms. Sci. Rep. 2021, 11, 27–29. [Google Scholar] [CrossRef]
- Kuffel, A.; Gray, A.; Daeid, N.N. Impact of Metal Ions on PCR Inhibition and RT-PCR Efficiency. Int. J. Leg. Med. 2021, 135, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Burillo, J.; Trobajo, R.; Vasselon, V.; Rimet, F.; Bouchez, A.; Mann, D.G. Evaluation and sensitivity analysis of diatom DNA metabarcoding for WFD bioassessment of Mediterranean rivers. Sci. Total Environ. 2020, 727, 138445. [Google Scholar] [CrossRef] [PubMed]
- Mora, D.; Abarca, N.; Proft, S.; Grau, J.H.; Enke, N.; Carmona, J.; Skibbe, O.; Jahn, R.; Zimmermann, J. Morphology and Metabarcoding: A Test with Stream Diatoms from Mexico Highlights the Complementarity of Identification Methods. Freshw. Sci. 2019, 38, 448–464. [Google Scholar] [CrossRef]
- Burfeid-Castellanos, A.M.; Mones, P.; Dani, M.; Beszteri, B. Non-Perennial Rivers and Streams in Extreme Hydrological Conditions—Comparing the Effectiveness of Amplicon Sequencing and Digital Microscopy for Diatom Biodiversity Appraisal. Front. Ecol. Evol. 2024, 12, 1355018. [Google Scholar] [CrossRef]
- Van Dam, H.; Mertens, A.; Sinkeldam, J. A coded checklist and ecological indicator values of freshwater diatoms from The Netherlands. Neth. J. Aquat. Ecol. 1994, 28, 117–133. [Google Scholar] [CrossRef]
- Burkholder, J.M.; Wetzel, R.G. Epiphytic Alkaline Phosphatase on Natural and Artificial Plants in an Oligotrophic Lake: Re-Evaluation of the Role of Macrophytes as a Phosphorus Source for Epiphytes. Limnol. Oceanogr. 1990, 35, 736–747. [Google Scholar] [CrossRef]
- Tapolczai, K.; Chonova, T.; Fidlerová, D.; Makovinská, J.; Mora, D.; Weigand, A.; Zimmermann, J. Molecular metrics to monitor ecological status of large rivers: Implementation of diatom DNA metabarcoding in the Joint Danube Survey 4. Ecol. Indic. 2024, 160, 111883. [Google Scholar] [CrossRef]
- Kelly, M.G.; Whitton, B.A. The Trophic Diatom Index: A New Index for Monitoring Eutrophication in Rivers. J. Appl. Phycol. 1995, 7, 433–444. [Google Scholar] [CrossRef]
- Kochoska, H.; Chardon, C.; Chonova, T.; Keck, F.; Kermarrec, L.; Larras, F.; Lefrancois, E.; Rivera, S.F.; Tapolczai, K.; Vasselon, V.; et al. Filling reference libraries with diatom environmental sequences: Strengths and weaknesses. Diatom Res. 2023, 38, 103–127. [Google Scholar] [CrossRef]
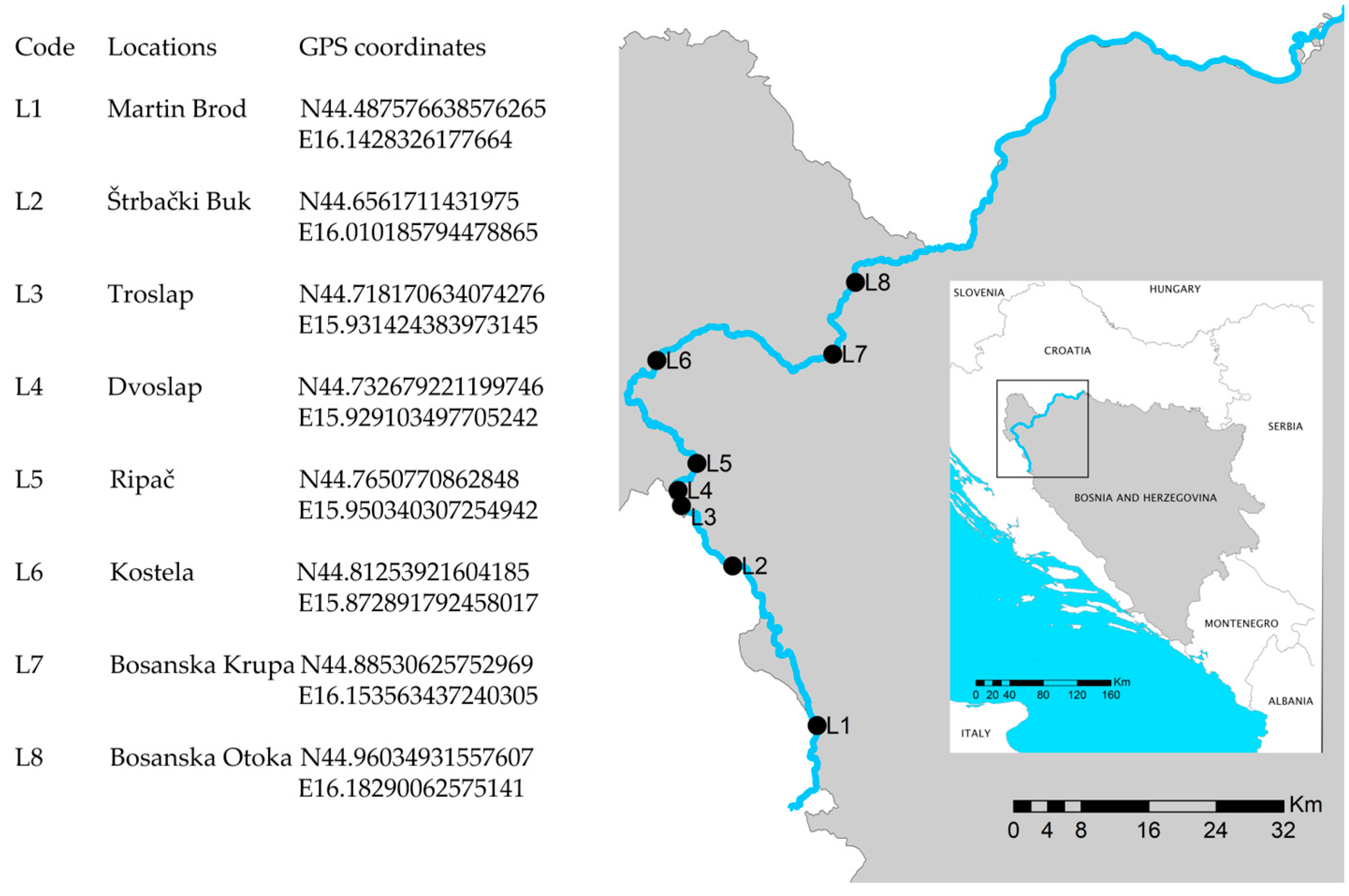
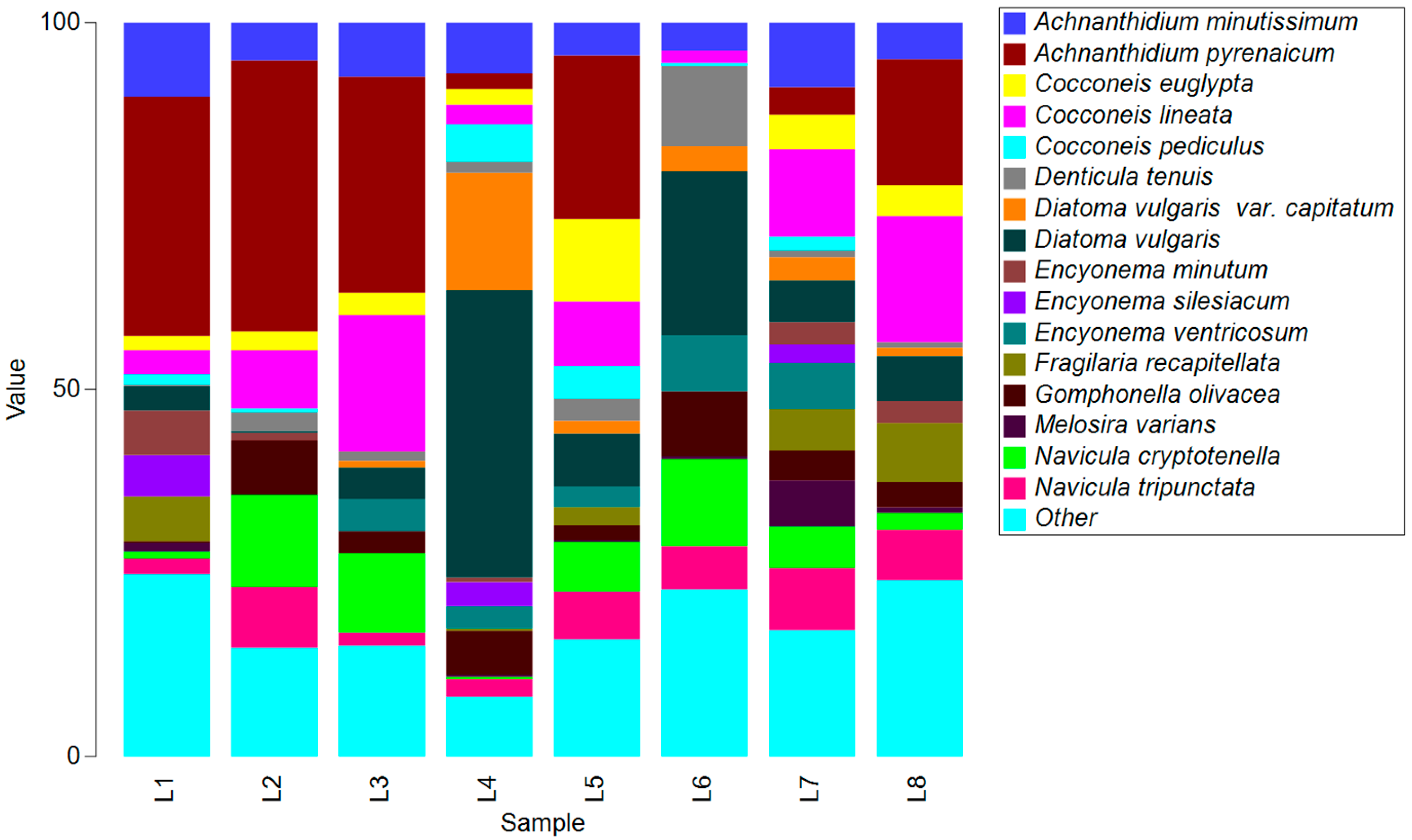
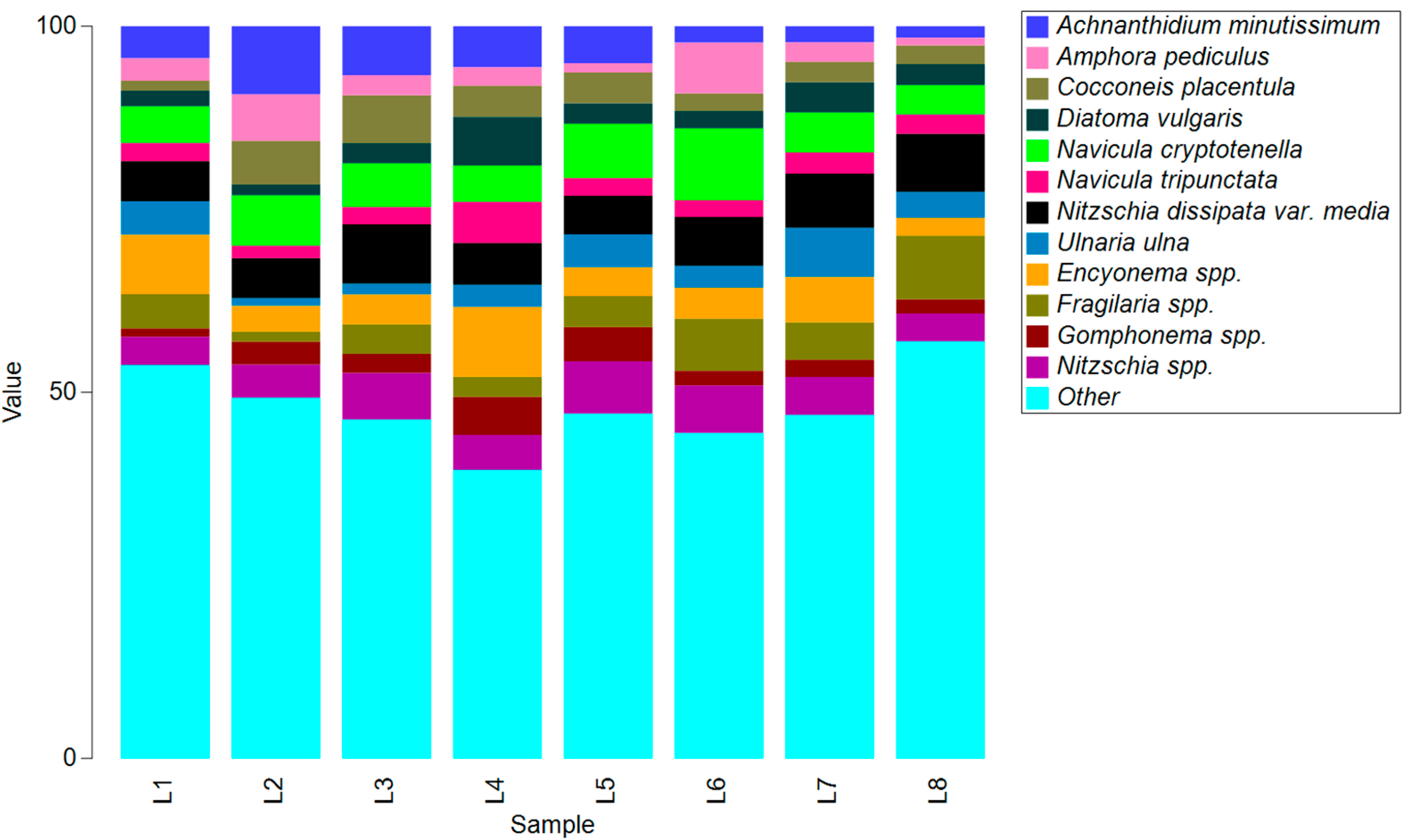
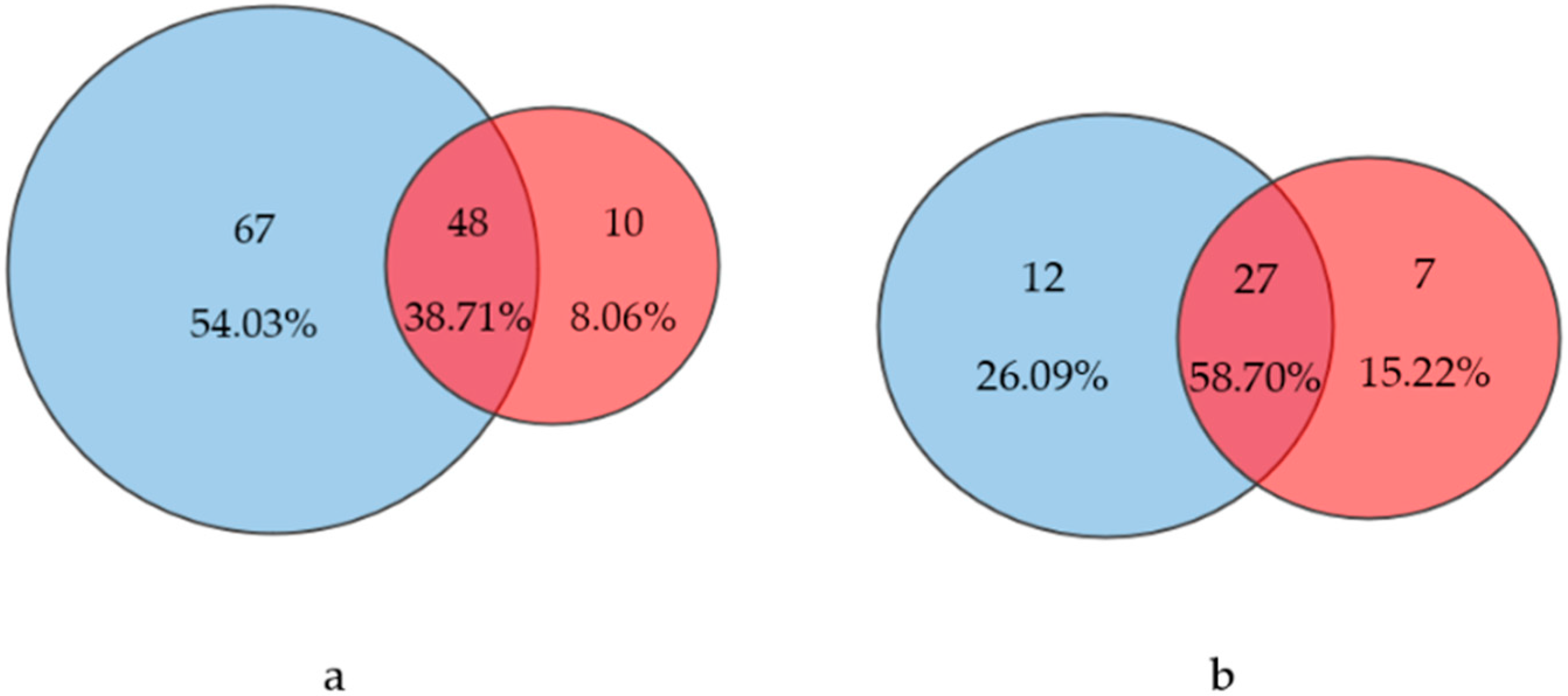
| Code | L1 | L2 | L3 | L4 | L5 | L6 | L7 | L8 |
|---|---|---|---|---|---|---|---|---|
| Name | Martin Brod | Štrbački Buk | Troslap | Dvoslap | Ripač | Kostela | Bosanska Krupa | Bosanska Otoka |
| River typology B&H * | 6 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| River typology HR * | HR-R_12 | HR-R_12 | HR-R_12 | HR-R_12 | HR-R_12 | HR-R_8 | HR-R_8 | HR-R_8 |
| Physical and chemical parameters | ||||||||
| Temperature °C | 11.4 | 13.3 | 11.7 | 12.3 | 12.7 | 14.1 | 15.0 | 15.9 |
| pH | 7.39 | 7.55 | 7.54 | 7.58 | 7.67 | 7.76 | 7.75 | 7.83 |
| Oxygen concentration (mg L−1) | 8.88 | 9.06 | 8.94 | 9.26 | 9.31 | 8.9 | 9.37 | 9.19 |
| Conductivity (µS cm−1) | 462 | 458 | 475 | 473 | 472 | 454 | 447 | 450 |
| Ammonium nitrogen, NH4+-N (mg L−1) | 0.024 | 0.025 | 0.009 | 0.019 | 0.019 | 0.028 | 0.037 | 0.038 |
| Ammonium, NH4+ (mg L−1) | 0.0309 | 0.0322 | 0.0116 | 0.0245 | 0.0245 | 0.0361 | 0.0477 | 0.0489 |
| Nitrite nitrogen, NO2−-N (mg L−1) | 0 | 0.0004 | 0 | 0.0001 | 0.0003 | 0.0014 | 0.0007 | 0.0009 |
| Nitrites, NO2− (mg L−1) | 0 | 0.0013 | 0 | 0.0003 | 0.001 | 0.0046 | 0.0023 | 0.003 |
| Nitrate nitrogen, NO3−-N (mg L−1) | 0.252 | 0.269 | 0.305 | 0.379 | 0.36 | 0.337 | 0.25 | 0.394 |
| Nitrates NO3− (mg L−1) | 1.115 | 1.1908 | 1.3501 | 1.6777 | 1.5936 | 1.4918 | 1.1067 | 1.7441 |
| Total nitrogen, TN (mg L−1) | 0.4 | 0.35 | 0.397 | 0.395 | 0.395 | 0.381 | 0.355 | 0.41 |
| Chemical oxygen demand, COD (mg L−1) | 12.5 | 25 | 18.8 | 6.25 | 12.5 | 6.25 | 12.5 | 6.25 |
| Biological oxygen demand, BOD (mg L−1) | 3.1 | 6.8 | 4.8 | 2.5 | 3.4 | 2.7 | 2.9 | 2.5 |
| Total phosphorus, TP (mg L−1) | 0.028 | 0.017 | 0.027 | 0.018 | 0.003 | 0.02 | 0.029 | 0.027 |
| Phosphates, PO43−-P (mg L−1) | 0.009 | 0.005 | 0.007 | 0.006 | 0.0011 | 0.01 | 0.011 | 0.008 |
| Total organic carbon, TOC (mg L−1) | 1.38 | 1.18 | 0.73 | 0.07 | 0 | 0 | 0 | 0.11 |
| Silicon dioxide, SiO2 (mg L−1) | 1.88 | 1.662 | 2.238 | 1.765 | 1.99 | 1.53 | 1.838 | 1.403 |
| Alkalinity as CaCO3 (mg L−1) | 214.9 | 227.3 | 238.7 | 231.4 | 233.5 | 225.2 | 227.3 | 225.2 |
| Turbidity (NTU) | 17.3 | 11.2 | 15 | 13.6 | 17.3 | 15.6 | 13.8 | 16.6 |
| Bicarbonates, HCO3− (mg L−1) | 246.4 | 261.1 | 275.7 | 256.9 | 258.6 | 248.9 | 258.6 | 251.3 |
| Ca2+ (mg L−1) | 80.2 | 97.5 | 91.2 | 92.8 | 83.4 | 84.9 | 81.8 | 84.9 |
| Mg2+ (mg L−1) | 19.5 | 0 | 13.8 | 3.817 | 19.1 | 15.3 | 12.4 | 9.5 |
| L1 | L2 | L3 | L4 | L5 | L6 | L7 | L8 | Total | |
|---|---|---|---|---|---|---|---|---|---|
| Read number | 42,602 | 44,938 | 54,131 | 43,975 | 37,312 | 40,172 | 34,402 | 35,787 | 333,319 |
| Number of detected ASVs | 79 | 160 | 101 | 47 | 103 | 100 | 75 | 86 | 265 |
| Number of ASVs assigned to the Phylum Bacillariophyta | 77 | 159 | 98 | 47 | 101 | 97 | 75 | 85 | 255 |
| Number of ASVs assigned to the Genus taxonomic level | 57 | 117 | 73 | 36 | 74 | 76 | 60 | 70 | 183 |
| Number of ASVs assigned to the Species taxonomic level | 41 | 88 | 54 | 25 | 50 | 53 | 44 | 53 | 133 |
| Number of unique Genera | 19 | 26 | 24 | 17 | 23 | 23 | 20 | 25 | 32 |
| Number of unique Species | 28 | 42 | 34 | 20 | 33 | 34 | 31 | 41 | 58 |
| Taxa Seen in Both Methods | Sum of Counts in OI | Sum of ASV Reads in MI | No. of ASVs | % of Similarity of ASVs | OI | MI | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |||||
| Achnanthidium minutissimum (Kützing) Czarnecki | 171 | 101,216 | 10 | 93.7 | ||||||||||||||||
| Achnanthidium pyrenaicum (Hustedt) H.Kobayasi | 504 | 2779 | 2 | 95.5 | ||||||||||||||||
| Amphora ovalis (Kützing) Kützing | 4 | 4519 | 2 | 94 | ||||||||||||||||
| Amphora pediculus (Kützing) Grunow | 39 | 1343 | 7 | 97.7 | ||||||||||||||||
| Caloneis fontinalis (Grunow) A.Cleve | 1 | 84 | 4 | 87.5 | ||||||||||||||||
| Cocconeis pediculus Ehrenberg | 50 | 594 | 1 | 94 | ||||||||||||||||
| Cocconeis placentula Ehrenberg | 2 | 2076 | 8 | 96.6 | ||||||||||||||||
| Cyclotella distinguenda Hustedt | 2 | 130 | 2 | 100 | ||||||||||||||||
| Cymatopleura elliptica (Brébisson) W.Smith | 1 | 50 | 1 | 87 | ||||||||||||||||
| Cymbella cymbiformis C.Agardh | 1 | 45 | 2 | 88.5 | ||||||||||||||||
| Cymbella lanceolata (C.Agardh) C.Agardh | 1 | 133 | 1 | 89 | ||||||||||||||||
| Denticula tenuis Kützing | 62 | 1086 | 2 | 100 | ||||||||||||||||
| Diatoma moniliformis (Kützing) D.M.Williams | 15 | 2585 | 1 | 94 | ||||||||||||||||
| Diatoma vulgaris Bory | 271 | 62,998 | 2 | 93 | ||||||||||||||||
| Diploneis subovalis Cleve | 2 | 434 | 3 | 99.6 | ||||||||||||||||
| Encyonema leibleinii (C.Agardh) W.J.Silva, R.Jahn, T.A.V.Ludwig, and M.Menezes | 2 | 56 | 1 | 100 | ||||||||||||||||
| Encyonema minutum (Hilse) D.G. Mann | 50 | 22 | 1 | 94 | ||||||||||||||||
| Encyonema silesiacum (Bleisch) D.G.Mann | 43 | 256 | 1 | 78 | ||||||||||||||||
| Eolimna minima (Grunow) Lange-Bertalot, nom. illeg. | 2 | 14 | 1 | 100 | ||||||||||||||||
| Epithemia sorex Kützing | 1 | 233 | 1 | 92 | ||||||||||||||||
| Eunotia arcus Ehrenberg | 3 | 21 | 1 | 100 | ||||||||||||||||
| Fistulifera saprophila (Lange-Bertalot and Bonik) Lange-Bertalot | 1 | 206 | 2 | 99.5 | ||||||||||||||||
| Fragilaria gracilis Østrup | 1 | 482 | 2 | 100 | ||||||||||||||||
| Frustulia vulgaris (Thwaites) De Toni | 2 | 20 | 1 | 99 | ||||||||||||||||
| Gomphonema pumilum (Grunow) E.Reichardt and Lange-Bertalot | 8 | 626 | 4 | 89.2 | ||||||||||||||||
| Gomphonema saprophilum (Lange-Bertalot and E.Reichardt) Abraca, R.Jahn, J.Zimmermann and Enke | 1 | 71 | 2 | 98.5 | ||||||||||||||||
| Gomphonema micropus Kützing | 1 | 13 | 1 | 100 | ||||||||||||||||
| Gomphonema tergestinum (Grunow) Fricke | 11 | 377 | 1 | 100 | ||||||||||||||||
| Karayevia ploenensis (Hustedt) Bukhtiyarova | 4 | 7 | 1 | 100 | ||||||||||||||||
| Melosira varians C.Agardh | 30 | 24,297 | 1 | 100 | ||||||||||||||||
| Navicula antonii Lange-Bertalot | 6 | 256 | 2 | 98 | ||||||||||||||||
| Navicula cryptotenella Lange-Bertalot | 158 | 5812 | 12 | 93.7 | ||||||||||||||||
| Navicula tripunctata (O.F.Müller) Bory | 138 | 23,424 | 2 | 92.5 | ||||||||||||||||
| Navicula capitatoradiata H.Germain ex Gasse | 5 | 523 | 1 | 100 | ||||||||||||||||
| Navicula gregaria Donkin | 2 | 71 | 1 | 100 | ||||||||||||||||
| Nitzschia dissipata (Kützing) Rabenhorst | 32 | 103 | 1 | 93 | ||||||||||||||||
| Nitzschia fonticola (Grunow) Grunow | 3 | 2144 | 4 | 94.5 | ||||||||||||||||
| Nitzschia gracilis Hantzsch | 2 | 79 | 1 | 100 | ||||||||||||||||
| Nitzschia linearis W.Smith | 1 | 588 | 4 | 98 | ||||||||||||||||
| Nitzschia palea (Kützing) W.Smith | 1 | 142 | 2 | 100 | ||||||||||||||||
| Nitzschia pusilla Grunow | 1 | 72 | 3 | 90 | ||||||||||||||||
| Nitzschia sigmoidea (Nitzsch) W.Smith | 1 | 691 | 3 | 94.6 | ||||||||||||||||
| Nitzschia tubicola Grunow | 2 | 8 | 1 | 97 | ||||||||||||||||
| Nitzschia dissipata var. media (Hantzsch) Grunow | 8 | 7149 | 9 | 92.6 | ||||||||||||||||
| Rhoicosphenia abbreviata (C.Agardh) Lange-Bertalot | 1 | 6 | 1 | 98 | ||||||||||||||||
| Sellaphora bacillum (Ehrenberg) D.G.Mann | 1 | 30 | 1 | 92 | ||||||||||||||||
| Surirella librile (Ehrenberg) Ehrenberg | 4 | 183 | 3 | 93.6 | ||||||||||||||||
| Ulnaria ulna (Nitzsch) Compère | 7 | 4998 | 3 | 98.3 | ||||||||||||||||
| Taxa seen in OI but not in MI | 0 | 0 | ||||||||||||||||||
| Achnanthidium anastasiae (Kaczmarska) Chaudev and Gololobova | 9 | 0 | ||||||||||||||||||
| Achnanthidium straubianum (Lange-Bertalot) Lange-Bertalot | 2 | 0 | ||||||||||||||||||
| Amphora copulata (Kützing) Schoeman and R.E.M. Archibald | 8 | 0 | ||||||||||||||||||
| Amphora inariensis Krammer | 15 | 0 | ||||||||||||||||||
| Amphora lange-bertalotii Levkov and Metzeltin | 1 | 0 | ||||||||||||||||||
| Aneumastus stroesei (Østrup) D.G.Mann | 3 | 0 | ||||||||||||||||||
| Caloneis bacillum (Grunow) Cleve | 3 | 0 | ||||||||||||||||||
| Caloneis lancettula (Schulz) Lange-Bertalot and Witkowski | 5 | 0 | ||||||||||||||||||
| Cocconeis euglypta Ehrenberg | 103 | 0 | ||||||||||||||||||
| Cocconeis lineata Ehrenberg | 219 | 0 | ||||||||||||||||||
| Cocconeis neodiminuta Krammer | 9 | 0 | ||||||||||||||||||
| Cocconeis placentula var. klinoraphis Geitler | 15 | 0 | ||||||||||||||||||
| Cocconeis pseudolineata (Geitler) Lange-Bertalot | 28 | 0 | ||||||||||||||||||
| Cymatopleura apiculata W.Smith | 2 | 0 | ||||||||||||||||||
| Cymatopleura solea (Brébisson) W.Smith | 3 | 0 | ||||||||||||||||||
| Cymbella compacta Østrup | 1 | 0 | ||||||||||||||||||
| Cymbella excisiformis Krammer | 1 | 0 | ||||||||||||||||||
| Cymbella lange-bertalotii Krammer | 1 | 0 | ||||||||||||||||||
| Diatoma vulgaris var. capitatum Grunow | 83 | 0 | ||||||||||||||||||
| Diploneis fontium Richardt and Lange-Bertalot | 1 | 0 | ||||||||||||||||||
| Diploneis krammeri Lange-Bertalot and E.Reichardt | 3 | 0 | ||||||||||||||||||
| Diploneis marginestriata Hustedt | 3 | 0 | ||||||||||||||||||
| Encyonema ventricosum (C.Agardh) Grunow | 69 | 0 | ||||||||||||||||||
| Encyonema vulgare Krammer | 3 | 0 | ||||||||||||||||||
| Encyonema caespitosum Kützing | 1 | 0 | ||||||||||||||||||
| Encyonopsis microcephala (Grunow) Krammer | 1 | 0 | ||||||||||||||||||
| Eunotia sp. Ehrenberg | 2 | 0 | ||||||||||||||||||
| Fallacia subhamulata (Grunow) D.G.Mann | 2 | 0 | ||||||||||||||||||
| Fragilaria mesolepta Rabenhorst | 1 | 0 | ||||||||||||||||||
| Fragilaria recapitellata Lange-Bertalot and Metzeltin | 76 | 0 | ||||||||||||||||||
| Fragilaria vaucheriae (Kützing) J.B.Petersen | 16 | 0 | ||||||||||||||||||
| Gomphonella olivacea (Hornemann) Rabenhorst | 108 | 0 | ||||||||||||||||||
| Gomphonella olivaceolacua (Lange-Bertalot and E.Reichart) R.Jahn and N.Abarca | 5 | 0 | ||||||||||||||||||
| Gomphonema clavatulum Reichardt | 1 | 0 | ||||||||||||||||||
| Gomphonema elegantissimum Reichardt and Lange-Bertalot | 6 | 0 | ||||||||||||||||||
| Gomphonema lateripunctatum E.Reichardt and Lange-Bertalot | 13 | 0 | ||||||||||||||||||
| Gomphonema minutum (C.Agardh) C.Agardh | 4 | 0 | ||||||||||||||||||
| Gomphonema parvulum (Kützing) Kützing | 3 | 0 | ||||||||||||||||||
| Gomphonema pseudotenellum Lange-Bertalot | 2 | 0 | ||||||||||||||||||
| Gomphonema sp. Ehrenberg | 1 | 0 | ||||||||||||||||||
| Gomphonema vibrio var. vibrio Ehrenberg | 1 | 0 | ||||||||||||||||||
| Gyrosigma attenuatum (Kützing) Rabenhorst | 7 | 0 | ||||||||||||||||||
| Gyrosigma sciotoense (W.S.Sullivant) Cleve | 6 | 0 | ||||||||||||||||||
| Gyrosigma obtusatum (Sullivant and Wormley) C.S.Boyer | 1 | 0 | ||||||||||||||||||
| Humidophila contenta (Grunow) Lowe, Kociolek, J.R.Johansen, Van de Vijver, Lange-Bertalot and Kopalová | 1 | 0 | ||||||||||||||||||
| Meridion circulare (Greville) C.Agardh | 21 | 0 | ||||||||||||||||||
| Navicula cryptofallax Lange-Bertalot and G.Hofmann | 2 | 0 | ||||||||||||||||||
| Navicula digitoconvergens Lange-Bertalot | 1 | 0 | ||||||||||||||||||
| Navicula hintzii Lange-Bertalot | 1 | 0 | ||||||||||||||||||
| Navicula oppugnata Hustedt | 6 | 0 | ||||||||||||||||||
| Navicula radiosa Kützing | 6 | 0 | ||||||||||||||||||
| Navicula reinhardtii (Grunow) Grunow | 7 | 0 | ||||||||||||||||||
| Navicula upsaliensis (Grunow) M.Peragallo | 4 | 0 | ||||||||||||||||||
| Navicula viridulacalcis Lange-Bertalot | 2 | 0 | ||||||||||||||||||
| Nitzschia intermedia Hantzsch | 10 | 0 | ||||||||||||||||||
| Nitzschia oligotraphenta (Lange-Bertalot) Lange-Bertalot | 2 | 0 | ||||||||||||||||||
| Nitzschia sublinearis Hustedt | 1 | 0 | ||||||||||||||||||
| Odontidium anceps (Ehrenberg) Ralfs | 5 | 0 | ||||||||||||||||||
| Odontidium mesodon (Kützing) Kützing | 2 | 0 | ||||||||||||||||||
| Placoneis sp. Mereschkowsky | 1 | 0 | ||||||||||||||||||
| Planothidium dubium (Grunow) Round and Bukhtiyarova | 1 | 0 | ||||||||||||||||||
| Planothidium lanceolatum (Brébisson ex Kützing) Lange-Bertalot | 1 | 0 | ||||||||||||||||||
| Psammothidium grischunum Bukhtiyarova and Round | 1 | 0 | ||||||||||||||||||
| Pseudostaurosira brevistriata (Grunow) D.M.Williams and Round | 1 | 0 | ||||||||||||||||||
| Reimeria sinuata (W.Gregory) Kociolek and Stoermer | 3 | 0 | ||||||||||||||||||
| Sellaphora laevissima (Kützing) D.G.Mann | 3 | 0 | ||||||||||||||||||
| Sellaphora sp. Mereschowsky | 1 | 0 | ||||||||||||||||||
| Surirella angusta Kützing | 1 | 0 | ||||||||||||||||||
| Surirella brebissonii var. kuetzingii Krammer and Lange-Bertalot | 2 | 0 | ||||||||||||||||||
| Ulnaria acus (Kützing) Aboal | 3 | 0 | ||||||||||||||||||
| Ulnaria capitata (Ehrenberg) Compère | 1 | 0 | ||||||||||||||||||
| Taxa seen in OI but not in MI | 0 | 0 | ||||||||||||||||||
| Achnanthidium spp. Kützing | 0 | 1806 | 1 | 100 | ||||||||||||||||
| Amphora spp. Ehrenberg ex Kützing | 0 | 743 | 2 | 99 | ||||||||||||||||
| Caloneis spp. Cleve | 0 | 2033 | 4 | 94.75 | ||||||||||||||||
| Cyclotella spp. (Kützing) Brébisson | 0 | 8 | 1 | 89 | ||||||||||||||||
| Cymbella spp. C.Agardh | 0 | 263 | 2 | 95.5 | ||||||||||||||||
| Diploneis spp. Ehrenberg ex Cleve | 0 | 48 | 1 | 100 | ||||||||||||||||
| Discostella nipponica (Skvortsov) A.Tuji and D.M.Williams | 0 | 42 | 1 | 78 | ||||||||||||||||
| Ellerbeckia sp. R.M.Crawford, | 0 | 7381 | 1 | 100 | ||||||||||||||||
| Encyonema spp. Kützing | 0 | 31,058 | 3 | 100 | ||||||||||||||||
| Encyonopsis spp. Krammer | 0 | 60 | 1 | 96 | ||||||||||||||||
| Fragilaria spp. Lyngbye | 0 | 6864 | 6 | 99.3 | ||||||||||||||||
| Fragilaria radians (Kützing) D.M.Williams and Round | 0 | 28 | 1 | 83 | ||||||||||||||||
| Gomphonema spp. Ehrenberg | 0 | 2820 | 5 | 96.4 | ||||||||||||||||
| Gomphonema affine Kützing | 0 | 78 | 1 | 76 | ||||||||||||||||
| Iconella spp. Jurilj | 0 | 168 | 2 | 100 | ||||||||||||||||
| Lindavia radiosa (Grunow) De Toni and Forti | 0 | 219 | 1 | 94 | ||||||||||||||||
| Mayamaea permitis (Hustedt) K.Bruder and Medlin | 0 | 18 | 1 | 100 | ||||||||||||||||
| Navicula spp. Bory | 0 | 215 | 4 | 88 | ||||||||||||||||
| Neidium spp. Pfitzer | 0 | 14 | 1 | 98 | ||||||||||||||||
| Nitzschia spp. Hassall | 0 | 1603 | 13 | 97.2 | ||||||||||||||||
| Nitzschia amphibia Grunow | 0 | 7 | 1 | 99 | ||||||||||||||||
| Nitzschia capitellata Hustedt | 0 | 11 | 1 | 98 | ||||||||||||||||
| Nitzschia draveillensis Coste and Ricard | 0 | 205 | 1 | 96 | ||||||||||||||||
| Sellaphora lanceolata D.G.Mann and S.Droop | 0 | 9 | 1 | 94 | ||||||||||||||||
| Staurosira spp. Ehrenberg | 0 | 308 | 3 | 100 | ||||||||||||||||
| Surirella spp. Turpin | 0 | 263 | 1 | 99 | ||||||||||||||||
| Locations Morphological Approach | L1 | L2 | L3 | L4 | L5 | L6 | L7 | L8 |
|---|---|---|---|---|---|---|---|---|
| TIDHR | 2.19 | 2.34 | 2.30 | 2.55 | 2.30 | 2.72 | 2.51 | 2.29 |
| EQRHR | 0.87 | 0.81 | 0.83 | 0.74 | 0.83 | 0.69 | 0.76 | 0.84 |
| SI | 1.79 | 1.77 | 1.93 | 2.06 | 1.80 | 1.88 | 1.91 | 1.88 |
| EQRSI | 0.78 | 0.78 | 0.69 | 0.61 | 0.76 | 0.72 | 0.70 | 0.72 |
| TDI | 42.32 | 52.19 | 50.3 | 76.12 | 55.52 | 73.79 | 69.24 | 55.03 |
| EQRTDI | 0.68 | 0.37 | 0.41 | 0.00 | 0.29 | 0.00 | 0.00 | 0.30 |
| IPS | 17.8 | 18.0 | 17.1 | 16.3 | 17.0 | 16.6 | 15.9 | 17.1 |
| EQRIPS | 0.86 | 0.88 | 0.82 | 0.77 | 0.81 | 0.79 | 0.74 | 0.82 |
| EQRB&H | 0.78 | 0.68 | 0.64 | 0.46 | 0.62 | 0.50 | 0.48 | 0.61 |
| Molecular approach | L1 | L2 | L3 | L4 | L5 | L6 | L7 | L8 |
| TIDHR | 2.44 | 2.70 | 2.64 | 2.43 | 2.48 | 2.80 | 2.74 | 2.62 |
| EQRHR | 0.78 | 0.69 | 0.70 | 0.78 | 0.76 | 0.67 | 0.69 | 0.73 |
| SI | 1.84 | 1.89 | 1.89 | 1.86 | 1.87 | 1.84 | 1.90 | 1.83 |
| EQRSI | 0.75 | 0.71 | 0.71 | 0.73 | 0.72 | 0.74 | 0.71 | 0.75 |
| TDI | 62.5 | 65.88 | 100 | 62.17 | 62.43 | 73.23 | 59.78 | 61.88 |
| EQRTDI | 0.20 | 0.05 | 0.00 | 0.14 | 0.13 | 0.00 | 0.19 | 0.14 |
| IPS | 14.3 | 14.4 | 15.3 | 15.3 | 14.6 | 13.2 | 15.0 | 14.3 |
| EQRIPS | 0.64 | 0.65 | 0.71 | 0.71 | 0.66 | 0.58 | 0.69 | 0.64 |
| EQRB&H | 0.53 | 0.47 | 0.47 | 0.52 | 0.51 | 0.44 | 0.53 | 0.51 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamberović, J.; Gligora Udovič, M.; Kulaš, A.; Tapolczai, K.; Orlić, S.; Jusufović, A.; Gajić, A.; Žutinić, P.; Ahmić, A.; Kalamujić Stroil, B. The Diatom Diversity and Ecological Status of a Tufa-Depositing River through eDNA Metabarcoding vs. a Morphological Approach—A Case Study of the Una River (Bosnia and Herzegovina). Microorganisms 2024, 12, 1722. https://doi.org/10.3390/microorganisms12081722
Kamberović J, Gligora Udovič M, Kulaš A, Tapolczai K, Orlić S, Jusufović A, Gajić A, Žutinić P, Ahmić A, Kalamujić Stroil B. The Diatom Diversity and Ecological Status of a Tufa-Depositing River through eDNA Metabarcoding vs. a Morphological Approach—A Case Study of the Una River (Bosnia and Herzegovina). Microorganisms. 2024; 12(8):1722. https://doi.org/10.3390/microorganisms12081722
Chicago/Turabian StyleKamberović, Jasmina, Marija Gligora Udovič, Antonija Kulaš, Kálmán Tapolczai, Sandi Orlić, Amela Jusufović, Almina Gajić, Petar Žutinić, Adisa Ahmić, and Belma Kalamujić Stroil. 2024. "The Diatom Diversity and Ecological Status of a Tufa-Depositing River through eDNA Metabarcoding vs. a Morphological Approach—A Case Study of the Una River (Bosnia and Herzegovina)" Microorganisms 12, no. 8: 1722. https://doi.org/10.3390/microorganisms12081722
APA StyleKamberović, J., Gligora Udovič, M., Kulaš, A., Tapolczai, K., Orlić, S., Jusufović, A., Gajić, A., Žutinić, P., Ahmić, A., & Kalamujić Stroil, B. (2024). The Diatom Diversity and Ecological Status of a Tufa-Depositing River through eDNA Metabarcoding vs. a Morphological Approach—A Case Study of the Una River (Bosnia and Herzegovina). Microorganisms, 12(8), 1722. https://doi.org/10.3390/microorganisms12081722








