Abstract
Japan has numerous vineyards with distinct geographical and climatic conditions. To the best of our knowledge, there is no comprehensive analysis of the diversity of yeasts associated with wine grapes from Japan. This study aimed to determine yeast diversity in wine grapes from four wine-producing regions in Japan and to evaluate the physicochemical characteristics of wines produced with indigenous Saccharomyces cerevisiae strains isolated from two regions. A total of 2648 strains were isolated from nine wine grape samples. MALDI-TOF MS and 26S rDNA sequence analyses revealed that the strains belonged to 21 non-Saccharomyces yeasts and 1 Saccharomyces yeast (S. cerevisiae). Non-Saccharomyces yeasts were found in high quantities and were highly distributed among the wine grape samples. Differences in the distribution of the identified yeast species were noted among the different wine grape varieties and regions. Indigenous S. cerevisiae strains of different genotypes from different regions exhibit distinct physiological traits. Our findings are expected to enhance our understanding of the local yeasts associated with Japanese vineyards and contribute to obtaining cultures that can provide region-specific organoleptic characteristics to local wines produced in Japan.
1. Introduction
One of the main sources of indigenous yeasts is the wine grape berry [1]. Numerous species/strains of indigenous yeasts are found on the berry surface and transferred to grape juice during winemaking. Indigenous yeasts are important for spontaneous fermentation and contribute to the complexity of wine production. S. cerevisiae and non-Saccharomyces strains such as Candida, Hanseniaspora, Metschnikowia, Pichia, Torulaspora, and Zygosaccharomyces have been isolated from wine grape berries and must from wine-producing regions [2,3,4,5,6,7,8,9,10,11,12,13]. The biodiversity and distribution patterns of wine grape yeasts vary by vineyard and region and are related to the geographical environment [1,14]. Moreover, indigenous wine grape yeasts may contribute to regional differences in wine and are part of terroirs, reflecting the typical characteristics of the region in which wine grapes are grown [15,16,17,18]. Terroir refers to the environment (soil, climate, topography, etc.) that gives a unique character to wine; it plays an important role in producing wines with region-specific organoleptic characteristics. Indigenous yeasts are also potential contributors to the unique characteristics and distinctive sensory attributes of wine.
The genetic diversity of S. cerevisiae associated with different vineyards contributes to the distinctiveness of wine phenotypes [19,20]. Several studies have reported the genetic diversity of S. cerevisiae and the preservation of bioresources for winemaking [21,22,23,24,25,26,27], both essential for maintaining the typical sensory properties of wines. This is because S. cerevisiae strains play a key role in providing distinctive organoleptic characteristics to wine produced via spontaneous fermentation.
Japan has numerous viticultural regions with distinct geographical and climatic conditions; thus, different yeast flora potentially exist in viticultural environments. Previous studies have reported the diversity of yeast strains isolated from wine grapes and must from several wine-producing areas of Japan, including Hiroshima, Okayama, Nagano, Yamanashi, Yamagata, and Iwate [28,29,30,31,32,33,34,35]. Yeast species in these wine-producing regions include the genera Candida, Cryptococcus, Hanseniaspora, Kloeckera, Metschnikowia, Pichia, Rhodotorula, and Saccharomyces. Indigenous yeasts from distinctive regions of Japan are considered parts of the terroir and can produce wines with region-specific organoleptic characteristics. However, to our knowledge, no comprehensive studies have been conducted on the yeast diversity in wine grapes from Japan. More information on the local yeast community in wine grapes is needed to evaluate the potential use of yeasts in winemaking and to produce high-quality wines with region-specific organoleptic characteristics.
Herein, we report the isolation, identification, and diversity of indigenous yeasts from wine grapes and spontaneous fermentation in vineyards located in four wine-producing regions in Japan. We also characterized the alcoholic fermentative profile of selected S. cerevisiae strains in laboratory fermentations. Our findings are expected to improve our understanding of yeast diversity and distribution in wine-producing regions in Japan and enable the preservation of potential wine yeasts for producing regionally differentiated wines, which may contribute to a better understanding of microbial terroirs in Japan.
2. Materials and Methods
2.1. Wine Grape Samples
In this study, nine grape samples of the 2021 vintage were collected from vineyards located in four regions of Japan (Figure 1). The following grape varieties were used: Yama Sauvignon (YS), Yama Blanc (YB), Zweigelt (ZG), Pinot Blanc (PB), Chardonnay (CH), Merlot (MR), and Cabernet Sauvignon (CS) (Table 1). Healthy, undamaged grapes were collected from each sample, aseptically stored in sterile plastic bags, cooled using an ice pack, and transported to the Institute of Enology and Viticulture, University of Yamanashi, Yamanashi, Japan. The enological parameters of the resulting grape juice (pH, sugar content, and total acidity) were determined following the standard method in Japan [36] and are shown in Table 2. The sugar content was determined via conversion from specific gravity data.
Figure 1.
Geographic information of vineyards sampled in Japan. Maps were downloaded from https://www.freemap.jp/ (accessed on 30 November 2023) free of charge and modified using Microsoft PowerPoint software ® 2402. Abbreviations: UR, Urausu (Hokkaido); UE, Ueda (Nagano Prefecture); SO, Suo-Oshima (Yamaguchi Prefecture); KN, Katsunuma (Yamanashi Prefecture).

Table 1.
Geographic information of sampling points from vineyards in Japan.

Table 2.
Physicochemical characteristics and number of yeast isolates obtained from the wine grape samples in this study.
2.2. Yeast Isolation
Yeast was isolated from the fermenting samples. Briefly, grape samples were manually destemmed and crushed in a sterile plastic bag. The must was fermented with the skin and seeds under microaerobic conditions in a sterilized 500 mL flask and incubated at 25 °C for 14–21 days. Samples (1 mL) were collected immediately after crushing the grapes and during spontaneous fermentation on days 2, 4, 7, 14, and 21. The fermenting samples were diluted with 0.85% NaCl (from 10−1 to 10−5). Then, 100 µL of these dilutions were spread onto YPD agar supplemented with 100 µg/mL chloramphenicol to inhibit bacterial growth. YPD agar plates were incubated at 25 °C until colonies formed. Colonies showing different morphologies were isolated from each YPD agar plate and then preserved at −80 °C in 10% glycerol.
2.3. Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry (MALDI-TOF MS) Analysis
Rapid and tentative identification of yeast isolates was performed using a MALDI-TOF mass spectrometer (Shimadzu, Kyoto, Japan). The isolates were cultivated for two days on a YPD agar plate at 25 °C, and one loop of a colony (approximately 10 mg) was suspended in 300 µL distilled water and 900 µL ethanol. The suspension was centrifuged for 2 min at 16,000× g. The supernatant was discarded, and the pellet was air-dried. For protein extraction, 50 µL of 70% formic acid was added to the pellet and mixed, and then 50 µL of acetonitrile was added to the resulting mixture. The centrifuge tube containing the dissolved pellet in acetonitrile was centrifuged for 2 min at 13,000 rpm, and 1 µL of the protein solution was spotted on a target plate (Shimadzu) and left to dry. Each spot was overlaid with 1 µL of α-cyano-4-hydroxycinnamic acid matrix solution (Shimadzu) and then air-dried at 25 °C before analysis. Mass spectra of each isolate were automatically acquired using an AXIMA Performance mass spectrometer (Shimadzu). Tentative identification of the yeast isolates was performed using SARAMIS software version 4.04 (AnagnosTec, Potsdam, Germany). To temporarily identify yeast isolates, the obtained mass spectra were compared with those of known yeast species available in the in-house library database of the Institute of Enology and Viticulture at the University of Yamanashi. Next, yeast isolates were preliminarily grouped according to their mass spectral patterns via cluster analysis using SARAMIS software version 4.04, and 139 isolates were selected for molecular identification based on their 26S rDNA D1/D2 domain sequences.
2.4. 26S rDNA D1/D2 Domain Sequence Analysis
Identification was confirmed via sequence analysis. Genomic DNA of the 139 representative isolates was extracted using PrepMan™ Ultra Sample Preparation Reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. The sequences of the D1/D2 domains of the 26S rDNA genes were amplified via PCR using primers NL1 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and NL4 (5′-GGAAGTAAAAGTCGTAACAAGG-3′) [37]. The PCR products were purified using a MonoFas DNA Purification Kit (GL Sciences, Tokyo, Japan) and sent to Fasmac Co., Ltd. (Kanagawa, Japan) for DNA sequence analysis with the same primers used for amplification. The obtained sequences were analyzed and compared with the sequences of the type strains using a BLASTN search (https://blast.ncbi.nlm.nih.gov/Blast.cgi (accessed on 11 December 2023)), considering an identity threshold of at least 98%.
2.5. Inter-Delta Analysis
In this study, 112 S. cerevisiae isolates (isolated from only two samples: MR in Katsunuma and CS in Ueda) were preliminarily classified based on their mass spectra patterns via cluster analysis using SARAMIS software version 4.04. A total of 23 representative isolates were selected for subsequent analysis. Genetic characterization of representative S. cerevisiae isolates was carried out via inter-delta analysis using a modified method with primers delta12 and delta21, based on a previous study [38]. The PCR products were separated via electrophoresis on 2.0% agarose gels, applied at 100 V for 130 min in 1× TBE buffer, and photographed under UV light. The band pattern obtained from each isolate was clustered using CLIQS 1D Pro analysis software version 1.5 (TotalLab Ltd., Newcastle upon Tyne, UK), and the unweighted pair group method using arithmetic averages was performed to construct a dendrogram. Finally, four S. cerevisiae isolates with different band patterns were selected and used for subsequent fermentation tests.
2.6. Laboratory-Scale Alcoholic Fermentation Using Selected Indigenous S. cerevisiae Isolates
To evaluate the enological traits of indigenous S. cerevisiae isolates, we carried out a fermentation test on four selected S. cerevisiae isolates in autoclaved grape juice derived from Muscat Bailey A (Vitis labrusca × Vitis vinifera), one the most popular Japanese wine grapes (pH 3.29; titratable acidity 6.56 g/L, as tartaric acid). The yeast assimilable nitrogen (YAN) of the grape juice was adjusted to 250 mg/L using diammonium phosphate and the sugar content was adjusted to 21% using sucrose. After adjusting the YAN and sugar content, the grape juice was autoclaved at 105 °C for 10 min. Yeast was cultured in sterile grape juice at 25 °C, and the culture was added to 100 mL of the grape juice (final cell density of approximately 106 cells/mL). The inoculated grape juice was fermented for two weeks using a Fermograph II-W instrument (ATTO, Tokyo, Japan) at 25 °C. The fermentation process was evaluated by monitoring the CO2 produced every 120 min for two weeks. A commercial strain, S. cerevisiae EC1118 (Lallemand Inc., Montreal, QC, Canada), was used as a control. The physicochemical characteristics of the wine were quantified at the end of the alcoholic fermentation. Each strain was fermented in triplicate.
2.7. Physicochemical Analysis of Wine Samples
The ethanol content, pH, and total acidity of the wine samples derived from the fermentation of each S. cerevisiae isolate were determined following the standard method in Japan [36].
2.8. Gas Chromatography (GC) and High-Performance Liquid Chromatography (HPLC)
Using the internal standard method, the low-boiling-point aroma components of the wine samples were analyzed by directly injecting the wine samples into a gas chromatograph (GC; GC2014, Shimadzu) equipped with a flame ionization detector (FID). A PEG600-packed column (15% Chromosorb W 60/80 mesh, 2 m × 3 mm i.d.) was used for separation. Nitrogen was used as the carrier gas, and the flow rate was kept constant at 30 mL/min. The temperature of the GC oven was set at 95 °C. The injector and detector temperatures were set at 180 °C and 185 °C, respectively. Briefly, 2 mL of the wine sample and 0.2 mL of an internal standard (acetaldehyde, ethyl acetate, n-propyl alcohol, isobutyl alcohol, isoamyl alcohol, or ethyl lactate) were added to a 5 mL test tube. The test tube was vortexed, and 1 µL of each sample was injected into the GC.
The intermediate- and high-boiling-point aroma components were quantified using the internal standard method by directly injecting wine samples into a GC (GC2014, Shimadzu) equipped with an FID and an autosampler. The aroma components were separated on a Pure-WAX column (GL Sciences Inc., Tokyo, Japan; 0.25 mm i.d. × 60 m, 0.25 µm film thickness) using helium carrier gas at a constant 1 mL/min flow rate. The temperature of the GC oven was initially set to 50 °C for 5 min, raised to 130 °C at a rate of 4 °C/min, further increased to 220 °C at a rate of 5 °C/min, and maintained at 220 °C for 12 min. The injector and detector temperatures were set to 250 °C and 260 °C, respectively. Briefly, 40 mL of the wine sample, 2 g ammonium sulfate, 0.4 mL of an internal standard (2-octanol, ethyl propanoate, ethyl butyrate, isoamyl acetate, ethyl hexanoate, n-hexyl acetate, ethyl lactate, n-hexyl alcohol, (Z)-3-hexen-1-ol(cis), ethyl octanoate, propionic acid, i-butyric acid, butanoic acid, ethyl decanoate, i-valeric acid, diethyl succinate, methionol, 2-phenethyl acetate, hexanoic acid, 2-phenethyl alcohol, octanoic acid, or decanoic acid), and 10 mL of pentane/diethyl ether (1:1) were added to a 50 mL glass centrifuge tube. The centrifuge tube was inverted for 5 min and then maintained at 15 °C for 5 min. After inversion twice, the sample was centrifuged at 1700× g for 20 min. The supernatant was transferred into a 2 mL vial, and 1.5 µL of the solution was injected into the GC. The split ratio was 1:15.
Organic acids in the wine samples were quantified using a Shimadzu LabSolutions HPLC system consisting of a Shim-pack SCR-102H column and an autosampler (AS-2000). The wine samples were diluted 1:10 with water (Milli-Q) and passed through a membrane filter (0.22 μm). The filtrate was transferred into a 1.5 mL vial tube. Samples were injected into the column at a 0.8 mL/min flow rate. Quantification was performed using calibration curves (R2 > 0.99) prepared with organic acid standards.
2.9. Statistical Analysis
The chemical compounds in the wine samples obtained via fermentation with indigenous S. cerevisiae strains were subjected to a one-way analysis of variance to evaluate the differences in physicochemical characteristics followed by the Tukey–Kramer test at a significance level of p < 0.01. The results are expressed as the means ± standard deviations.
3. Results
3.1. Isolation and Identification of Yeast Species
A total of 2648 yeast isolates were collected from nine grape samples collected from four different regions of Japan. The number of isolates and the physicochemical characteristics of the grape samples are listed in Table 2. The pH levels ranged from 2.91 to 3.84, the sugar contents ranged from 15.9 to 20.2%, and the total acidity ranged from 3.96 to 8.28 g/L.
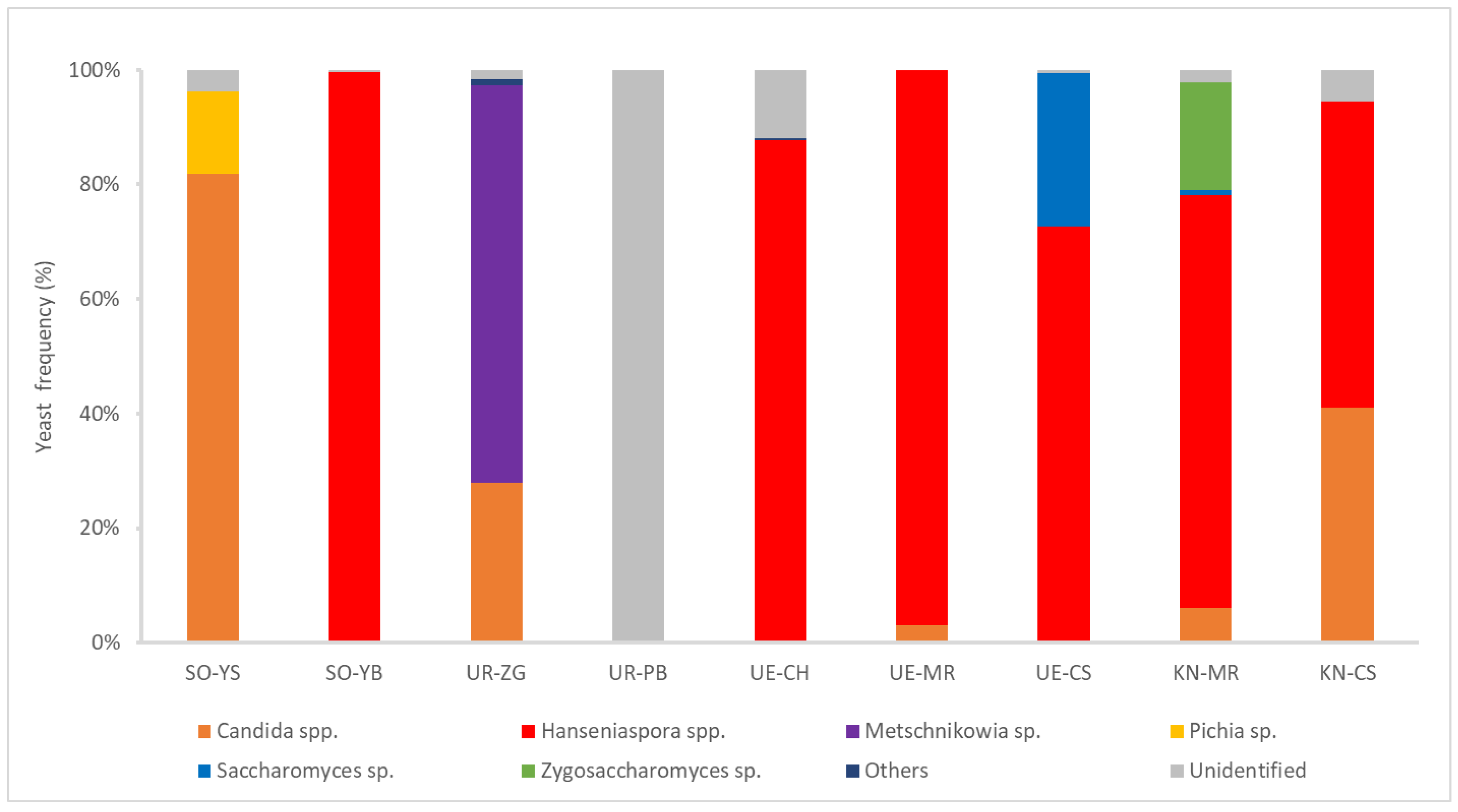
All yeast isolates were analyzed via MALDI-TOF MS for rapid and tentative identification. In this study, 157 yeast isolates from PB in Urausu could not be identified because their MS data did not correspond to the reference data. The yeast genera observed included Candida, Hanseniaspora, Metschnikowia, Pichia, Saccharomyces, and Zygosaccharomyces (Figure 2). Hanseniaspora was the most abundant and frequently isolated genus, identified in six samples. Saccharomyces was detected in only two samples (CS from Ueda and MR from Katsunuma). Metschnikowia, Pichia, and Zygosaccharomyces were also detected in only one sample. Although these results were tentative, they revealed a high overall diversity of non-Saccharomyces yeasts in wine grapes collected from different regions of Japan.
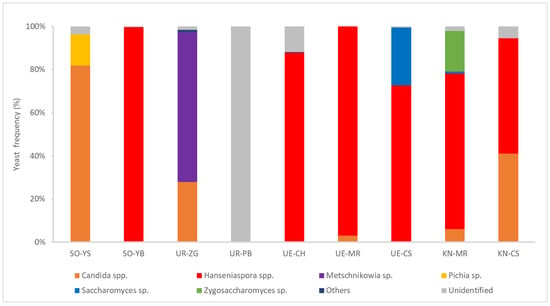
Figure 2.
Distribution frequencies of yeasts isolated from wine grapes in four wine-producing regions, as determined using MALDI-TOF MS.
Table S1 shows the identification results for the 139 representative yeast isolates based on 26S rDNA D1/D2 domain sequence analysis. Most yeast isolates were identified to have a high similarity (>99%) with the type strains of known species. A total of 22 species (16 genera) were detected: Aureobasidium pullulans, Clavispora sp., Hanseniaspora guilliermondii, Hanseniaspora opuntiae, Hanseniaspora uvarum, Hanseniaspora valbyensis, Hanseniaspora vineae, Lachancea thermotolerans, Martiniozyma asiatica, Metschnikowia shanxiensis, Meyerozyma caribbica, Papiliotrema laurentii, Pichia manshurica, Rhodotorula graminis, Rhodotorula nothofagi, Saccharomyces cerevisiae, Saturnispora diversa, Sporidiobolus pararoseus, Starmerella apicola, Starmerella bacillaris (syn. Candida zemplinina), Torulaspora delbrueckii, and Zygosaccharomyces bailii (Table 3). The PB isolates from Urausu, which could not be identified via MALDI-TOF MS, were identified as R. nothofagi and S. pararoseus based on 26S rDNA sequence analysis.

Table 3.
Yeast species and their percentage of frequency (%) isolated from wine grapes from four wine-producing regions in Japan.
Differences in the distribution of the identified yeast species were observed among different grape varieties collected from the same region. In Suo-Oshima, M. caribbica was the dominant species detected in YS samples, while H. guilliermondii was the most frequently isolated species in YB samples. In Urausu, the isolates from ZG and PB were also diverse. The most frequently isolated species from ZG was M. shanxiensis (69.4%), followed by T. delbrueckii (30.6%) and R. nothofagi (99.4%). In Ueda, the species most frequently isolated from CH, MR, and CS samples was H. uvarum (87.46%, 63.7%, and 62.47%, respectively). The second most frequently isolated species was R. nothofagi for CH (9.90%), H. valbyensis for MR (25.8%), and S. cerevisiae for CS (26.91%). The wine grapes from Katsunuma (MR and CS) also had a high abundance of H. uvarum (56.89% and 46.87%, respectively), followed by Z. bailii in MR (20.05%) and S. bacillaris in CS (41.60%). Sixteen species (A. pullulans, Clavispora sp., H. opuntiae, H. valbyensis, L. thermotolerans, M. asiatica, M. shanxiensis, M. caribbica, P. laurentii, P. manshurica, R. graminis, S. diversa, S. pararoseus, S. apicola, T. delbrueckii, and Z. bailii) detected in this study were exclusively found in only one sample. Our data also showed that the diversity of isolated indigenous yeasts in Japan differed between wine-producing regions. Although H. uvarum was the most abundant species in MR samples from two different regions (Ueda and Katsunuma), H. valbyensis was only the second most abundant species in MR samples from Ueda (25.8%); it was not found in the MR samples from Katsunuma. In addition, H. uvarum was the most abundant species in the CS from Ueda and Katsunuma, whereas S. bacillaris was isolated at a frequency of 41.60% from Katsunuma and only 0.25% from Ueda.
3.2. Inter-Delta Analysis of Indigenous S. cerevisiae Isolates
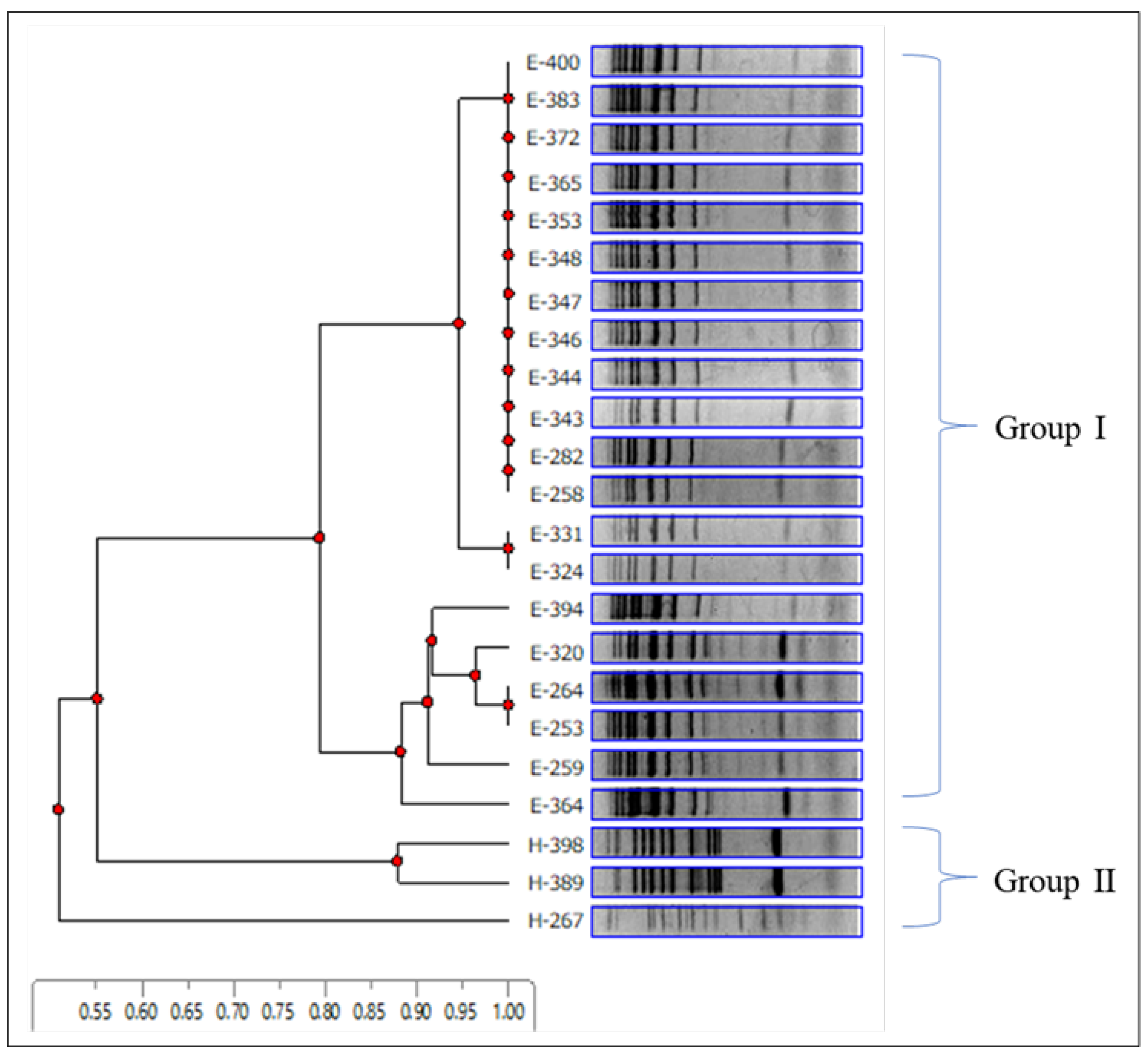
In this study, 112 isolates identified as S. cerevisiae were collected from wine grapes from Ueda and Katsunuma. Specifically, 23 representative isolates (20 isolates from a vineyard in Ueda and 3 isolates from Katsunuma) were selected for inter-delta analysis to determine their genetic diversity. Ten distinct genotypes were identified in this study. The band pattern data from the inter-delta analysis were used to construct a dendrogram (Figure 3). The cluster analysis showed that the 23 isolates diverged into two major groups at a 0.55 distance, suggesting that the two wine-producing regions have S. cerevisiae strains of different genotypes. Group I comprised 20 isolates originating from Ueda with seven genotypes, and group II was composed of isolates from Katsunuma with three genotypes.
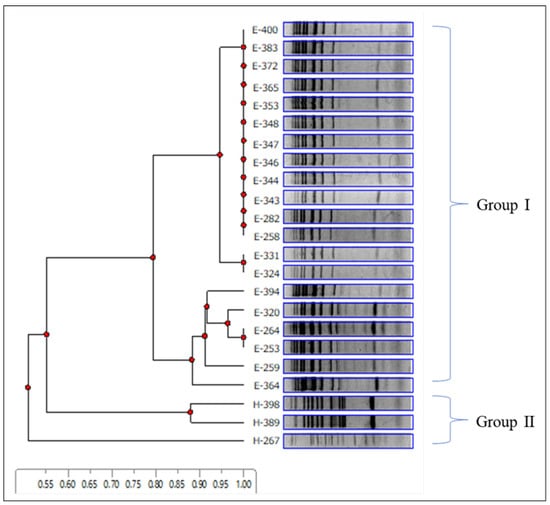
Figure 3.
Inter-delta analysis of representative Saccharomyces cerevisiae strains isolated from Ueda and Katsunuma vineyards. Electrophoretic patterns and cluster analyses were performed using delta12 and delta21 primers, as described by Legras and Karst (2003) [38]. The dendrogram was constructed using an unweighted pair group method with an arithmetic mean. Group I comprised 20 isolates originating from Ueda with seven genotypes, and group II was composed of isolates from Katsunuma with three genotypes.
3.3. Physicochemical Properties of Wines Produced Using Indigenous Yeasts
Four representative indigenous S. cerevisiae isolates with different genotypes, E-253 and E-400 isolated from a vineyard in Ueda and H-267 and H-389 from Katsunuma, were used for fermentation tests with sterilized Muscat Bailey A juice to evaluate the enological traits of these isolates. S. cerevisiae EC1118 (Lallemand, Inc.) was used as a control. Table 4 shows the physicochemical characteristics of the wines produced via fermentation with these isolates. Our results showed that the physicochemical characteristics varied depending on the strain. A comparison of wines produced using indigenous S. cerevisiae isolates with different genotypes from Ueda (E-253 and E-400) and Katsunuma (H-267 and H-389) revealed that the concentrations of 16 of the 28 wine components investigated were significantly different (p < 0.01). Among these components, the concentrations of lactic acid, acetic acid, isobutyl alcohol, ethyl octanoate, and decanoic acid significantly differed between the wines produced using the two Katsunuma isolates and those produced using the two Ueda isolates.

Table 4.
Physicochemical characteristics of and concentrations of aroma components in wines produced by fermentation with indigenous Saccharomyces cerevisiae E-253 and E-400 isolated from Ueda (Nagano) and H-267 and H-389 isolated from Katsunuma (Yamanashi). S. cerevisiae EC1118 was used as a control.
The alcohol content and pH values of the wines obtained after fermentation for two weeks were similar. In contrast, the total acidity and lactic and acetic acid concentrations significantly differed between wines produced with isolates from Ueda (E-253 and E-400) and those from Katsunuma (H-267 and H-389). The total acidity and lactic acid concentrations in wines produced via fermentation with the Katsunuma isolates (H-267 and H-389) were significantly higher than those produced with EC1118. Similarly, the acetic acid concentration in wines fermented with the two Ueda isolates (E-253 and E-400) was significantly higher than in wines fermented with EC1118.
Nineteen aromatic components were identified and quantified using GC. Significant differences also occurred in the low-boiling-point aroma components of the wines produced. Wines produced with the Ueda isolates (E-253 and E-400) had higher acetaldehyde concentrations than those derived from Katsunuma isolates (H-267 and H-389), and significant differences in acetaldehyde concentrations were noted between wines produced with E-253 and H-389 and those produced with E-400 and H-389. All wines produced using the indigenous isolates had lower acetaldehyde and ethyl acetate concentrations than those produced using EC1118. The concentrations of isobutyl and isoamyl alcohols in the wines produced with H-267 were significantly higher than those in the other wines. All low-boiling-point aroma components in the wines produced using E-253 and E-400 had lower concentrations than those produced using EC1118.
Regarding the intermediate- and high-boiling-point aroma components, the ethyl octanoate and decanoic acid concentrations significantly differed between wines obtained via fermentation with the two Katsunuma isolates and those obtained via fermentation with the two Ueda isolates. Most intermediate- and high-boiling-point aroma components in wines produced with indigenous isolates were more highly concentrated than those in the control wine. The concentration of isoamyl acetate, known for its fruity aroma, was higher in wines fermented with H-267 and H-389 than in those fermented with EC1118. The wine produced by fermentation with H-389 had a higher concentration of ethyl hexanoate than that produced by fermentation with EC1118. Wines produced using all four indigenous isolates (E-253, E-400, H-267, and H-389) had higher 2-phenethyl alcohol concentrations than those produced using EC1118.
4. Discussion
Yeast species from the genera Hanseniaspora, Metschnikowia, and Saccharomyces, particularly H. uvarum, M. pulcherrima, and S. cerevisiae, respectively, are commonly found in wine grapes and wines in Japan [28,30,32,33,34] but have not been well-studied in the past 20 years. In this study, 2648 isolates were collected from nine grape samples collected from four wine-producing regions in Japan. All isolates were initially analyzed via MALDI-TOF MS for rapid identification, revealing that the isolates belonged to six major yeast genera. Non-Saccharomyces species were detected at high frequencies in all grape samples, whereas Saccharomyces species were detected in only two samples.
The identification of 22 yeast species belonging to 16 genera isolated from nine grape samples via 26S rDNA sequence analysis (Table 3) indicated that non-Saccharomyces yeast species were dominant, consistent with previous studies in Japan [28,32] and other countries [2,5,8,39,40]. Although non-Saccharomyces yeasts are less capable of fermenting sugars in juice than Saccharomyces yeasts [41,42], they can produce secondary metabolites that contribute to the diversity and complexity of wine, and research has focused on their application to winemaking [41,43]. Recently, commercial starter cultures containing non-Saccharomyces species and strains have been used in winemaking [44].
H. uvarum is the most abundant species isolated from wine grapes in Ueda and Katsunuma. H. uvarum is known to be frequently isolated from wine grapes [1,4,8,32,45]. The effect of H. uvarum on the physicochemical characteristics of wine has also been studied [45,46]. Exploring the potential of starter cultures containing indigenous H. uvarum strains in the local winemaking industry is important. H. vineae is also commonly isolated from wine grape samples in Ueda and Katsunuma. H. vineae improves wine aroma [47]. A previous sensory analysis demonstrated a significant increase in the fruity aroma evoking bananas, pears, apples, citrus fruits, and guava in wine produced using H. vineae [48]. The H. vineae strains isolated in this study should be further analyzed for their potential to produce wines with a characteristic aroma and flavor. Other isolated Hanseniaspora species include H. guilliermondii, H. opuntiae, and H. valbyensis. This is the first study to identify H. opuntiae and H. valbyensis in wine grapes in Japan.
In contrast, S. bacillaris (synonym: C. zemplinina) was the predominant species in CS samples from Katsunuma. C. zemplinina has been previously isolated from wine grapes in Japan [34], and its application in winemaking has been investigated [49,50]. We are the first to isolate and identify Z. bailii from wine grape samples from Katsunuma. Although this species is a spoilage yeast responsible for off-flavors and sediment formation [51], it produces high levels of esters and shows potential as a co-starter during fermentation with S. cerevisiae [52]. M. caribbica is the most abundant yeast species in YS samples from Suo-Oshima and is a new species found in wine grapes in Japan. M. caribbica is commonly found in wine grapes in Italy, Spain, and other regions [5,8,53]. M. shanxiensis was isolated from only one wine grape sample (UR-ZG). This species was first isolated from the surfaces of jujube fruits collected in China [54]. This is the first report of M. shanxiensis isolated from wine grapes and wine-producing environments.
T. delbrueckii was isolated only from wine grapes in Urausu. One of the most well-studied non-Saccharomyces yeasts, T. delbrueckii is commercially available for wine production [44]. R. nothofagi was the dominant species isolated from PB samples from Urausu, a region with a cold climate. The basidiomycetous yeast genus Rhodotorula is found in wine grapes in Japan (Rhodotorula glutinis and R. minuta) [32]. R. nothofagi was the third Rhodotorula species isolated from wine grapes in Japan. In addition, A. pullulans, Clavispora sp., L. thermotolerans, P. laurentii, R. graminis, S. diversa, S. pararoseus, and S. apicola were isolated at very low frequencies (0.49%, 2.5%, 0.33%, 0.6%, 1.65%, 0.25%, 0.6%, and 0.25%, respectively) from one sample only, which led us to speculate that their presence on wine grapes was probably incidental. These results show the data of a single year’s isolation. To have more comprehensive data on the yeast diversity at the species level in wine grapes in Japan, further studies on the isolation and identification of yeasts carried out in different years and different regions will be necessary.
Our study offers new insights into yeast diversity in the wine-producing regions of Japan. Future work should evaluate the contribution of indigenous non-Saccharomyces isolates from Japanese wine grapes to wine aroma and discover new strains with excellent enological traits. The wine-grape-associated yeast community is altered by climatic conditions, soil, and cultivars [1,14]. Our finding that indigenous yeast species differ by region indicates that yeast diversity in wine grapes from Japan may depend on the grape variety or vineyard location. Different yeast distributions in different regions may contribute to producing wine with distinctive flavors [15,55].
Non-Saccharomyces yeasts were found in high quantities and were widely distributed in the wine grape samples examined in this study. In contrast, S. cerevisiae isolates were found in only two grape samples (CS in Ueda and MR in Katsunuma). S. cerevisiae has an extremely low abundance in wine grapes [8,56,57]. In contrast, S. cerevisiae was the predominant species when self-enrichment (spontaneous fermentation) was performed [53]. Although self-enrichment was performed in this study, S. cerevisiae was still detected in low abundance. Different approaches should be considered to increase the abundance of isolated S. cerevisiae in the future, such as adding SO2, fermentation temperature control, and maintaining anaerobic conditions to suppress non-Saccharomyces yeast from becoming dominant [35].
In a previous study, S. cerevisiae strains were isolated from vineyard soils, grapes, grape juices, and grape pomace in the Yamanashi area and classified into 35 karyotypes [33]. In this study, indigenous S. cerevisiae isolates were chosen for further genotypic characterization and the evaluation of their enological traits. The dendrogram from the inter-delta analysis suggested that the two wine-producing regions contained S. cerevisiae strains of different genotypes. The S. cerevisiae isolates in this study resulted from a single year of isolation, and further isolation in consecutive vintages is needed for reliable genotypic characterization, similar to other studies conducted using consecutive vintages [10,26]. In addition, these strains were not isolated from the same grape varieties; therefore, the influence of the grape variety should also be considered. Further studies are required to determine whether these indigenous S. cerevisiae isolates contribute to the typicity of wines from these regions.
The isolated S. cerevisiae strains exhibited interesting physiological characteristics. Our results show, for the first time, that indigenous yeasts with different genotypes originating from different wine-producing regions in Japan have distinctive enological traits. In this study, we prefer to focus on investigating the differences in the enological traits of indigenous S. cerevisiae strains rather than enological points for our strains being suitable for application in winemaking. Therefore, our results suggest preliminary data and initial steps for application in winemaking. It will be needed to measure reducing sugars and glycerol, which are important parameters for wines, to add SO2, and to investigate fermentation dynamics. Further analyses are necessary to verify the use of these strains in winemaking with final sensorial evaluation. Moreover, further studies to investigate the organoleptic characteristics that native yeasts bring to wines produced in different regions using must from various grape varieties and the grapes from which the yeasts were isolated are required. Non-Saccharomyces strains were not included in the fermentation tests in this study. Genotypic and phenotypic characterizations of non-Saccharomyces strains are needed to improve our understanding of yeast diversity in Japan and the preservation of potential wine yeasts, considering their high quantity and high distribution among the samples. Further investigations on the enological traits of non-Saccharomyces strains will be necessary.
5. Conclusions
We studied the diversity and distribution of yeast associated with wine grapes from vineyards in Japan. There is a lack of information on such yeast, so our findings provide new information on the yeast flora in the grape-growing regions of Japan. Wine grapes in Japan are the principal source of different yeast species, particularly non-Saccharomyces species. However, the evaluation of indigenous S. cerevisiae isolates revealed that these strains have different enological traits, and region-specific yeast strains may exist and contribute to the wine terroir. Our findings will contribute to a better understanding of local yeasts associated with these terroirs and the use of starter cultures to provide region-specific organoleptic characteristics to local wines.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms12091769/s1, Table S1: Representative isolates and identified yeast species based on the sequencing of the 26S rDNA D1/D2 domain.
Author Contributions
Conceptualization, M.O. and K.S.; methodology, M.O. and K.S.; software, K.K.; validation, K.K., K.S. and M.O.; formal analysis, K.K.; investigation, K.S. and K.K.; resources, M.O.; data curation, M.O. and K.S.; writing—original draft preparation, K.S.; writing—review and editing, M.O.; visualization, K.K.; supervision, M.O.; project administration, M.O.; funding acquisition, M.O. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by JSPS KAKENHI (grant number: 21K05873) and the Institute for Fermentation, Osaka (IFO), Japan (grant number: G-2023-1-013).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article or Supplementary Material.
Acknowledgments
We thank Yui Okumura for technical assistance during sampling and for identifying various isolates.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Barata, A.; Malfeito-Ferreira, M.; Loureiro, V. The microbial ecology of wine grape berries. Int. J. Food Microbiol. 2012, 153, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Li, S.S.; Cheng, C.; Li, Z.; Chen, J.Y.; Yan, B.; Han, B.Z.; Reeves, M. Yeast species associated with wine grapes in China. Int. J. Food Microbiol. 2010, 138, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Guo, J.; Liu, F.; Liu, Y. Identification of indigenous yeast flora isolated from the five winegrape varieties harvested in Xiangning, China. Antonie Van Leeuwenhoek 2014, 105, 533–540. [Google Scholar] [CrossRef] [PubMed]
- Brysch-Herzberg, M.; Seidel, M. Yeast diversity on grapes in two German wine growing regions. Int. J. Food Microbiol. 2015, 214, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Kachalkin, A.V.; Abdullabekova, D.A.; Magomedova, E.S.; Magomedov, G.G.; Chernov, I.Y. Yeasts of the vineyards in Dagestan and other regions. Microbiology 2015, 84, 425–432. [Google Scholar] [CrossRef]
- Bučková, M.; Puškárová, A.; Ženišová, K.; Kraková, L.; Piknová, Ľ.; Kuchta, T.; Pangallo, D. Novel insights into microbial community dynamics during the fermentation of Central European ice wine. Int. J. Food Microbiol. 2018, 266, 42–51. [Google Scholar] [CrossRef]
- Bougreau, M.; Ascencio, K.; Bugarel, M.; Nightingale, K.; Loneragan, G. Yeast species isolated from Texas High Plains vineyards and dynamics during spontaneous fermentations of Tempranillo grapes. PLoS ONE 2019, 14, e0216246. [Google Scholar] [CrossRef]
- Vaudano, E.; Quinterno, G.; Costantini, A.; Pulcini, L.; Pessione, E.; Garcia-Moruno, E. Yeast distribution in Grignolino grapes growing in a new vineyard in Piedmont and the technological characterization of indigenous Saccharomyces spp. strains. Int. J. Food Microbiol. 2019, 289, 154–161. [Google Scholar] [CrossRef]
- Franco, W.; Benavides, S.; Valencia, P.; Ramirez, C.; Urtubia, A. Native yeasts and lactic acid bacteria isolated from spontaneous fermentation of seven grape cultivars from the Maule Region (Chile). Foods 2021, 10, 1737. [Google Scholar] [CrossRef]
- Gonzalez-Alonso, I.; Walker, M.E.; Pascual-Vallejo, M.E.; Naharro-Carrasco, G.; Jiranek, V. Capturing yeast associated with grapes and spontaneous fermentations of the Negro Sauri minority variety from an experimental vineyard near Leon. Sci. Rep. 2021, 11, 3748. [Google Scholar] [CrossRef]
- Zhao, Y.; Sun, Q.; Zhu, S.; Du, F.; Mao, R.; Liu, L.; Tian, B.; Zhu, Y. Biodiversity of non-Saccharomyces yeasts associated with spontaneous fermentation of Cabernet Sauvignon wines from Shangri-La wine region, China. Sci. Rep. 2021, 11, 5150. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Jiang, J.; Song, Y.; Zang, X.; Wang, G.; Pei, Y.; Song, Y.; Qin, Y.; Liu, Y. Yeast diversity during spontaneous fermentations and oenological characterisation of indigenous Saccharomyces cerevisiae for potential as wine starter cultures. Microorganisms 2022, 10, 1455. [Google Scholar] [CrossRef] [PubMed]
- Gurakan, G.C.; Aktuna, I.; Seyedmonir, E. Diversity of wild yeasts during spontaneous fermentation of wines from local grape varieties in Turkey. Am. J. Enol. Vitic. 2022, 73, 308–320. [Google Scholar] [CrossRef]
- Drumonde-Neves, J.; Franco-Duarte, R.; Lima, T.; Schuller, D.; Pais, C. Association between grape yeast communities and the vineyard ecosystems. PLoS ONE 2017, 12, e0169883. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. USA 2014, 111, E139–E148. [Google Scholar] [CrossRef]
- Gilbert, J.A.; van der Lelie, D.; Zarraonaindia, I. Microbial terroir for wine grapes. Proc. Natl. Acad. Sci. USA 2014, 111, 5–6. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, P.; Chen, D.; Howell, K. From the vineyard to the winery: How microbial ecology drives regional distinctiveness of wine. Front. Microbiol. 2019, 10, 2679. [Google Scholar] [CrossRef]
- Alexandre, H. Wine yeast terroir: Separating the wheat from the chaff-for an open debate. Microorganisms 2020, 8, 787. [Google Scholar] [CrossRef]
- Knight, S.; Goddard, M.R. Quantifying separation and similarity in a Saccharomyces cerevisiae metapopulation. ISME J. 2015, 9, 361–370. [Google Scholar] [CrossRef]
- Knight, S.; Klaere, S.; Fedrizzi, B.; Goddard, M.R. Regional microbial signatures positively correlate with differential wine phenotypes: Evidence for a microbial aspect to terroir. Sci. Rep. 2015, 5, 14233. [Google Scholar] [CrossRef]
- Gayevskiy, V.; Goddard, M.R. Geographic delineations of yeast communities and populations associated with vines and wines in New Zealand. ISME J. 2012, 6, 1281–1290. [Google Scholar] [CrossRef]
- Tofalo, R.; Perpetuini, G.; Fasoli, G.; Schirone, M.; Corsetti, A.; Suzzi, G. Biodiversity study of wine yeasts belonging to the “terroir” of Montepulciano d’Abruzzo “Colline Teramane” revealed Saccharomyces cerevisiae strains exhibiting atypical and unique 5.8S-ITS restriction patterns. Food Microbiol. 2014, 39, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Capece, A.; Granchi, L.; Guerrini, S.; Mangani, S.; Romaniello, R.; Vincenzini, M.; Romano, P. diversity of Saccharomyces cerevisiae strains isolated from two Italian wine-producing regions. Front. Microbiol. 2016, 7, 1018. [Google Scholar] [CrossRef] [PubMed]
- Viel, A.; Legras, J.L.; Nadai, C.; Carlot, M.; Lombardi, A.; Crespan, M.; Migliaro, D.; Giacomini, A.; Corich, V. The geographic distribution of Saccharomyces cerevisiae isolates within three Italian neighboring winemaking regions reveals strong differences in yeast abundance, genetic diversity and industrial strain dissemination. Front. Microbiol. 2017, 8, 1595. [Google Scholar] [CrossRef] [PubMed]
- Granchi, L.; Ganucci, D.; Buscioni, G.; Mangani, S.; Guerrini, S. The biodiversity of Saccharomyces cerevisiae in spontaneous wine fermentation: The occurrence and persistence of winery-strains. Fermentation 2019, 5, 86. [Google Scholar] [CrossRef]
- Chalvantzi, I.; Banilas, G.; Tassou, C.; Nisiotou, A. Patterns of genetic diversity and the invasion of commercial starters in Saccharomyces cerevisiae vineyard populations of Santorini Island. Foods 2020, 9, 561. [Google Scholar] [CrossRef]
- Zabukovec, P.; Cadez, N.; Cus, F. Isolation and identification of indigenous wine yeasts and their use in alcoholic fermentation. Food Technol. Biotechnol. 2020, 58, 337–347. [Google Scholar] [CrossRef]
- Shimatani, Y.; Nagata, Y. Studies on microflora related to wine making (1): Mould and yeast flora in a vineyard. J. Ferment. Technol. 1967, 45, 179–184. [Google Scholar]
- Shimatani, Y.; Nonaka, M. Studies on microflora related to wine making (2): Distribution of film yeasts in vineyards and wine making processes in five grape and wine producing districts of Japan. J. Ferment. Technol. 1967, 45, 185–190. [Google Scholar]
- Goto, S.; Yokotsuka, I. Wild yeast populations in fresh grape musts of different harvest times. J. Ferment. Technol. 1977, 55, 417–422. [Google Scholar]
- Goto, S. Changes in the wild yeast flora of sulfited grape musts. J. Inst. Enol. Vitic. Yamanashi Univ. 1980, 15, 29–32. [Google Scholar]
- Yanagida, F.; Ichinose, F.; Shinohara, T.; Goto, S. Distribution of wild yeasts in the white grape varieties at Central Japan. J. Gen. Appl. Microbiol. 1992, 38, 501–504. [Google Scholar] [CrossRef][Green Version]
- Shinohara, T.; Furuya, H.; Yanagida, F.; Miki, T. Ecological distribution and phenotypic diversity of Saccharomyces cerevisiae strains from the wine-producing area in Yamanashi, Japan. Microbiol. Cult. Coll. 2003, 19, 69–80. [Google Scholar]
- Shiga, T.; Otoguro, M.; Yamada, S.; Anzo, M.; Omura, H.; Kishimoto, M. Effect of scale-up method in spontaneous fermentation on wine quality. J. ASEV Jpn. 2020, 31, 133–139. [Google Scholar]
- Shimizu, H.; Kamada, A.; Koyama, K.; Iwashita, K.; Goto-Yamamoto, N. Yeast diversity during the spontaneous fermentation of wine with only the microbiota on grapes cultivated in Japan. J. Biosci. Bioeng. 2023, 136, 35–43. [Google Scholar] [CrossRef]
- National Research Institute of Brewing: Standard Analytical Methods of the National Research Institute of Brewing, Japan. 2017. Available online: https://www.nrib.go.jp/bun/nribanalysis.html (accessed on 16 May 2022).
- Kurtzman, C.P.; Robnett, C.J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek 1998, 73, 331–371. [Google Scholar] [CrossRef]
- Legras, J.L.; Karst, F. Optimisation of interdelta analysis for Saccharomyces cerevisiae strain characterisation. FEMS Microbiol. Lett. 2003, 221, 249–255. [Google Scholar] [CrossRef]
- Chavan, P.; Mane, S.; Kulkarni, G.; Shaikh, S.; Ghormade, V.; Nerkar, D.P.; Shouche, Y.; Deshpande, M.V. Natural yeast flora of different varieties of grapes used for wine making in India. Food Microbiol. 2009, 26, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Castrillo Cachón, D.; Rabuñal Crego, E.; Neira González, N.; Blanco Camba, P. Yeast diversity on grapes from Galicia, NW Spain: Biogeographical patterns and the influence of the farming system. OENO One 2019, 53, 573–587. [Google Scholar] [CrossRef]
- Jolly, N.P.; Varela, C.; Pretorius, I.S. Not your ordinary yeast: Non-Saccharomyces yeasts in wine production uncovered. FEMS Yeast Res. 2014, 14, 215–237. [Google Scholar] [CrossRef]
- Grangeteau, C.; Gerhards, D.; von Wallbrunn, C.; Alexandre, H.; Rousseaux, S.; Guilloux-Benatier, M. Persistence of Two Non-Saccharomyces Yeasts (Hanseniaspora and Starmerella) in the Cellar. Front. Microbiol. 2016, 7, 268. [Google Scholar] [CrossRef] [PubMed]
- Romano, P.; Fiore, C.; Paraggio, M.; Caruso, M.; Capece, A. Function of yeast species and strains in wine flavour. Int. J. Food Microbiol. 2003, 86, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Roudil, L.; Russo, P.; Berbegal, C.; Albertin, W.; Spano, G.; Capozzi, V. Non-Saccharomyces commercial starter cultures: Scientific trends, recent patents and innovation in the wine sector. Recent Pat. Food Nutr. Agric. 2020, 11, 27–39. [Google Scholar] [CrossRef] [PubMed]
- Mendoza, L.M.; Vega-Lopez, G.A.; Fernández de Ullivarri, M.; Raya, R.R. Population and oenological characteristics of non-Saccharomyces yeasts associated with grapes of Northwestern Argentina. Arch. Microbiol. 2019, 201, 235–244. [Google Scholar] [CrossRef]
- Tristezza, M.; Tufariello, M.; Capozzi, V.; Spano, G.; Mita, G.; Grieco, F. The oenological potential of Hanseniaspora uvarum in simultaneous and sequential co-fermentation with Saccharomyces cerevisiae for industrial wine production. Front. Microbiol. 2016, 7, 670. [Google Scholar] [CrossRef]
- Martin, V.; Valera, M.; Medina, K.; Boido, E.; Carrau, F. Oenological impact of the Hanseniaspora/Kloeckera yeast genus on wines—A review. Fermentation 2018, 4, 76. [Google Scholar] [CrossRef]
- Medina, K.; Boido, E.; Fariña, L.; Gioia, O.; Gomez, M.E.; Barquet, M.; Gaggero, C.; Dellacassa, E.; Carrau, F. Increased flavour diversity of Chardonnay wines by spontaneous fermentation and co-fermentation with Hanseniaspora vineae. Food Chem. 2013, 141, 2513–2521. [Google Scholar] [CrossRef]
- Nisiotou, A.; Sgouros, G.; Mallouchos, A.; Nisiotis, C.S.; Michaelidis, C.; Tassou, C.; Banilas, G. The use of indigenous Saccharomyces cerevisiae and Starmerella bacillaris strains as a tool to create chemical complexity in local wines. Food Res. Int. 2018, 111, 498–508. [Google Scholar] [CrossRef]
- Binati, R.L.; Lemos Junior, W.J.F.; Luzzini, G.; Slaghenaufi, D.; Ugliano, M.; Torriani, S. Contribution of non-Saccharomyces yeasts to wine volatile and sensory diversity: A study on Lachancea thermotolerans, Metschnikowia spp. and Starmerella bacillaris strains isolated in Italy. Int. J. Food Microbiol. 2020, 318, 108470. [Google Scholar] [CrossRef]
- Loureiro, V.; Malfeito-Ferreira, M. Spoilage yeasts in the wine industry. Int. J. Food Microbiol. 2003, 86, 23–50. [Google Scholar] [CrossRef]
- Garavaglia, J.; Schneider, R.d.C.d.S.; Camargo Mendes, S.D.; Welke, J.E.; Zini, C.A.; Caramão, E.B.; Valente, P. Evaluation of Zygosaccharomyces bailii BCV 08 as a co-starter in wine fermentation for the improvement of ethyl esters production. Microbiol. Res. 2015, 173, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Cordero-Bueso, G.; Vigentini, I.; Foschino, R.; Maghradze, D.; Ruiz-Munoz, M.; Benitez-Trujillo, F.; Cantoral, J.M. Culturable Yeast Diversity of Grape Berries from Vitis vinifera ssp. sylvestris (Gmelin) Hegi. J. Fungi 2022, 8, 410. [Google Scholar] [CrossRef]
- Xue, M.L.; Zhang, L.Q.; Wang, Q.M.; Zhang, J.S.; Bai, F.Y. Metschnikowia sinensis sp. nov., Metschnikowia zizyphicola sp. nov. and Metschnikowia shanxiensis sp. nov., novel yeast species from jujube fruit. Int. J. Syst. Evol. Microbiol. 2006, 56, 2245–2250. [Google Scholar] [CrossRef][Green Version]
- Liu, D.; Legras, J.L.; Zhang, P.; Chen, D.; Howell, K. Diversity and dynamics of fungi during spontaneous fermentations and association with unique aroma profiles in wine. Int. J. Food Microbiol. 2021, 338, 108983. [Google Scholar] [CrossRef] [PubMed]
- Raspor, P.; Milek, D.M.; Polanc, J.; Mozina, S.S.; Cadez, N. Yeasts isolated from three varieties of grapes cultivated in different locations of the Dolenjska vine-growing region, Slovenia. Int. J. Food Microbiol. 2006, 109, 97–102. [Google Scholar] [CrossRef]
- Silva, G.A.; Agustini, B.C.; de Mello, L.M.R.; Tonietto, J. Autochthonous yeast populations from different brazilian geographic indications. BIO Web Conf. 2016, 7, 02030. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).