Virulence Factors and Susceptibility to Ciprofloxacin, Vancomycin, Triclosan, and Chlorhexidine among Enterococci from Clinical Specimens, Food, and Wastewater
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of Strains
2.2. Isolation and Identification
2.3. Antimicrobial Susceptibility Testing
2.4. Triclosan and Chlorhexidine Susceptibility Testing–Minimum Inhibitory Concentration
2.5. Biofilm Formation
2.6. Cell Surface Hydrophobicity
2.7. Assessment of β-Hemolytic Activity
2.8. Statistical Analysis
3. Results
3.1. Antimicrobial Susceptibility of Enterococcus spp. to Vancomycin and Ciprofloxacin according to Origin
3.2. Antimicrobial Susceptibility of Enterococcus spp. to Triclosan and Chlorhexidine
3.3. Biofilm Formation Ability
3.3.1. E. faecalis—Association between Biofilm-Forming Strength and CIP and VAN Susceptibility
3.3.2. E. faecium—Association between Biofilm-Forming Capacity and CIP and VAN Susceptibility
3.3.3. E. faecalis—Association between Biofilm-Forming Strength and CHX and TCL MICs
3.3.4. E. faecium—Association between Biofilm-Forming Strength and CHX and TCL MICs
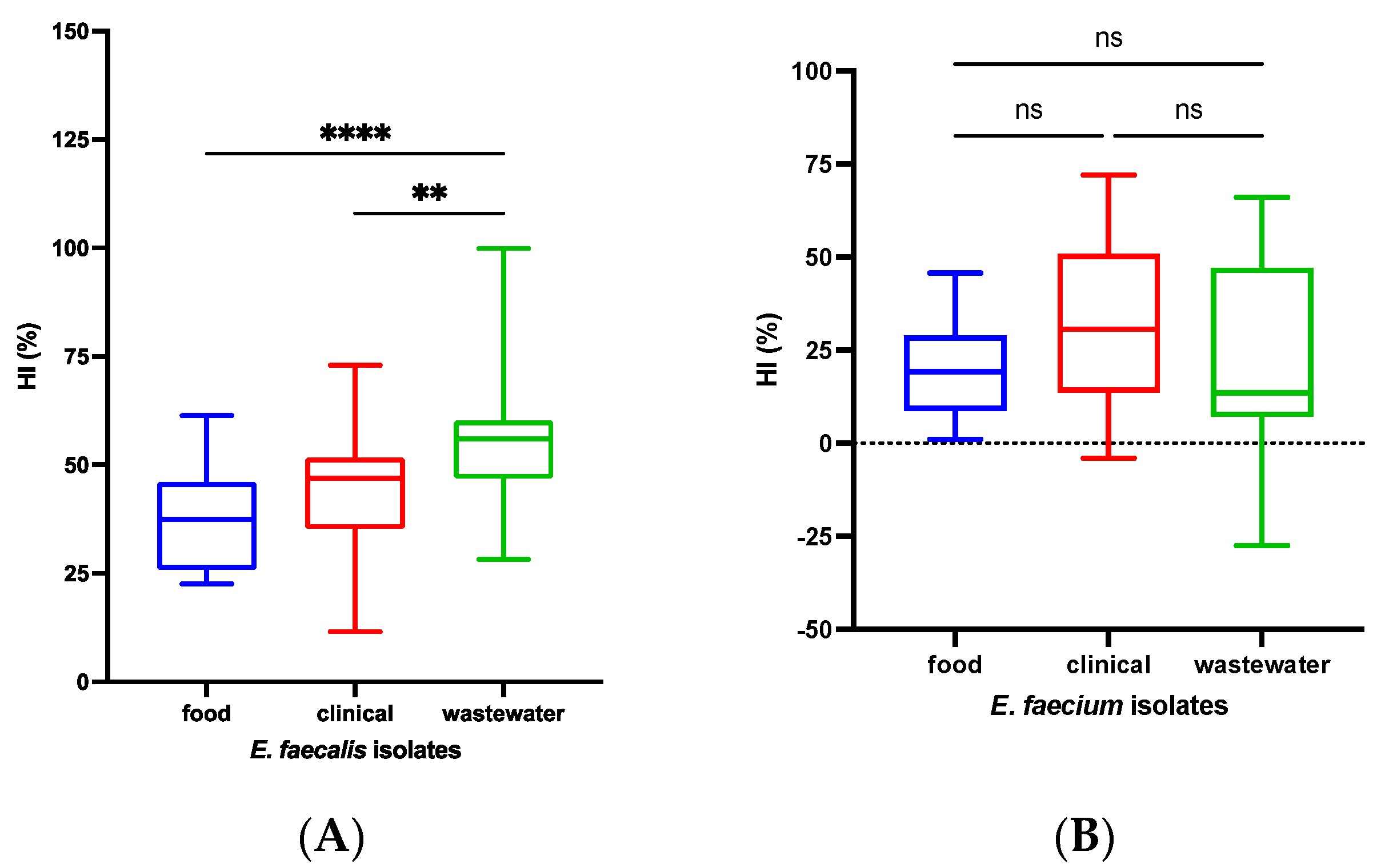
3.4. Cell Surface Hydrophobicity Index
3.5. Hemolytic Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Parte, A.C.; Sardà Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic Names with Standing in Nomenclature (LPSN) Moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef] [PubMed]
- Batt, C.; Tortorello, M. Encyclopedia of Food Microbiology, 2nd ed.; Academic Press Elsevier: Amsterdam, The Netherlands, 2014. [Google Scholar]
- Flores, C.; Loureiro, L.; Bessa, L.; Costa, P. Presence of Multidrug-Resistant E. coli, Enterococcus spp. and Salmonella spp. in Lakes and Fountains of Porto, Portugal. J. Water Resour. Prot. 2013, 5, 1117–1126. [Google Scholar] [CrossRef]
- Poh, C.H.; Oh, H.M.L.; Tan, A.L. Epidemiology and Clinical Outcome of Enterococcal Bacteraemia in an Acute Care Hospital. J. Infect. 2006, 52, 383–386. [Google Scholar] [CrossRef] [PubMed]
- Fisher, K.; Phillips, C. The Ecology, Epidemiology and Virulence of Enterococcus. Microbiology 2009, 155, 1749–1757. [Google Scholar] [CrossRef]
- Brinkwirth, S.; Ayobami, O.; Eckmanns, T.; Markwart, R. Hospital-Acquired Infections Caused by Enterococci: A Systematic Review and Meta-Analysis, WHO European Region, 1 January 2010 to 4 February 2020. Euro Surveill. 2021, 26, 2001628. [Google Scholar] [CrossRef] [PubMed]
- Tambić Andrašević, A.; Lucić, S.; Tambić, T. Rezistencija Na Antibiotike u Hrvatskoj [Antibiotic Resistance in Croatia]. Med. Flum. 2018, 54, 312–321. [Google Scholar] [CrossRef]
- Maillard, J.Y. Bacterial Target Sites for Biocide Action. J. Appl. Microbiol. 2022, 92, 16S–27S. [Google Scholar] [CrossRef]
- Fraise, A. Susceptibility of Antibiotic-Resistant Cocci to Biocides. J. Appl. Microbiol. 2002, 92, 158S–162S. [Google Scholar] [CrossRef]
- Lebreton, F.; Willems, R.; Gilmore, M. Enterococcus Diversity, Origins in Nature, and Gut Colonization. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M., Clewell, D., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Murray, P.R.; Rosenthal, K.; Pfaller, M.A. Medical Microbiology, 9th ed.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 207–209. ISBN 9780323674508. [Google Scholar]
- Hollenbeck, B.L.; Rice, L.B. Intrinsic and Acquired Resistance Mechanisms in Enterococcus. Virulence 2012, 3, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Tahmourespour, A.; Kermanshahi, R.K.; Salehi, R.; Nabinejad, A. The Relationship between Cell Surface Hydrophobicity and Antibiotic Resistance of Streptococcal Strains Isolated from Dental Plaque and Caries. Iran. J. Basic. Med. Sci. 2008, 10, 251–255. [Google Scholar] [CrossRef]
- Foulquié Moreno, M.R.; Sarantinopoulos, P.; Tsakalidou, E.; De Vuyst, L. The Role and Application of Enterococci in Food and Health. Int. J. Food Microbiol. 2006, 106, 1–24. [Google Scholar] [CrossRef]
- Barbosa, J.; Gibbs, P.A.; Teixeira, P. Virulence Factors among Enterococci Isolated from Traditional Fermented Meat Products Produced in the North of Portugal. Food Control 2010, 21, 651–656. [Google Scholar] [CrossRef]
- Van Tyne, D.; Martin, M.J.; Gilmore, M.S. Structure, Function, and Biology of the Enterococcus faecalis Cytolysin. Toxins 2013, 5, 895–911. [Google Scholar] [CrossRef] [PubMed]
- Koch, S.; Hufnagel, M.; Theilacker, C.; Huebner, J. Enterococcal Infections: Host Response, Therapeutic, and Prophylactic Possibilities. Vaccine 2004, 22, 822–830. [Google Scholar] [CrossRef]
- Russell, A. Introduction of Biocides into Clinical Practice and the Impact on Antibiotic-Resistant Bacteria. J. Appl. Microbiol. 2002, 92, 121S–135S. [Google Scholar] [CrossRef] [PubMed]
- Walsh, C.; Fanning, S. Antimicrobial Resistance in Foodborne Pathogens—A Cause for Concern. Curr. Drug Targets 2008, 9, 808–815. [Google Scholar] [CrossRef] [PubMed]
- Gomez Escalada, M.; Russell, A.D.; Maillard, J.Y.; Ochs, D. Triclosan-Bacteria Interactions: Single or Multiple Target Sites? Lett. Appl. Microbiol. 2005, 41, 476–481. [Google Scholar] [CrossRef] [PubMed]
- Maillard, J.Y. Bacterial Resistance to Biocides in the Healthcare Environment: Should It Be of Genuine Concern? J. Hosp. Infect. 2007, 65, 60–72. [Google Scholar] [CrossRef]
- Levy, S. Active Efflux, a Common Mechanism for Biocide and Antibiotic Resistance. J. Appl. Microbiol. 2022, 92, 65S–71S. [Google Scholar] [CrossRef]
- Schwaiger, K.; Harms, K.S.; Bischoff, M.; Preikschat, P.; Mölle, G.; Bauer-Unkauf, I.; Lindorfer, S.; Thalhammer, S.; Bauer, J.; Hölzel, C.S. Insusceptibility to Disinfectants in Bacteria from Animals, Food and Humans—Is There a Link to Antimicrobial Resistance? Front. Microbiol. 2014, 5, 88. [Google Scholar] [CrossRef]
- Braoudaki, M.; Hilton, A.C. Adaptive Resistance to Biocides in Salmonella enterica and Escherichia coli O157 and Cross-Resistance to Antimicrobial Agents. J. Clin. Microbiol. 2004, 42, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, P.; McBain, A.J. Potential Impact of Increased Use of Biocides in Consumer Products on Prevalence of Antibiotic Resistance. Clin. Microbiol. Rev. 2003, 16, 189–208. [Google Scholar] [CrossRef] [PubMed]
- Rozman, U.; Pušnik, M.; Kmetec, S.; Duh, D.; Turk, S.Š. Reduced Susceptibility and Increased Resistance of Bacteria against Disinfectants: A Systematic Review. Microorganisms 2021, 9, 2550. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, I.; Oggioni, M.R.; Knight, D.; Curiao, T.; Coque, T.; Kalkanci, A.; Martinez, J.L.; Baldassarri, L.; Orefici, G.; Yetiş, Ü.; et al. Evaluation of Epidemiological Cut-off Values Indicates That Biocide Resistant Subpopulations Are Uncommon in Natural Isolates of Clinically-Relevant Microorganisms. PLoS ONE 2014, 9, e86669. [Google Scholar] [CrossRef]
- Bhatia, M.; Mishra, B.; Thakur, A.; Dogra, V.; Loomba, P.S. Evaluation of Susceptibility of Glycopeptide-Resistant and Glycopeptide-Sensitive Enterococci to Commonly Used Biocides in a Super-Speciality Hospital: A Pilot Study. J. Nat. Sci. Biol. Med. 2017, 8, 199–202. [Google Scholar] [CrossRef] [PubMed]
- Beier, R.C.; Duke, S.E.; Ziprin, R.L.; Harvey, R.B.; Hume, M.E.; Poole, T.L.; Scott, H.M.; Highfield, L.D.; Alali, W.Q.; Andrews, K.; et al. Antibiotic and Disinfectant Susceptibility Profiles of Vancomycin-Resistant Enterococcus faecium (VRE) Isolated from Community Wastewater in Texas. Bull. Environ. Contam. Toxicol. 2008, 80, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Ohlsson, A.K.; Ullberg, M.; Özenci, V. Evaluation of Species-Specific PCR, Bruker MS, VITEK MS and the VITEK 2 System for the Identification of Clinical Enterococcus Isolates. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 3073–3077. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Wu, B.; Li, Y.; Jin, X.; Wang, X. Identification and Safety Assessment of Enterococcus thailandicus TC1 Isolated from Healthy Pigs. PLoS ONE 2021, 16, e0254081. [Google Scholar] [CrossRef]
- Eucast. EUCAST. Available online: https://www.eucast.org/ (accessed on 25 July 2024).
- Eucast. Clinical Breakpoints and Dosing of Antibiotics. Available online: https://www.eucast.org/clinical_breakpoints (accessed on 25 July 2024).
- ISO 20776-1:2020; Hrvatski zavod za norme Ispitivanje Osjetljivosti Infektivnih Agensa i Procjena Svojstava Antimikrobne Osjetljivosti Ispitnih Proizvoda-1. Dio: Referentna Metoda Mikrorazrjeđivanja Bujonom Za Ispitivanje in Vitro Djelovanja Antimikrobnih Agensa Protiv Brzog Rasta Aerobnih Bakterija Koje Uzrokuju Infektivne Bolesti (ISO 20776-1:2019, Ispravljena Verzija 2019-12; EN ISO 20776-1:2020). ISO: Geneva, Switzerland, 2020. Available online: https://repozitorij.hzn.hr/norm/HRN+EN+ISO+20776-1%3A2020 (accessed on 16 May 2024).
- Stepanović, S.; Vuković, D.; Hola, V.; Di Bonaventura, G.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of Biofilm in Microtiter Plates: Overview of Testing Conditions and Practical Recommendations for Assessment of Biofilm Production by Staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef]
- Stępień-Pyśniak, D.; Hauschild, T.; Kosikowska, U.; Dec, M.; Urban-Chmiel, R. Biofilm Formation Capacity and Presence of Virulence Factors among Commensal Enterococcus spp. from Wild Birds. Sci. Rep. 2019, 9, 11204. [Google Scholar] [CrossRef]
- Semedo, T.; Santos, M.A.; Martins, P.; Silva Lopes, M.F.; Figueiredo Marques, J.J.; Tenreiro, R.; Barreto Crespo, M.T. Comparative Study Using Type Strains and Clinical and Food Isolates to Examine Hemolytic Activity and Occurrence of the Cyl Operon in Enterococci. J. Clin. Microbiol. 2003, 41, 2569–2576. [Google Scholar] [CrossRef]
- Jasp, T. JASP, Version 0.18.3 [Computer Software]. Available online: https://jasp-stats.org/faq/how-do-i-cite-jasp/ (accessed on 16 May 2024).
- Velazquez-Meza, M.E.; Galarde-López, M.; Carrillo-Quiróz, B.; Alpuche-Aranda, C.M. Antimicrobial Resistance: One Health Approach. Vet. World 2022, 15, 743. [Google Scholar] [CrossRef] [PubMed]
- Arshadi, M.; Mahmoudi, M.; Motahar, M.S.; Soltani, S.; Pourmand, M.R. Virulence Determinants and Antimicrobial Resistance Patterns of Vancomycin-Resistant Enterococcus faecium Isolated from Different Sources in Southwest Iran. Iran J. Public Health 2018, 47, 264. [Google Scholar]
- Sinel, C.; Cacaci, M.; Meignen, P.; Guérin, F.; Davies, B.W.; Sanguinetti, M.; Giard, J.C.; Cattoir, V. Subinhibitory Concentrations of Ciprofloxacin Enhance Antimicrobial Resistance and Pathogenicity of Enterococcus faecium. Antimicrob. Agents Chemother. 2017, 61, e02763-16. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.B.; Seo, H.J.; Seo, K.W.; Jeon, H.Y.; Kim, D.K.; Kim, S.W.; Lim, S.K.; Lee, Y.J. Characteristics of High-Level Ciprofloxacin-Resistant Enterococcus faecalis and Enterococcus faecium from Retail Chicken Meat in Korea. J. Food Prot. 2018, 81, 1357–1363. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.C.; Woo, G.J. Characterization of Antimicrobial Resistance and Quinolone Resistance Factors in High-Level Ciprofloxacin-Resistant Enterococcus faecalis and Enterococcus faecium Isolates Obtained from Fresh Produce and Fecal Samples of Patients. J. Sci. Food Agric. 2017, 97, 2858–2864. [Google Scholar] [CrossRef] [PubMed]
- Sobhanipoor, M.H.; Ahmadrajabi, R.; Nave, H.H.; Saffari, F. Reduced Susceptibility to Biocides among Enterococci from Clinical and Non-Clinical Sources. Infect. Chemother 2021, 53, 696. [Google Scholar] [CrossRef] [PubMed]
- Jones, I.A.; Joshi, L.T. Biocide Use in the Antimicrobial Era: A Review. Molecules 2021, 26, 2276. [Google Scholar] [CrossRef]
- Geraldes, C.; Tavares, L.; Gil, S.; Oliveira, M. Enterococcus Virulence and Resistant Traits Associated with Its Permanence in the Hospital Environment. Antibiotics 2022, 11, 857. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, H.P. Triclosan: A Widely Used Biocide and Its Link to Antibiotics. FEMS Microbiol. Lett. 2001, 202, 1–7. [Google Scholar] [CrossRef]
- Cameron, A.; Barbieri, R.; Read, R.; Church, D.; Adator, E.H.; Zaheer, R.; McAllister, T.A. Functional Screening for Triclosan Resistance in a Wastewater Metagenome and Isolates of Escherichia coli and Enterococcus spp. from a Large Canadian Healthcare Region. PLoS ONE 2019, 14, e0211144. [Google Scholar] [CrossRef] [PubMed]
- Abbood, H.M.; Hijazi, K.; Gould, I.M. Chlorhexidine Resistance or Cross-Resistance, That Is the Question. Antibiotics 2023, 12, 798. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, P.; Hans, A.; Ruikar, K.; Guan, Z.; Palmer, K.L. Reduced Chlorhexidine and Daptomycin Susceptibility in Vancomycin-Resistant Enterococcus faecium after Serial Chlorhexidine Exposure. Antimicrob. Agents Chemother 2018, 62, e01235-17. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.P.; Antunes, P.; Willems, R.; Corander, J.; Coque, T.M.; Peixe, L.; Freitas, A.R.; Novais, C. Evolution of Chlorhexidine Susceptibility and of the EfrEF Operon among Enterococcus faecalis from Diverse Environments, Clones, and Time Spans. Microbiol. Spectr. 2022, 10, e0117622. [Google Scholar] [CrossRef] [PubMed]
- Goudarzi, M.; Mobarez, A.; Njar-Peerayeh, S.; Mirzaee, M. Prevalence of Biofilm Formation and Vancomycin-Resistant Genes among Enterococcus faecium Isolated from Clinical and Environmental Specimens in Lorestan Hospitals—PubMed. Iran. J. Microbiol. 2018, 10, 74–81. [Google Scholar] [PubMed]
- Mladenović, K.G.; Grujović, M.; Nikodijević, D.D.; čOmić, L.R. The Hydrophobicity of Enterobacteria and Their Co-Aggregation with Enterococcus faecalis Isolated from Serbian Cheese. Biosci. Microbiota Food Health 2020, 39, 227. [Google Scholar] [CrossRef]
- Cho, J.A.; Roh, Y.J.; Son, H.R.; Choi, H.; Lee, J.W.; Kim, S.J.; Lee, C.H. Assessment of the Biofilm-Forming Ability on Solid Surfaces of Periprosthetic Infection-Associated Pathogens. Sci. Rep. 2022, 12, 18669. [Google Scholar] [CrossRef]
- Elsner, H.A.; Sobottka, I.; Mack, D.; Claussen, M.; Laufs, R.; Wirth, R. Virulence Factors of Enterococcus faecalis and Enterococcus faecium Blood Culture Isolates. Eur. J. Clin. Microbiol. Infect. Dis. 2000, 19, 39–42. [Google Scholar] [CrossRef] [PubMed]
- Coburn, P.S.; Gilmore, M.S. The Enterococcus faecalis Cytolysin: A Novel Toxin Active against Eukaryotic and Prokaryotic Cells. Cell. Microbiol. 2003, 5, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Vergis, E.N.; Shankar, N.; Chow, J.W.; Hayden, M.K.; Snydman, D.R.; Zervos, M.J.; Linden, P.K.; Wagener, M.M.; Muder, R.R. Association between the Presence of Enterococcal Virulence Factors Gelatinase, Hemolysin, and Enterococcal Surface Protein and Mortality among Patients with Bacteremia Due to Enterococcus Faecalis. Clin. Infect. Dis. 2002, 35, 570–575. [Google Scholar] [CrossRef]
| Clinical n (%) | Food n (%) | Wastewater n (%) | p-Value | |||
|---|---|---|---|---|---|---|
| E. faecalis | CIP | R | 12 (54.55) | 6 (27.27) | 4 (18.18) | 0.094 |
| S | 17 (25.37) | 24 (35.82) | 26 (38.81) | 0.368 | ||
| VAN | R | 3 (27.27) | 1 (9.09) | 7 (63.64) | 0.078 | |
| S | 26 (33.33) | 29 (37.18) | 23 (29.49) | 0.707 | ||
| E. faecium | CIP | R | 30 (53.57) | 11 (19.64) | 15 (26.79) | 0.005 |
| S | 0 (0.0) | 19 (55.88) | 15 (44.12) | 0.493 | ||
| VAN | R | 25 (75.76) | 2 (6.06) | 6 (18.18) | <0.001 | |
| S | 5 (8.77) | 28 (49.12) | 24 (42.11) | <0.001 |
| Triclosan (mg/L) | Chlorhexidine (mg/L) | ||||
|---|---|---|---|---|---|
| Species | Isolation Source | MIC50 | MIC90 | MIC50 | MIC90 |
| E. faecalis | Clinical specimens | 8 | 8 | 16 | 32 |
| Food | 8 | 16 | 4 | 8 | |
| Wastewater | 16 | 32 | 8 | 16 | |
| E. faecium | Clinical specimens | 8 | 8 | 8 | 16 |
| Food | 8 | 16 | 2 | 4 | |
| Wastewater | 4 | 8 | 2 | 4 | |
| Biofilm Strength | E. faecalis n (%) | E. faecium n (%) | ||||
|---|---|---|---|---|---|---|
| Food | Clinical Specimens | Wastewater | Food | Clinical Specimens | Wastewater | |
| No biofilm (I) | 0 (0.00) | 0 (0.00) | 2 (6.67) | 4 (13.33) | 1 (3.33) | 20 (66.67) |
| Weak (II) | 0 (0.00) | 2 (6.67) | 2 (6.67) | 0 (0.00) | 0 (0.00) | 6 (20.00) |
| Moderate (III) | 1 (3.45) | 6 (20.00) | 10 (33.33) | 4 (13.33) | 5 (16.67) | 4 (13.33) |
| Strong (IV) | 28 (96.55) | 22 (73.33) | 16 (53.33) | 22 (73.33) | 24 (80.00) | 0 (0.00) |
| Total biofilm producers (II, III, IV) | 29 (100.00) | 30 (100.00) | 28 (93.33) | 26 (86.67) | 29 (96.67) | 10 (33.33) |
| Biofilm Strength | CIP-R n (%) | CIP-S n (%) | p-Value |
|---|---|---|---|
| No biofilm (I) | 1 (50.00) | 1 (50.00) | |
| Weak (II) | 0 (0.00) | 4 (100.00) | NA |
| Moderate (III) | 5 (29.41) | 12 (70.59) | 0.009 |
| Strong (IV) | 16 (24.24) | 50 (75.76) | <0.001 |
| Total biofilm producers (II, III, IV) | 21 (24.14) | 66 (75.86) | <0.001 |
| Biofilm strength | VAN-R n (%) | VAN-S n (%) | p-value |
| No biofilm (I) | 0 (0.00) | 2 (100.00) | |
| Weak (II) | 0 (0.00) | 4 (100.00) | NA |
| Moderate (III) | 3 (17.65) | 14 (82.35) | 0.008 |
| Strong (IV) | 8 (12.12) | 58 (87.88) | <0.001 |
| Total biofilm producers (II, III, IV) | 11 (12.64) | 76 (87.36) | <0.001 |
| Biofilm Strength | CIP-R n (%) | CIP-S n (%) | p-Value |
|---|---|---|---|
| No biofilm (I) | 12 (48.00) | 13 (52.00) | |
| Weak (II) | 4 (66.67) | 2 (33.33) | NA |
| Moderate (III) | 9 (69.23) | 4 (30.77) | 0.166 |
| Strong (IV) | 31 (67.39) | 15 (32.61) | 0.018 * |
| Total biofilm producers (II, III, IV) | 44 (67.69) | 21 (32.31) | 0.004 * |
| Biofilm strength | VAN-R n (%) | VAN-S n (%) | p-value |
| No biofilm (I) | 8 (32.00) | 17 (68.00) | |
| Weak (II) | 2 (33.33) | 4 (66.67) | NA |
| Moderate (III) | 4 (30.77) | 9 (69.23) | 0.166 |
| Strong (IV) | 19 (41.30) | 27 (58.70) | 0.238 |
| Total biofilm producers (II, III, IV) | 25 (38.46) | 40 (61.54) | 0.063 |
| E. faecalis n (%) | E. faecium n (%) | ||||||
|---|---|---|---|---|---|---|---|
| β-Hemolysis | Food | Clinical Specimens | Wastewater | Food | Clinical Specimens | Wastewater | |
| Aerobic incubation | No | 27 (90.00) | 26 (89.66) | 24 (80.00) | 27 (90.00) | 30 (100.00) | 30 (100.00) |
| Yes | 3 (10.00) | 3 (10.34) | 6 (20.00) | 3 (10.00) | 0 (0.00) | 0 (0.00) | |
| Total | 30 (100.00) | 29 (100.00) | 30 (100.00) | 30 (100.00) | 30 (100.00) | 30 (100.00) | |
| Anaerobic incubation | No | 26 (86.67) | 23 (79.31) | 23 (76.67) | 27 (90.00) | 30 (100.00) | 30 (100.00) |
| Yes | 4 (13.33) | 6 (20.69) | 7 (23.33) | 3 (10.00) | 0 (0.00) | 0 (0.00) | |
| Total | 30 (100.00) | 29 (100.00) | 30 (100.00) | 30 (100.00) | 30 (100.00) | 30 (100.00) | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gorski, D.B.; Vlainić, J.; Škrlec, I.; Novak, S.; Novosel, Ž.; Biloglav, Z.; Plečko, V.; Kosalec, I. Virulence Factors and Susceptibility to Ciprofloxacin, Vancomycin, Triclosan, and Chlorhexidine among Enterococci from Clinical Specimens, Food, and Wastewater. Microorganisms 2024, 12, 1808. https://doi.org/10.3390/microorganisms12091808
Gorski DB, Vlainić J, Škrlec I, Novak S, Novosel Ž, Biloglav Z, Plečko V, Kosalec I. Virulence Factors and Susceptibility to Ciprofloxacin, Vancomycin, Triclosan, and Chlorhexidine among Enterococci from Clinical Specimens, Food, and Wastewater. Microorganisms. 2024; 12(9):1808. https://doi.org/10.3390/microorganisms12091808
Chicago/Turabian StyleGorski, Diana Brlek, Josipa Vlainić, Ivana Škrlec, Silvia Novak, Željka Novosel, Zrinka Biloglav, Vanda Plečko, and Ivan Kosalec. 2024. "Virulence Factors and Susceptibility to Ciprofloxacin, Vancomycin, Triclosan, and Chlorhexidine among Enterococci from Clinical Specimens, Food, and Wastewater" Microorganisms 12, no. 9: 1808. https://doi.org/10.3390/microorganisms12091808
APA StyleGorski, D. B., Vlainić, J., Škrlec, I., Novak, S., Novosel, Ž., Biloglav, Z., Plečko, V., & Kosalec, I. (2024). Virulence Factors and Susceptibility to Ciprofloxacin, Vancomycin, Triclosan, and Chlorhexidine among Enterococci from Clinical Specimens, Food, and Wastewater. Microorganisms, 12(9), 1808. https://doi.org/10.3390/microorganisms12091808









