The Future Exploring of Gut Microbiome-Immunity Interactions: From In Vivo/Vitro Models to In Silico Innovations
Abstract
:1. Introduction
- production of metabolites such as short-chain fatty acids (SCFAs), bile acids (BAs), and tryptophan metabolites, which influence the activity of immune cells by promoting the production of regulatory T cells (Tregs) and effector T cells involved in the maintenance of immune tolerance and the prevention of excessive inflammatory responses [2,3,4];
- interaction with intestinal epithelial cells, stimulating the synthesis of antimicrobial molecules such as defensins, which help maintain the integrity of the intestinal barrier and prevent the invasion of pathogenic microbes [5];
- synthesis of lipopolysaccharides and peptidoglycans that activate Toll-like receptors (TLRs) expressed by innate immune cells, modulating cytokine production and influencing inflammatory responses [6];
- physical and functional maturation of the IS in the early years of life [7].
2. In Vivo Models for Studying the Microbiota-Immune System Axis
2.1. Vertebrates
- Rodents
- Rats
- Mice
- Guinea Pigs
- Rabbit (Oryctolagus cuniculus)
- Pigs
- Other Mammals
- Zebrafish (Danio rerio)
2.2. Invertebrates
- Galleria mellonella
- Caenorhabditis elegans
- Drosophila melanogaster
| Animal Model | Advantages | Disadvantages | Applications |
|---|---|---|---|
| Rats | Closer physiology to humans; larger size allows complex studies; available disease models; easy maintenance; great overlap with the human microbiome. | Different diet/living conditions from humans; less microbiota variability. | Gut dysbiosis [27,28,29,30,31]; dietary/pharmaceutical effects [27]; microbiota analysis across GI tract [33]; age-related changes [34] |
| Mice (M. musculus) | Anatomical/physiological similarity to humans; similar bacterial composition at the phylum level. | Dietary and anatomical differences affect microbiota; some unique bacteria (e.g., SFB, Deferribacteres) not found in humans. | Diet-pathogens interactions and genotype effects on GM [35]; comparative microbial analysis [15,48]; immune system maturation [45,46,47] |
| Guinea pigs (Cavia porcellus) | Intestinal E-cadherin similarity; comparable microbial phyla and immune responses to humans. | Limited research; genus-level microbiota differences. | Human diseases models [53]; microbiota-immune axis exploration [53] |
| Rabbit (Oryctolagus cuniculus) | Immune system similar to human; intermediate size; well-characterized gut microbiota. | Higher costs and maintenance. | Infectious diseases (e.g., syphilis, TB) [60]; immune system research [60,62,63]; GM studies across life stages [64,65,66] |
| Pigs | Similar size and GI structure to humans; well-characterized microbiota; stable human microbiota colonization; gnotobiotic models. | Size and cost; complex genetic manipulation. | GI physiology and immune ontogeny [70,71]; diet-induced obesity; human microbiota colonization and immune responses [75,76,77,79,80]; vaccine efficacy and enteric immunity [83] |
| Non-Human Primates | High physiological and immune similarity to humans; similar gut microbiota. | High cost and size; ethical considerations. | Immune responses studies [96]; host-microbiome interactions [97] |
| Dogs (Canis familiaris) | Similar GI structure and immune responses to humans; comparable chronic inflammatory conditions (e.g., IBD). | Unique immune traits; high cost; ethical concerns. | GM and immune responses research [87,89]; comparative immune system studies [90,91,92,93]. |
| Zebrafish (Danio rerio) | Genetic and structural parallels to humans; optical transparency; separate innate/adaptive immune systems. | Limited immune complexity; challenges in clinical translation. | Microbiota in immune system development [109,110,111,113]; SCFA and intestinal inflammation [114,115]. |
| Galleria mellonella | No ethical constraints; short life cycle; similar immune system to mammals | Differences in the adaptive immunity compared to mammals. | Pathogens virulence [116,125,126,127]; immune memory [129]; antimicrobial studies [131]; microbiota research [132,133]. |
| Caenorhabditis elegans (C. elegans) | Transparency for studies; simple nervous system; genetically tractable; conserved innate immunity. | Lack of adaptive immunity and mobile immune cells. | Microbiota-immunity interactions [134,139,140,141]; probiotic research [142,143,144]. |
| Drosophila melanogaster | Low maintenance; simple genetics; well-documented immune pathways; simple microbiota dominated by Proteobacteria and Firmicutes. | Lacks of adaptive immunity. | Pathogens immune response [149,156,157,158]; GM and immune signalling studies [166,167,168,170,171,172,173,174,175,176]. |
3. In Vitro Models
3.1. 2D Models
3.2. 3D Models
3.3. Microfluidic/On-Chip Models
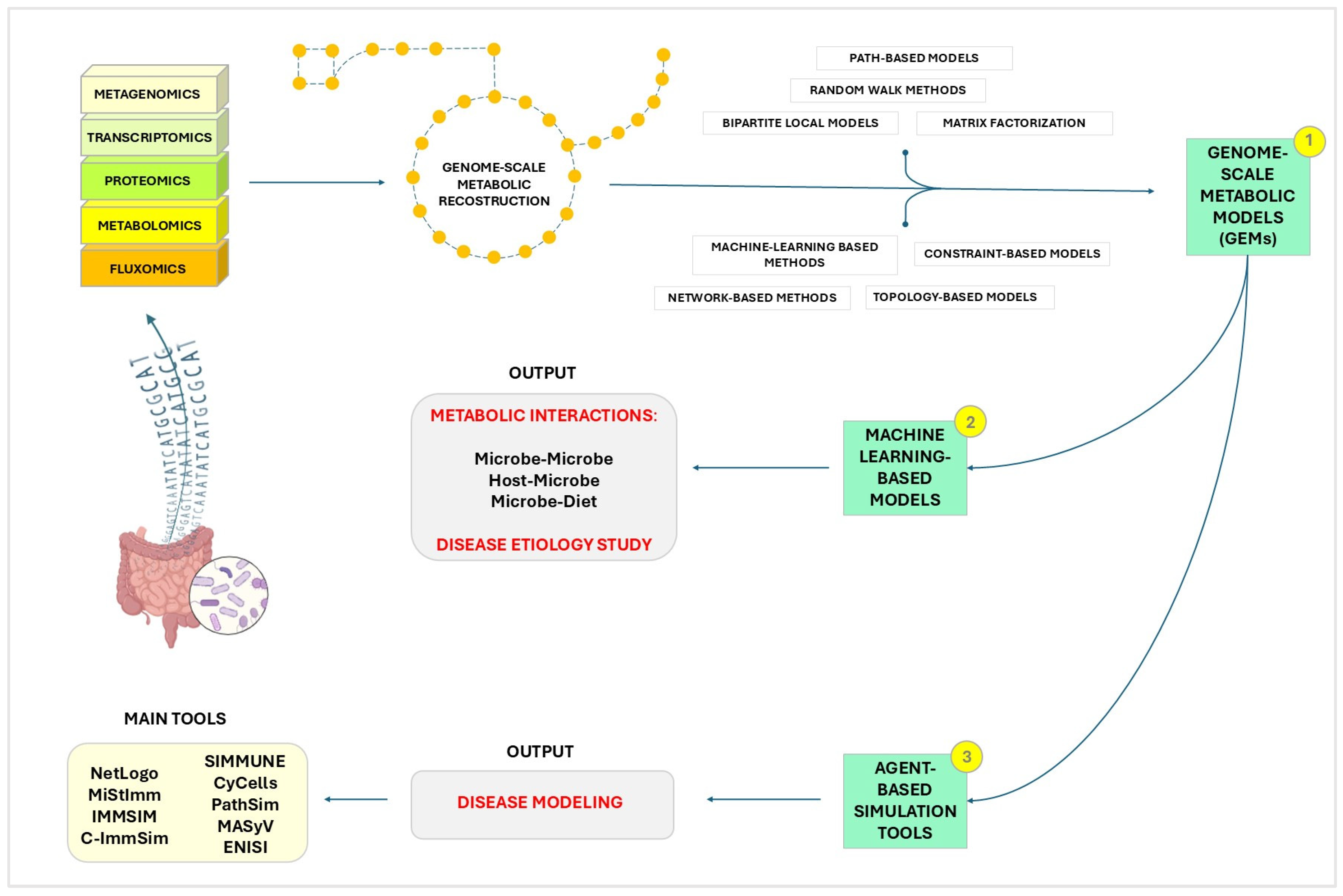
4. In Silico Models
- Simulation of Microbial Ecosystems: In-silico models allow scientists to simulate the complex GM ecosystem. By using computational techniques, researchers can model the growth, interaction, and metabolic processes of diverse microbial communities, providing insights that are challenging to obtain through traditional experimental methods [219,220].
- Immune system modulation: understanding how GM influences the host’s IS is crucial for developing therapeutic strategies. In-silico models can simulate immune responses to various stimuli, helping to identify potential targets for immunomodulation [223].
- Disease modeling: computational models are essential for exploring how disruptions in the GM contribute to inflammatory and autoimmune diseases [224]. They enable the identification of microbial signatures associated with these diseases and predict the effects of potential treatments.
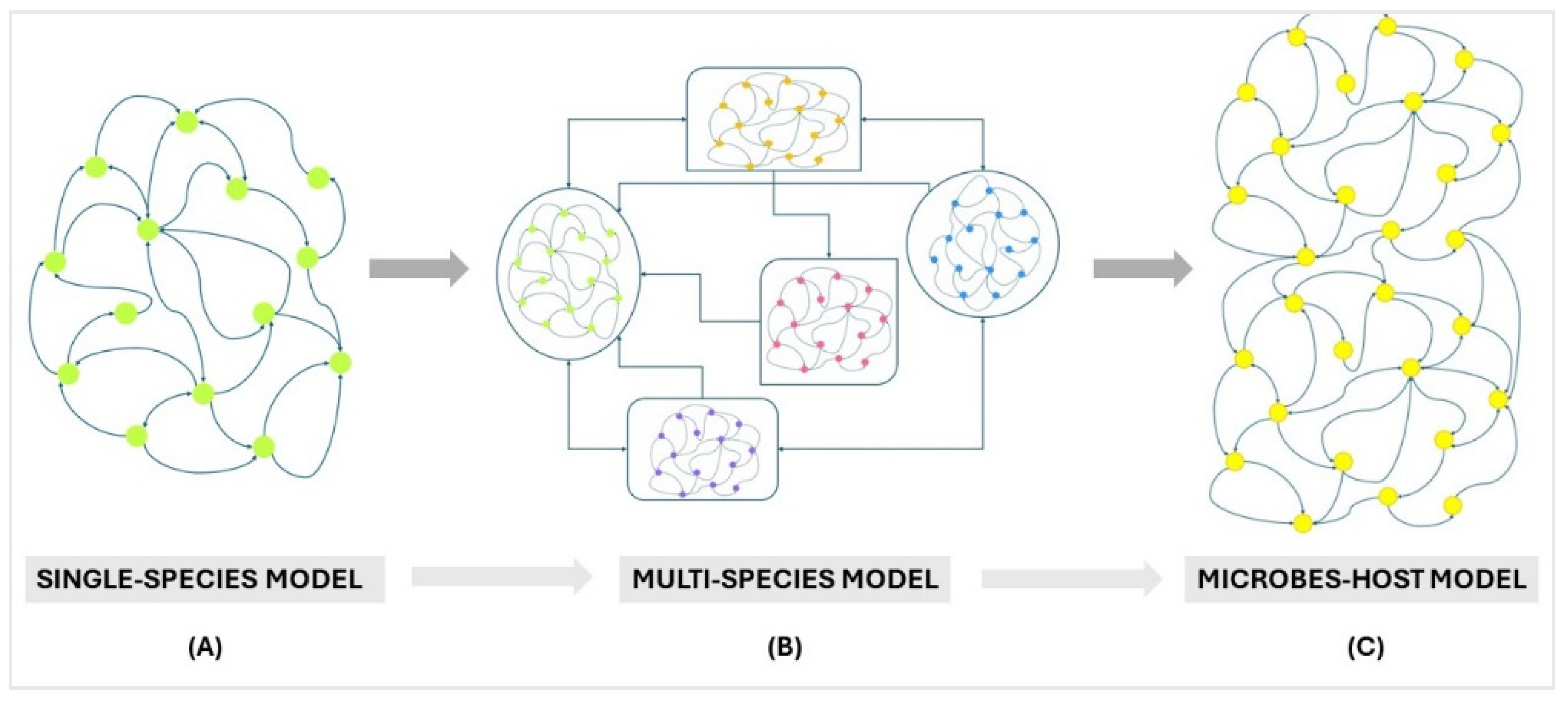
4.1. Multi-Species Ecosystem Models
4.2. Machine Learning-Based Models
4.3. Agent-Based Simulation Tools
5. Conclusions
6. Search Strategy from Repository
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sommer, F.; Bäckhed, F. The Gut Microbiota-Masters of Host Development and Physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Louis, P.; Hold, G.L.; Flint, H.J. The Gut Microbiota, Bacterial Metabolites and Colorectal Cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef] [PubMed]
- Rothhammer, V.; Mascanfroni, I.D.; Bunse, L.; Takenaka, M.C.; Kenison, J.E.; Mayo, L.; Chao, C.C.; Patel, B.; Yan, R.; Blain, M.; et al. Type i Interferons and Microbial Metabolites of Tryptophan Modulate Astrocyte Activity and Central Nervous System Inflammation via the Aryl Hydrocarbon Receptor. Nat. Med. 2016, 22, 586–597. [Google Scholar] [CrossRef] [PubMed]
- Michaudel, C.; Sokol, H. The Gut Microbiota at the Service of Immunometabolism. Cell Metab. 2020, 32, 514–523. [Google Scholar] [CrossRef]
- Kaur, H.; Ali, S.A.; Yan, F. Interactions between the Gut Microbiota-Derived Functional Factors and Intestinal Epithelial Cells—Implication in the Microbiota-Host Mutualism. Front. Immunol. 2022, 13, 1006081. [Google Scholar] [CrossRef]
- Semin, I.; Ninnemann, J.; Bondareva, M.; Gimaev, I.; Kruglov, A.A. Interplay Between Microbiota, Toll-Like Receptors and Cytokines for the Maintenance of Epithelial Barrier Integrity. Front. Med. 2021, 8, 644333. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Cui, H.L.; Tzianabos, A.O.; Kasper, D.L. An Immunomodulatory Molecule of Symbiotic Bacteria Directs Maturation of the Host Immune System. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef]
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in Health and Diseases. Signal Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, N.; Su, X.; Gao, Y.; Yang, R. Gut-Microbiota-Derived Metabolites Maintain Gut and Systemic Immune Homeostasis. Cells 2023, 12, 793. [Google Scholar] [CrossRef]
- Vandamme, T.F. Use of Rodents as Models of Human Diseases. J. Pharm. Bioallied Sci. 2014, 6, 2–9. [Google Scholar] [CrossRef]
- Hugenholtz, F.; de Vos, W.M. Mouse Models for Human Intestinal Microbiota Research: A Critical Evaluation. Cell Mol. Life Sci. 2018, 75, 149–160. [Google Scholar] [CrossRef]
- Faith, J.J.; Rey, F.E.; O’Donnell, D.; Karlsson, M.; McNulty, N.P.; Kallstrom, G.; Goodman, A.L.; Gordon, J.I. Creating and Characterizing Communities of Human Gut Microbes in Gnotobiotic Mice. ISME J. 2010, 4, 1094–1098. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Liu, M.; Chan, X.Y.; Tan, S.Y.; Subramaniam, S.; Fan, Y.; Loh, E.; Tou, K.; Chang, E.; Tan, T.C.; et al. Uncovering the Mystery of Opposite Circadian Rhythms between Mouse and Human Leukocytes in Humanized Mice. Blood 2017, 130, 1995–2005. [Google Scholar] [CrossRef]
- Dijk, D.-J.; Archer, S.N. Light, Sleep, and Circadian Rhythms: Together again. PLoS Biol. 2009, 7, e1000145. [Google Scholar] [CrossRef]
- Nguyen, T.L.A.; Vieira-Silva, S.; Liston, A.; Raes, J. How Informative Is the Mouse for Human Gut Microbiota Research? Dis. Model. Mech. 2015, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Nagpal, R.; Wang, S.; Solberg Woods, L.C.; Seshie, O.; Chung, S.T.; Shively, C.A.; Register, T.C.; Craft, S.; McClain, D.A.; Yadav, H. Comparative Microbiome Signatures and Short-Chain Fatty Acids in Mouse, Rat, Non-Human Primate, and Human Feces. Front. Microbiol. 2018, 9, 2897. [Google Scholar] [CrossRef] [PubMed]
- Franklin, C.L.; Ericsson, A.C. Microbiota and Reproducibility of Rodent Models. Lab. Anim. 2017, 46, 114–122. [Google Scholar] [CrossRef]
- Fritz, J.V.; Desai, M.S.; Shah, P.; Schneider, J.G.; Wilmes, P. From Meta-Omics to Causality: Experimental Models for Human Microbiome Research. Microbiome 2013, 1, 14. [Google Scholar] [CrossRef]
- Brower, M.; Grace, M.; Kotz, C.M.; Koya, V. Comparative Analysis of Growth Characteristics of Sprague Dawley Rats Obtained from Different Sources. Lab. Anim. Res. 2015, 31, 166–173. [Google Scholar] [CrossRef]
- Prodinger, P.M.; Bürklein, D.; Foehr, P.; Kreutzer, K.; Pilge, H.; Schmitt, A.; Eisenhart-Rothe, R.V.; Burgkart, R.; Bissinger, O.; Tischer, T. Improving Results in Rat Fracture Models: Enhancing the Efficacy of Biomechanical Testing by a Modification of the Experimental Setup. BMC Musculoskelet. Disord. 2018, 19, 243. [Google Scholar] [CrossRef]
- Singhal, A.; Aliouat, E.M.; Hervé, M.; Mathys, V.; Kiass, M.; Creusy, C.; Delaire, B.; Tsenova, L.; Fleurisse, L.; Bertout, J.; et al. Experimental Tuberculosis in the Wistar Rat: A Model for Protective Immunity and Control of Infection. PLoS ONE 2011, 6, e18632. [Google Scholar] [CrossRef] [PubMed]
- Maronpot, R.R.; Nyska, A.; Foreman, J.E.; Ramot, Y. The Legacy of the F344 Rat as a Cancer Bioassay Model (a Retrospective Summary of Three Common F344 Rat Neoplasms). Crit. Rev. Toxicol. 2016, 46, 641–675. [Google Scholar] [CrossRef]
- Cadoni, C. Fischer 344 and Lewis Rat Strains as a Model of Genetic Vulnerability to Drug Addiction. Front. Neurosci. 2016, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Coppens, C.M.; de Boer, S.F.; Buwalda, B.; Koolhaas, J.M. Aggression and Aspects of Impulsivity in Wild-Type Rats. Aggress. Behav. 2014, 40, 300–308. [Google Scholar] [CrossRef] [PubMed]
- Vidal, J.; Buwalda, B.; Koolhaas, J.M. Male Wistar Rats Are More Susceptible to Lasting Social Anxiety than Wild-Type Groningen Rats Following Social Defeat Stress during Adolescence. Behav. Process. 2011, 88, 76–80. [Google Scholar] [CrossRef] [PubMed]
- van den Brandt, J.; Fischer, H.J.; Walter, L.; Hünig, T.; Klöting, I.; Reichardt, H.M. Type 1 Diabetes in BioBreeding Rats Is Critically Linked to an Imbalance between Th17 and Regulatory T Cells and an Altered TCR Repertoire. J. Immunol. 2010, 185, 2285–2294. [Google Scholar] [CrossRef] [PubMed]
- Rose, W.A.; Sakamoto, K.; Leifer, C.A. Multifunctional Role of Dextran Sulfate Sodium for In Vivo Modeling of Intestinal Diseases. BMC Immunol. 2012, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Garg, A.; Garg, S.; Singh, V. Animal Model Used for Experimental Study of Diabetes Mellitus: An Overview Review Article Animal Model Used for Experimental Study of Diabetes Mellitus: An Overview. Asian J. Biomater. Res. 2016, 2, 99–110. [Google Scholar]
- Qi, Z.; Lyu, M.; Yang, L.; Yuan, H.; Cao, Y.; Zhai, L.; Dang, W.; Liu, J.; Yang, F.; Li, Y. A Novel and Reliable Rat Model of Autism. Front. Psychiatry 2021, 12, 549810. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, M.M.; Campos, F.L.; Coimbra, B.; Pêgo, J.M.; Rodrigues, C.; Lima, R.; Rodrigues, A.J.; Sousa, N.; Salgado, A.J. Behavioral Characterization of the 6-Hydroxidopamine Model of Parkinson’s Disease and Pharmacological Rescuing of Non-Motor Deficits. Mol. Neurodegener. 2013, 8, 14. [Google Scholar] [CrossRef]
- Miao, M.; Peng, M.; Chen, H.; Liu, B. Effects of Baihe Dihuang Powder on Chronic Stress Depression Rat Models. Saudi J. Biol. Sci. 2019, 26, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Guo, R.; Zhu, J.; Wang, Q.; Ju, Y.; Xie, Y.; Zheng, Y.; Wang, Z.; Li, T.; Liu, Z.; et al. A Gene Catalogue of the Sprague-Dawley Rat Gut Metagenome. Gigascience 2018, 7, giy055. [Google Scholar] [CrossRef]
- Li, D.; Chen, H.; Mao, B.; Yang, Q.; Zhao, J.; Gu, Z.; Zhang, H.; Chen, Y.Q.; Chen, W. Microbial Biogeography and Core Microbiota of the Rat Digestive Tract. Sci. Rep. 2017, 8, 45840. [Google Scholar] [CrossRef] [PubMed]
- Flemer, B.; Gaci, N.; Borrel, G.; Sanderson, I.R.; Chaudhary, P.P.; Tottey, W.; O’Toole, P.W.; Brugère, J.-F. Fecal Microbiota Variation across the Lifespan of the Healthy Laboratory Rat. Gut Microbes 2017, 8, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Beck, J.A.; Lloyd, S.; Hafezparast, M.; Lennon-Pierce, M.; Eppig, J.T.; Festing, M.F.; Fisher, E.M. Genealogies of Mouse Inbred Strains. Nat. Genet. 2000, 24, 23–25. [Google Scholar] [CrossRef]
- Ghoshal, N.G.; Bal, H.S. Comparative Morphology of the Stomach of Some Laboratory Mammals. Lab. Anim. 1989, 23, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Tannock, G.W.; Tangerman, A.; Van Schaik, A.; McConnell, M.A. Deconjugation of Bile Acids by Lactobacilli in the Mouse Small Bowel. Appl. Env. Microbiol. 1994, 60, 3419–3420. [Google Scholar] [CrossRef]
- Benson, A.K.; Kelly, S.A.; Legge, R.; Ma, F.; Low, S.J.; Kim, J.; Zhang, M.; Oh, P.L.; Nehrenberg, D.; Hua, K.; et al. Individuality in Gut Microbiota Composition Is a Complex Polygenic Trait Shaped by Multiple Environmental and Host Genetic Factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18933–18938. [Google Scholar] [CrossRef]
- Walter, J. Ecological Role of Lactobacilli in the Gastrointestinal Tract: Implications for Fundamental and Biomedical Research. Appl. Environ. Microbiol. 2008, 74, 4985–4996. [Google Scholar] [CrossRef] [PubMed]
- Frese, S.A.; Benson, A.K.; Tannock, G.W.; Loach, D.M.; Kim, J.; Zhang, M.; Oh, P.L.; Heng, N.C.K.; Patil, P.B.; Juge, N.; et al. The Evolution of Host Specialization in the Vertebrate Gut Symbiont Lactobacillus Reuteri. PLoS Genet. 2011, 7, e1001314. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial Ecology: Human Gut Microbes Associated with Obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Rawls, J.F.; Mahowald, M.A.; Ley, R.E.; Gordon, J.I. Reciprocal Gut Microbiota Transplants from Zebrafish and Mice to Germ-Free Recipients Reveal Host Habitat Selection. Cell 2006, 127, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Bik, E.M.; Eckburg, P.B.; Gill, S.R.; Nelson, K.E.; Purdom, E.A.; Francois, F.; Perez-Perez, G.; Blaser, M.J.; Relman, D.A. Molecular Analysis of the Bacterial Microbiota in the Human Stomach. Proc. Natl. Acad. Sci. USA 2006, 103, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Bolotin, A.; de Wouters, T.; Schnupf, P.; Bouchier, C.; Loux, V.; Rhimi, M.; Jamet, A.; Dervyn, R.; Boudebbouze, S.; Blottière, H.M.; et al. Genome Sequence of “Candidatus Arthromitus” Sp. Strain SFB-Mouse-NL, a Commensal Bacterium with a Key Role in Postnatal Maturation of Gut Immune Functions. Genome Announc. 2014, 2, e00705-14. [Google Scholar] [CrossRef]
- Suzuki, K.; Meek, B.; Doi, Y.; Muramatsu, M.; Chiba, T.; Honjo, T.; Fagarasan, S. Aberrant Expansion of Segmented Filamentous Bacteria in IgA-Deficient Gut. Proc. Natl. Acad. Sci. USA 2004, 101, 1981–1986. [Google Scholar] [CrossRef]
- Gaboriau-Routhiau, V.; Rakotobe, S.; Lécuyer, E.; Mulder, I.; Lan, A.; Bridonneau, C.; Rochet, V.; Pisi, A.; De Paepe, M.; Brandi, G.; et al. The Key Role of Segmented Filamentous Bacteria in the Coordinated Maturation of Gut Helper T Cell Responses. Immunity 2009, 31, 677–689. [Google Scholar] [CrossRef]
- Ivanov, I.I.; Atarashi, K.; Manel, N.; Brodie, E.L.; Shima, T.; Karaoz, U.; Wei, D.; Goldfarb, K.C.; Santee, C.A.; Lynch, S.V.; et al. Induction of Intestinal Th17 Cells by Segmented Filamentous Bacteria. Cell 2009, 139, 485–498. [Google Scholar] [CrossRef]
- Krych, L.; Hansen, C.H.F.; Hansen, A.K.; van den Berg, F.W.J.; Nielsen, D.S. Quantitatively Different, yet Qualitatively Alike: A Meta-Analysis of the Mouse Core Gut Microbiome with a View towards the Human Gut Microbiome. PLoS ONE 2013, 8, e62578. [Google Scholar] [CrossRef]
- Hildebrand, F.; Nguyen, T.L.A.; Brinkman, B.; Yunta, R.G.; Cauwe, B.; Vandenabeele, P.; Liston, A.; Raes, J. Inflammation-Associated Enterotypes, Host Genotype, Cage and Inter-Individual Effects Drive Gut Microbiota Variation in Common Laboratory Mice. Genome Biol. 2013, 14, R4. [Google Scholar] [CrossRef]
- Sakaguchi, E.; Nabata, A. Comparison of Fibre Digestion and Digesta Retention Time between Nutrias (Myocaster coypus) and Guinea-Pigs (Cavia porcellus). Comp. Biochem. Physiol. Comp. Physiol. 1992, 103, 601–604. [Google Scholar] [CrossRef]
- Hildebrand, F.; Ebersbach, T.; Nielsen, H.B.; Li, X.; Sonne, S.B.; Bertalan, M.; Dimitrov, P.; Madsen, L.; Qin, J.; Wang, J.; et al. A Comparative Analysis of the Intestinal Metagenomes Present in Guinea Pigs (Cavia porcellus) and Humans (Homo sapiens). BMC Genom. 2012, 13, 514. [Google Scholar] [CrossRef]
- Crowley, E.J.; King, J.M.; Wilkinson, T.; Worgan, H.J.; Huson, K.M.; Rose, M.T.; McEwan, N.R. Comparison of the Microbial Population in Rabbits and Guinea Pigs by next Generation Sequencing. PLoS ONE 2017, 12, e0165779. [Google Scholar] [CrossRef] [PubMed]
- Padilla-Carlin, D.J.; McMurray, D.N.; Hickey, A.J. The Guinea Pig as a Model of Infectious Diseases. Comp. Med. 2008, 58, 324–340. [Google Scholar] [PubMed]
- Owen, E.C.; West, D.W.; Coates, M.E. Metabolism of Riboflavine in Germ-Free and Conventional Rabbits. Br. J. Nutr. 1970, 24, 259–267. [Google Scholar] [CrossRef]
- Webb, D.R. Animal Models of Human Disease: Inflammation. Biochem. Pharmacol. 2014, 87, 121–130. [Google Scholar] [CrossRef]
- De, S.N.; Chatterje, D.N. An Experimental Study of the Mechanism of Action of Vibrio Cholerae on the Intestinal Mucous Membrane. J. Pathol. Bacteriol. 1953, 66, 559–562. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.; Maltby, M.P.; Payne, J.M. Factors Influencing the Response of Ligated Rabbit-Gut Segments to Injected Escherichia Coli. J. Pathol. Bacteriol. 1958, 76, 491–499. [Google Scholar] [CrossRef]
- Duncan, C.L.; Sugiyama, H.; Strong, D.H. Rabbit Ileal Loop Response to Strains of Clostridium Perfringens. J. Bacteriol. 1968, 95, 1560–1566. [Google Scholar] [CrossRef]
- Arm, H.G.; Floyd, T.M.; Faber, J.E.; Hayes, J.R. Use of ligated segments of rabbit small intestine in experimental shigellosis. J. Bacteriol. 1965, 89, 803–809. [Google Scholar] [CrossRef]
- Esteves, P.J.; Abrantes, J.; Baldauf, H.-M.; BenMohamed, L.; Chen, Y.; Christensen, N.; González-Gallego, J.; Giacani, L.; Hu, J.; Kaplan, G.; et al. The Wide Utility of Rabbits as Models of Human Diseases. Exp. Mol. Med. 2018, 50, 1–10. [Google Scholar] [CrossRef]
- Jiménez-García, A.; Balongo-García, R.; Alconero, F.F.; Araji, O.A.; Martínez, G.J.; Haba, M.G.; Morales, L.C.; Beviá, J.M.O.; Martínez, J.C. Intestinal Wall Damage in Simple Ileus in Rabbits: Immune-Modulator Role of Somatostatin. Hepatogastroenterology 2004, 51, 1030–1036. [Google Scholar]
- Neves, F.; Abrantes, J.; Almeida, T.; de Matos, A.L.; Costa, P.P.; Esteves, P.J. Genetic Characterization of Interleukins (IL-1α, IL-1β, IL-2, IL-4, IL-8, IL-10, IL-12A, IL-12B, IL-15 and IL-18) with Relevant Biological Roles in Lagomorphs. Innate Immun. 2015, 21, 787–801. [Google Scholar] [CrossRef]
- Perkins, H.D.; van Leeuwen, B.H.; Hardy, C.M.; Kerr, P.J. The Complete CDNA Sequences of IL-2, IL-4, IL-6 AND IL-10 from the European Rabbit (Oryctolagus cuniculus). Cytokine 2000, 12, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Abecia, L.; Fondevila, M.; Balcells, J.; McEwan, N.R. The Effect of Lactating Rabbit Does on the Development of the Caecal Microbial Community in the Pups They Nurture. J. Appl. Microbiol. 2007, 103, 557–564. [Google Scholar] [CrossRef] [PubMed]
- Combes, S.; Michelland, R.J.; Monteils, V.; Cauquil, L.; Soulié, V.; Tran, N.U.; Gidenne, T.; Fortun-Lamothe, L. Postnatal Development of the Rabbit Caecal Microbiota Composition and Activity. FEMS Microbiol. Ecol. 2011, 77, 680–689. [Google Scholar] [CrossRef] [PubMed]
- Cotozzolo, E.; Cremonesi, P.; Curone, G.; Menchetti, L.; Riva, F.; Biscarini, F.; Marongiu, M.L.; Castrica, M.; Castiglioni, B.; Miraglia, D.; et al. Characterization of Bacterial Microbiota Composition along the Gastrointestinal Tract in Rabbits. Animals 2021, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Monteils, V.; Cauquil, L.; Combes, S.; Godon, J.-J.; Gidenne, T. Potential Core Species and Satellite Species in the Bacterial Community within the Rabbit Caecum. FEMS Microbiol. Ecol. 2008, 66, 620–629. [Google Scholar] [CrossRef]
- Gutierrez, K.; Dicks, N.; Glanzner, W.G.; Agellon, L.B.; Bordignon, V. Efficacy of the Porcine Species in Biomedical Research. Front. Genet. 2015, 6, 293. [Google Scholar] [CrossRef]
- Köhn, F. The Minipig in Biomedical Research; CRC Press: Boca Raton, FL, USA, 2012. [Google Scholar]
- Ryu, J.; Prather, R.S.; Lee, K. Use of Gene-Editing Technology to Introduce Targeted Modifications in Pigs. J. Anim. Sci. Biotechnol. 2018, 9, 5. [Google Scholar] [CrossRef]
- Sinkora, M.; Butler, J.E. Progress in the Use of Swine in Developmental Immunology of B and T Lymphocytes. Dev. Comp. Immunol. 2016, 58, 1–17. [Google Scholar] [CrossRef]
- Pedersen, R.; Ingerslev, H.-C.; Sturek, M.; Alloosh, M.; Cirera, S.; Christoffersen, B.Ø.; Moesgaard, S.G.; Larsen, N.; Boye, M. Characterisation of Gut Microbiota in Ossabaw and Göttingen Minipigs as Models of Obesity and Metabolic Syndrome. PLoS ONE 2013, 8, e56612. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An Obesity-Associated Gut Microbiome with Increased Capacity for Energy Harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Furet, J.-P.; Firmesse, O.; Gourmelon, M.; Bridonneau, C.; Tap, J.; Mondot, S.; Doré, J.; Corthier, G. Comparative Assessment of Human and Farm Animal Faecal Microbiota Using Real-Time Quantitative PCR. FEMS Microbiol. Ecol. 2009, 68, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, R.; Andersen, A.D.; Mølbak, L.; Stagsted, J.; Boye, M. Changes in the Gut Microbiota of Cloned and Non-Cloned Control Pigs during Development of Obesity: Gut Microbiota during Development of Obesity in Cloned Pigs. BMC Microbiol. 2013, 13, 30. [Google Scholar] [CrossRef]
- Wang, M.; Donovan, S.M. Human Microbiota-Associated Swine: Current Progress and Future Opportunities. ILAR J. 2015, 56, 63–73. [Google Scholar] [CrossRef]
- Chung, H.; Pamp, S.J.; Hill, J.A.; Surana, N.K.; Edelman, S.M.; Troy, E.B.; Reading, N.C.; Villablanca, E.J.; Wang, S.; Mora, J.R.; et al. Gut Immune Maturation Depends on Colonization with a Host-Specific Microbiota. Cell 2012, 149, 1578–1593. [Google Scholar] [CrossRef]
- Pang, X.; Hua, X.; Yang, Q.; Ding, D.; Che, C.; Cui, L.; Jia, W.; Bucheli, P.; Zhao, L. Inter-Species Transplantation of Gut Microbiota from Human to Pigs. ISME J. 2007, 1, 156–162. [Google Scholar] [CrossRef]
- Che, C.; Pang, X.; Hua, X.; Zhang, B.; Shen, J.; Zhu, J.; Wei, H.; Sun, L.; Chen, P.; Cui, L.; et al. Effects of Human Fecal Flora on Intestinal Morphology and Mucosal Immunity in Human Flora-Associated Piglet. Scand. J. Immunol. 2009, 69, 223–233. [Google Scholar] [CrossRef]
- Sang, Y.; Ruchala, P.; Lehrer, R.I.; Ross, C.R.; Rowland, R.R.R.; Blecha, F. Antimicrobial Host Defense Peptides in an Arteriviral Infection: Differential Peptide Expression and Virus Inactivation. Viral Immunol. 2009, 22, 235–242. [Google Scholar] [CrossRef]
- Dawson, H.D. A Comparative Assessment of the Pig, Mouse and Human Genomes. Minipig Biomed. Res. 2011, 1, 323–342. [Google Scholar]
- Kapetanovic, R.; Fairbairn, L.; Beraldi, D.; Sester, D.P.; Archibald, A.L.; Tuggle, C.K.; Hume, D.A. Pig Bone Marrow-Derived Macrophages Resemble Human Macrophages in Their Response to Bacterial Lipopolysaccharide. J. Immunol. 2012, 188, 3382–3394. [Google Scholar] [CrossRef]
- Twitchell, E.L.; Tin, C.; Wen, K.; Zhang, H.; Becker-Dreps, S.; Azcarate-Peril, M.A.; Vilchez, S.; Li, G.; Ramesh, A.; Weiss, M.; et al. Modeling Human Enteric Dysbiosis and Rotavirus Immunity in Gnotobiotic Pigs. Gut Pathog. 2016, 8, 51. [Google Scholar] [CrossRef]
- Meurens, F.; Summerfield, A.; Nauwynck, H.; Saif, L.; Gerdts, V. The Pig: A Model for Human Infectious Diseases. Trends Microbiol. 2012, 20, 50–57. [Google Scholar] [CrossRef]
- Fairbairn, L.; Kapetanovic, R.; Sester, D.P.; Hume, D.A. The Mononuclear Phagocyte System of the Pig as a Model for Understanding Human Innate Immunity and Disease. J. Leukoc. Biol. 2011, 89, 855–871. [Google Scholar] [CrossRef]
- Song, S.J.; Lauber, C.; Costello, E.K.; Lozupone, C.A.; Humphrey, G.; Berg-Lyons, D.; Caporaso, J.G.; Knights, D.; Clemente, J.C.; Nakielny, S.; et al. Cohabiting Family Members Share Microbiota with One Another and with Their Dogs. eLife 2013, 2, e00458. [Google Scholar] [CrossRef] [PubMed]
- Swanson, K.S.; Dowd, S.E.; Suchodolski, J.S.; Middelbos, I.S.; Vester, B.M.; Barry, K.A.; Nelson, K.E.; Torralba, M.; Henrissat, B.; Coutinho, P.M.; et al. Phylogenetic and Gene-Centric Metagenomics of the Canine Intestinal Microbiome Reveals Similarities with Humans and Mice. ISME J. 2011, 5, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Ericsson, A.C. The Use of Non-Rodent Model Species in Microbiota Studies. Lab. Anim. 2019, 53, 259–270. [Google Scholar] [CrossRef]
- Vázquez-Baeza, Y.; Hyde, E.R.; Suchodolski, J.S.; Knight, R. Dog and Human Inflammatory Bowel Disease Rely on Overlapping yet Distinct Dysbiosis Networks. Nat. Microbiol. 2016, 1, 16177. [Google Scholar] [CrossRef] [PubMed]
- Foltz, J.A.; Somanchi, S.S.; Yang, Y.; Aquino-Lopez, A.; Bishop, E.E.; Lee, D.A. NCR1 Expression Identifies Canine Natural Killer Cell Subsets with Phenotypic Similarity to Human Natural Killer Cells. Front. Immunol. 2016, 7, 521. [Google Scholar] [CrossRef]
- Graves, S.S.; Gyurkocza, B.; Stone, D.M.; Parker, M.H.; Abrams, K.; Jochum, C.; Gallo, S.; Saad, M.; Johnson, M.M.; Rosinski, S.L.; et al. Development and Characterization of a Canine-Specific Anti-CD94 (KLRD-1) Monoclonal Antibody. Vet. Immunol. Immunopathol. 2019, 211, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Bergeron, L.M.; McCandless, E.E.; Dunham, S.; Dunkle, B.; Zhu, Y.; Shelly, J.; Lightle, S.; Gonzales, A.; Bainbridge, G. Comparative Functional Characterization of Canine IgG Subclasses. Vet. Immunol. Immunopathol. 2014, 157, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-H.; Shin, D.-J.; Kim, Y.; Kim, C.-J.; Lee, J.-J.; Yoon, M.S.; Uong, T.N.T.; Yu, D.; Jung, J.-Y.; Cho, D.; et al. Comparison of Phenotypic and Functional Characteristics Between Canine Non-B, Non-T Natural Killer Lymphocytes and CD3+CD5dimCD21− Cytotoxic Large Granular Lymphocytes. Front. Immunol. 2018, 9, 841. [Google Scholar] [CrossRef]
- Dow, S. A Role for Dogs in Advancing Cancer Immunotherapy Research. Front. Immunol. 2020, 10, 2935. [Google Scholar] [CrossRef] [PubMed]
- Davenport, E.R.; Sanders, J.G.; Song, S.J.; Amato, K.R.; Clark, A.G.; Knight, R. The Human Microbiome in Evolution. BMC Biol. 2017, 15, 127. [Google Scholar] [CrossRef] [PubMed]
- Buse, E. Development of the Immune System in the Cynomolgus Monkey: The Appropriate Model in Human Targeted Toxicology? J. Immunotoxicol. 2005, 2, 211–216. [Google Scholar] [CrossRef]
- Li, H.-Z.; Li, N.; Wang, J.-J.; Li, H.; Huang, X.; Guo, L.; Zheng, H.-W.; He, Z.-L.; Zhao, Y.; Yang, Z.-N.; et al. Dysbiosis of Gut Microbiome Affecting Small Intestine Morphology and Immune Balance: A Rhesus Macaque Model. Zool. Res. 2020, 41, 20–31. [Google Scholar] [CrossRef]
- Harding, J.D. Nonhuman Primates and Translational Research: Progress, Opportunities, and Challenges. ILAR J. 2017, 58, 141–150. [Google Scholar] [CrossRef]
- Phillips, K.A.; Bales, K.L.; Capitanio, J.P.; Conley, A.; Czoty, P.W.; ’t Hart, B.A.; Hopkins, W.D.; Hu, S.-L.; Miller, L.A.; Nader, M.A.; et al. Why Primate Models Matter. Am. J. Primatol. 2014, 76, 801–827. [Google Scholar] [CrossRef]
- Parichy, D.M. The Natural History of Model Organisms: Advancing Biology through a Deeper Understanding of Zebrafish Ecology and Evolution. eLife 2015, 4, e05635. [Google Scholar] [CrossRef]
- Yossa, R.; Sarker, P.K.; Vandenberg, G.W. Preliminary Evidence of the Contribution of the Intestinal Microflora to Biotin Supply in Zebrafish Danio rerio (Hamilton-Buchanan). Zebrafish 2011, 8, 221–227. [Google Scholar] [CrossRef]
- Gonzales, J.M.; Law, S.H.W. Feed and Feeding Regime Affect Growth Rate and Gonadosomatic Index of Adult Zebrafish (Danio Rerio). Zebrafish 2013, 10, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Sathkumara, H.D.; Eaton, J.L.; Field, M.A.; Govan, B.L.; Ketheesan, N.; Kupz, A. A Murine Model of Tuberculosis/Type 2 Diabetes Comorbidity for Investigating the Microbiome, Metabolome and Associated Immune Parameters. Anim. Model. Exp. Med. 2021, 4, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The Zebrafish Reference Genome Sequence and Its Relationship to the Human Genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef]
- Lickwar, C.R.; Camp, J.G.; Weiser, M.; Cocchiaro, J.L.; Kingsley, D.M.; Furey, T.S.; Sheikh, S.Z.; Rawls, J.F. Genomic Dissection of Conserved Transcriptional Regulation in Intestinal Epithelial Cells. PLoS Biol. 2017, 15, e2002054. [Google Scholar] [CrossRef] [PubMed]
- Meijer, A.H.; van der Vaart, M.; Spaink, H.P. Real-Time Imaging and Genetic Dissection of Host-Microbe Interactions in Zebrafish. Cell Microbiol. 2014, 16, 39–49. [Google Scholar] [CrossRef]
- Trede, N.S.; Langenau, D.M.; Traver, D.; Look, A.T.; Zon, L.I. The Use of Zebrafish to Understand Immunity. Immunity 2004, 20, 367–379. [Google Scholar] [CrossRef]
- Page, D.M.; Wittamer, V.; Bertrand, J.Y.; Lewis, K.L.; Pratt, D.N.; Delgado, N.; Schale, S.E.; Mcgue, C.; Jacobsen, B.H.; Doty, A.; et al. An Evolutionarily Conserved Program of B-Cell Development and Activation in Zebrafish. Blood 2013, 122, e1–e11. [Google Scholar] [CrossRef]
- Murdoch, C.C.; Rawls, J.F. Commensal Microbiota Regulate Vertebrate Innate Immunity-Insights from the Zebrafish. Front. Immunol. 2019, 10, 2100. [Google Scholar] [CrossRef]
- Masud, S.; Torraca, V.; Meijer, A.H. Modeling Infectious Diseases in the Context of a Developing Immune System. In Current Topics in Developmental Biology; Academic Press Inc.: Cambridge, MA, USA, 2017; Volume 124, pp. 277–329. [Google Scholar]
- Bates, J.M.; Akerlund, J.; Mittge, E.; Guillemin, K. Intestinal Alkaline Phosphatase Detoxifies Lipopolysaccharide and Prevents Inflammation in Zebrafish in Response to the Gut Microbiota. Cell Host Microbe 2007, 2, 371–382. [Google Scholar] [CrossRef]
- Murdoch, C.C.; Espenschied, S.T.; Matty, M.A.; Mueller, O.; Tobin, D.M.; Rawls, J.F. Intestinal Serum Amyloid a Suppresses Systemic Neutrophil Activation and Bactericidal Activity in Response to Microbiota Colonization. PLoS Pathog. 2019, 15, e1007381. [Google Scholar] [CrossRef]
- Brugman, S.; Schneeberger, K.; Witte, M.; Klein, M.R.; van den Bogert, B.; Boekhorst, J.; Timmerman, H.M.; Boes, M.L.; Kleerebezem, M.; Nieuwenhuis, E.E.S. T Lymphocytes Control Microbial Composition by Regulating the Abundance of Vibrio in the Zebrafish Gut. Gut Microbes 2014, 5, 737–747. [Google Scholar] [CrossRef] [PubMed]
- Cholan, P.M.; Han, A.; Woodie, B.R.; Watchon, M.; Kurz, A.R.M.; Laird, A.S.; Britton, W.J.; Ye, L.; Holmes, Z.C.; McCann, J.R.; et al. Conserved Anti-Inflammatory Effects and Sensing of Butyrate in Zebrafish. Gut Microbes 2020, 12, 1824563. [Google Scholar] [CrossRef] [PubMed]
- Morales Fénero, C.; Amaral, M.A.; Xavier, I.K.; Padovani, B.N.; Paredes, L.C.; Takiishi, T.; Lopes-Ferreira, M.; Lima, C.; Colombo, A.; Saraiva Câmara, N.O. Short Chain Fatty Acids (SCFAs) Improves TNBS-Induced Colitis in Zebrafish. Curr. Res. Immunol. 2021, 2, 142–154. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.J.Y.; Loh, J.M.S.; Proft, T. Galleria mellonella Infection Models for the Study of Bacterial Diseases and for Antimicrobial Drug Testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef]
- Kavanagh, K.; Fallon, J.P. Galleria mellonella Larvae as Models for Studying Fungal Virulence. Fungal Biol. Rev. 2010, 24, 79–83. [Google Scholar] [CrossRef]
- Champion, O.L.; Wagley, S.; Titball, R.W. Galleria mellonella as a Model Host for Microbiological and Toxin Research. Virulence 2016, 7, 840–845. [Google Scholar] [CrossRef]
- Binder, U.; Maurer, E.; Lass-Flörl, C. Galleria mellonella: An Invertebrate Model to Study Pathogenicity in Correctly Defined Fungal Species. Fungal Biol. 2016, 120, 288–295. [Google Scholar] [CrossRef]
- Wittwer, D.; Franchini, A.; Ottaviani, E.; Wiesner, A. Presence of IL-1- and TNF-like Molecules in Galleria mellonella (Lepidoptera) Haemocytes and in an Insect Cell Line from Estigmene acraea (Lepidoptera). Cytokine 1999, 11, 637–642. [Google Scholar] [CrossRef]
- Browne, N.; Heelan, M.; Kavanagh, K. An Analysis of the Structural and Functional Similarities of Insect Hemocytes and Mammalian Phagocytes. Virulence 2013, 4, 597–603. [Google Scholar] [CrossRef]
- Lavine, M.D.; Strand, M.R. Insect Hemocytes and Their Role in Immunity. Insect Biochem. Mol. Biol. 2002, 32, 1295–1309. [Google Scholar] [CrossRef]
- Pech, L.L. Strand MR Granular Cells Are Required for Encapsulation of Foreign Targets by Insect Haemocytes. J. Cell Sci. 1996, 109, 2053–2060. [Google Scholar] [CrossRef]
- Kavanagh, K.; Reeves, E.P. Exploiting the Potential of Insects for in Vivo Pathogenicity Testing of Microbial Pathogens. FEMS Microbiol. Rev. 2004, 28, 101–112. [Google Scholar] [CrossRef]
- Bolouri Moghaddam, M.R.; Tonk, M.; Schreiber, C.; Salzig, D.; Czermak, P.; Vilcinskas, A.; Rahnamaeian, M. The Potential of the Galleria mellonella Innate Immune System Is Maximized by the Co-Presentation of Diverse Antimicrobial Peptides. Biol. Chem. 2016, 397, 939–945. [Google Scholar] [CrossRef]
- Bergin, D.; Reeves, E.P.; Renwick, J.; Wientjes, F.B.; Kavanagh, K. Superoxide Production in Galleria mellonella Hemocytes: Identification of Proteins Homologous to the NADPH Oxidase Complex of Human Neutrophils. Infect. Immun. 2005, 73, 4161–4170. [Google Scholar] [CrossRef]
- Krishnan, N.; Hyršl, P.; Šimek, V. Nitric Oxide Production by Hemocytes of Larva and Pharate Prepupa of Galleria mellonella in Response to Bacterial Lipopolysaccharide: Cytoprotective or Cytotoxic? Comp. Biochem. Physiol.-C Toxicol. Pharmacol. 2006, 142, 103–110. [Google Scholar] [CrossRef] [PubMed]
- Bergin, D.; Murphy, L.; Keenan, J.; Clynes, M.; Kavanagh, K. Pre-Exposure to Yeast Protects Larvae of Galleria mellonella from a Subsequent Lethal Infection by Candida Albicans and Is Mediated by the Increased Expression of Antimicrobial Peptides. Microbes Infect. 2006, 8, 2105–2112. [Google Scholar] [CrossRef] [PubMed]
- Sułek, M.; Kordaczuk, J.; Mak, P.; Śmiałek-Bartyzel, J.; Hułas-Stasiak, M.; Wojda, I. Immune Priming Modulates Galleria mellonella and Pseudomonas entomophila Interaction. Antimicrobial Properties of Kazal Peptide Pr13a. Front. Immunol. 2024, 15, 1358247. [Google Scholar] [CrossRef] [PubMed]
- Kordaczuk, J.; Sułek, M.; Mak, P.; Zdybicka-Barabas, A.; Śmiałek, J.; Wojda, I. Cationic Protein 8 Plays Multiple Roles in Galleria mellonella Immunity. Sci. Rep. 2022, 12, 11737. [Google Scholar] [CrossRef]
- Gallorini, M.; Marinacci, B.; Pellegrini, B.; Cataldi, A.; Dindo, M.L.; Carradori, S.; Grande, R. Immunophenotyping of Hemocytes from Infected Galleria mellonella Larvae as an Innovative Tool for Immune Profiling, Infection Studies and Drug Screening. Sci. Rep. 2024, 14, 759. [Google Scholar] [CrossRef]
- Upfold, J.; Rejasse, A.; Nielsen-Leroux, C.; Jensen, A.B.; Sanchis-Borja, V. The Immunostimulatory Role of an Enterococcus-Dominated Gut Microbiota in Host Protection against Bacterial and Fungal Pathogens in Galleria mellonella Larvae. Front. Insect Sci. 2023, 3, 1260333. [Google Scholar] [CrossRef]
- Ruiz Barrionuevo, J.M.; Vilanova-Cuevas, B.; Alvarez, A.; Martín, E.; Malizia, A.; Galindo-Cardona, A.; de Cristóbal, R.E.; Occhionero, M.A.; Chalup, A.; Monmany-Garzia, A.C.; et al. The Bacterial and Fungal Gut Microbiota of the Greater Wax Moth, Galleria mellonella L. Consuming Polyethylene and Polystyrene. Front. Microbiol. 2022, 13, 918861. [Google Scholar] [CrossRef]
- Portal-Celhay, C.; Bradley, E.R.; Blaser, M.J. Control of Intestinal Bacterial Proliferation in Regulation of Lifespan in Caenorhabditis Elegans. BMC Microbiol. 2012, 12, 49. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.D.; Luallen, R.J. An Organismal Understanding of C. Elegans Innate Immune Responses, from Pathogen Recognition to Multigenerational Resistance. Semin. Cell Dev. Biol. 2024, 154, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Irazoqui, J.E.; Urbach, J.M.; Ausubel, F.M. Evolution of Host Innate Defence: Insights from Caenorhabditis Elegans and Primitive Invertebrates. Nat. Rev. Immunol. 2010, 10, 47–58. [Google Scholar] [CrossRef]
- Pukkila-Worley, R.; Ausubel, F.M. Immune Defense Mechanisms in the Caenorhabditis Elegans Intestinal Epithelium. Curr. Opin. Immunol. 2012, 24, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, J.; Obeng, N.; Yang, W.; Pees, B.; Petersen, C.; Waschina, S.; Kissoyan, K.A.; Aidley, J.; Hoeppner, M.P.; Bunk, B.; et al. The Functional Repertoire Contained within the Native Microbiota of the Model Nematode Caenorhabditis Elegans. ISME J. 2020, 14, 26–38. [Google Scholar] [CrossRef]
- Singh, A.; Luallen, R.J. Understanding the Factors Regulating Host-Microbiome Interactions Using Caenorhabditis Elegans. Philos. Trans. R. Soc. B Biol. Sci. 2024, 379, 20230059. [Google Scholar] [CrossRef]
- Griem-Krey, H.; Petersen, C.; Hamerich, I.K.; Schulenburg, H. The Intricate Triangular Interaction between Protective Microbe, Pathogen and Host Determines Fitness of the Metaorganism. Proc. Biol. Sci. 2023, 290, 20232193. [Google Scholar] [CrossRef]
- Pees, B.; Peters, L.; Treitz, C.; Hamerich, I.K.; Kissoyan, K.A.B.; Tholey, A.; Dierking, K. The Caenorhabditis Elegans Proteome Response to Two Protective Pseudomonas Symbionts. mBio 2024, 15, e0346323. [Google Scholar] [CrossRef]
- Goyache, I.; Yavorov-Dayliev, D.; Milagro, F.I.; Aranaz, P. Caenorhabditis Elegans as a Screening Model for Probiotics with Properties against Metabolic Syndrome. Int. J. Mol. Sci. 2024, 25, 1321. [Google Scholar] [CrossRef]
- Ali, M.S.; Ahmed, S.; Takeuchi, S.; Wada, T.; Kage-Nakadai, E. Improvement of Locomotion Caused by Lactococcus Lactis Subsp. Lactis in the Model Organism Caenorhabditis Elegans. Nutrients 2023, 15, 4482. [Google Scholar] [CrossRef]
- Komura, T.; Takemoto, A.; Kosaka, H.; Suzuki, T.; Nishikawa, Y. Prolonged Lifespan, Improved Perception, and Enhanced Host Defense of Caenorhabditis Elegans by Lactococcus Cremoris Subsp. Cremoris. Microbiol. Spectr. 2022, 10, e0045421. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Luo, F.; Xu, Y.; Zhang, Y.; Jin, L.H. Drosophila Innate Immunity Involves Multiple Signaling Pathways and Coordinated Communication Between Different Tissues. Front. Immunol. 2022, 13, 905370. [Google Scholar] [CrossRef]
- Hanson, M.A.; Lemaitre, B. New Insights on Drosophila Antimicrobial Peptide Function in Host Defense and Beyond. Curr. Opin. Immunol. 2020, 62, 22–30. [Google Scholar] [CrossRef]
- Brogden, K.A. Antimicrobial Peptides: Pore Formers or Metabolic Inhibitors in Bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.; Gallo, R.L. AMPed up Immunity: How Antimicrobial Peptides Have Multiple Roles in Immune Defense. Trends Immunol. 2009, 30, 131–141. [Google Scholar] [CrossRef]
- Joo, H.S.; Fu, C.I.; Otto, M. Bacterial Strategies of Resistance to Antimicrobial Peptides. Philos. Trans. R. Soc. B Biol. Sci. 2016, 371, 20150292. [Google Scholar] [CrossRef]
- Bulet, P.; Dimarcq, J.L.; Hetru, C.; Lagueux, M.; Charlet, M.; Hegy, G.; Van Dorsselaer, A.; Hoffmann, J.A. A Novel Inducible Antibacterial Peptide of Drosophila Carries an O-Glycosylated Substitution. J. Biol. Chem. 1993, 268, 14893–14897. [Google Scholar] [CrossRef]
- Lanot, R.; Zachary, D.; Holder, F.; Meister, M. Postembryonic Hematopoiesis in Drosophila. Dev. Biol. 2001, 230, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.J.; Hartenstein, V.; Banerjee, U. Thicker Than Blood: Conserved Mechanisms in Drosophila and Vertebrate Hematopoiesis. Dev. Cell 2003, 5, 673–690. [Google Scholar] [CrossRef]
- Banerjee, U.; Girard, J.R.; Goins, L.M.; Spratford, C.M. Drosophila as a Genetic Model for Hematopoiesis. Genetics 2019, 211, 367–417. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Luo, F.; Jin, L.H. The Drosophila Lymph Gland Is an Ideal Model for Studying Hematopoiesis. Dev. Comp. Immunol. 2018, 83, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Kurucz, É.; Váczi, B.; Márkus, R.; Laurinyecz, B.; Vilmos, P.; Zsámboki, J.; Csorba, K.; Gateff, E.; Hultmark, D.; Andó, I. Definition of Drosophila Hemocyte Subsets by Cell-Type Specific Antigens. Acta Biol. Hung. 2007, 58, 95–111. [Google Scholar] [CrossRef] [PubMed]
- Hetru, C.; Hoffmann, J.A. NF-KappaB in the Immune Response of Drosophila. Cold Spring Harb. Perspect. Biol. 2009, 1, a000232. [Google Scholar] [CrossRef]
- Chen, Q.; Giedt, M.; Tang, L.; Harrison, D.A. Tools and Methods for Studying the Drosophila JAK/STAT Pathway. Methods 2014, 68, 160–172. [Google Scholar] [CrossRef]
- Tafesh-Edwards, G.; Eleftherianos, I. JNK Signaling in Drosophila Immunity and Homeostasis. Immunol. Lett. 2020, 226, 7–11. [Google Scholar] [CrossRef]
- Tafesh-Edwards, G.; Eleftherianos, I. The Role of Drosophila Microbiota in Gut Homeostasis and Immunity. Gut Microbes 2023, 15, 2208503. [Google Scholar] [CrossRef]
- Storelli, G.; Defaye, A.; Erkosar, B.; Hols, P.; Royet, J.; Leulier, F. Lactobacillus Plantarum Promotes Drosophila Systemic Growth by Modulating Hormonal Signals through TOR-Dependent Nutrient Sensing. Cell Metab. 2011, 14, 403–414. [Google Scholar] [CrossRef]
- Scott, K.; Fischer, C.N.; Trautman, E.P.; Crawford, J.M.; Stabb, E.V.; Handelsman, J.; Broderick, N.A. Metabolite Exchange between Microbiome Members Produces Compounds That Influence Drosophila Behavior. eLife 2017, 6, e18855. [Google Scholar] [CrossRef]
- Huang, J.H.; Douglas, A.E. Consumption of Dietary Sugar by Gut Bacteria Determines Drosophila Lipid Content. Biol. Lett. 2015, 11, 20150469. [Google Scholar] [CrossRef]
- Wong, A.C.N.; Dobson, A.J.; Douglas, A.E. Gut Microbiota Dictates the Metabolic Response of Drosophila to Diet. J. Exp. Biol. 2014, 217, 1894–1901. [Google Scholar] [CrossRef] [PubMed]
- Yamauchi, T.; Oi, A.; Kosakamoto, H.; Akuzawa-Tokita, Y.; Murakami, T.; Mori, H.; Miura, M.; Obata, F. Gut Bacterial Species Distinctively Impact Host Purine Metabolites during Aging in Drosophila. iScience 2020, 23, 101477. [Google Scholar] [CrossRef] [PubMed]
- Blum, J.E.; Fischer, C.N.; Miles, J.; Handelsman, J. Frequent Replenishment Sustains the Beneficial Microbiome of Drosophila Melanogaster. mBio 2013, 4, e00860-13. [Google Scholar] [CrossRef]
- Liu, X.; Hodgson, J.J.; Buchon, N. Drosophila as a Model for Homeostatic, Antibacterial, and Antiviral Mechanisms in the Gut. PLoS Pathog. 2017, 13, e1006277. [Google Scholar] [CrossRef] [PubMed]
- Limmer, S.; Quintin, J.; Hetru, C.; Ferrandon, D. Virulence on the Fly: Drosophila Melanogaster as a Model Genetic Organism to Decipher Host-Pathogen Interactions. Curr. Drug Targets 2011, 12, 978–999. [Google Scholar] [CrossRef]
- Fauvarque, M.O. Small Flies to Tackle Big Questions: Assaying Complex Bacterial Virulence Mechanisms Using Drosophila Melanogaster. Cell Microbiol. 2014, 16, 824–833. [Google Scholar] [CrossRef]
- Erkosar, B.; Leulier, F. Transient Adult Microbiota, Gut Homeostasis and Longevity: Novel Insights from the Drosophila Model. FEBS Lett. 2014, 588, 4250–4257. [Google Scholar] [CrossRef]
- Buchon, N.; Broderick, N.A.; Chakrabarti, S.; Lemaitre, B. Invasive and Indigenous Microbiota Impact Intestinal Stem Cell Activity through Multiple Pathways in Drosophila. Genes Dev. 2009, 23, 2333–2344. [Google Scholar] [CrossRef]
- Lee, K.A.; Kim, S.H.; Kim, E.K.; Ha, E.M.; You, H.; Kim, B.; Kim, M.J.; Kwon, Y.; Ryu, J.H.; Lee, W.J. Bacterial-Derived Uracil as a Modulator of Mucosal Immunity and Gut-Microbe Homeostasis in Drosophila. Cell 2013, 153, 797–811. [Google Scholar] [CrossRef]
- Luo, H.; Li, M.; Wang, F.; Yang, Y.; Wang, Q.; Zhao, Y.; Du, F.; Chen, Y.; Shen, J.; Zhao, Q.; et al. The Role of Intestinal Stem Cell within Gut Homeostasis: Focusing on Its Interplay with Gut Microbiota and the Regulating Pathways. Int. J. Biol. Sci. 2022, 18, 5185–5206. [Google Scholar] [CrossRef]
- Leulier, F.; Parquet, C.; Pili-Floury, S.; Ryu, J.H.; Caroff, M.; Lee, W.J.; Mengin-Lecreulx, D.; Lemaitre, B. The Drosophila Immune System Detects Bacteria through Specific Peptidoglycan Recognition. Nat. Immunol. 2003, 4, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Buchon, N.; Silverman, N.; Cherry, S. Immunity in Drosophila Melanogaster-from Microbial Recognition to Whole-Organism Physiology. Nat. Rev. Immunol. 2014, 14, 796–810. [Google Scholar] [CrossRef] [PubMed]
- Buchon, N.; Broderick, N.A.; Poidevin, M.; Pradervand, S.; Lemaitre, B. Drosophila Intestinal Response to Bacterial Infection: Activation of Host Defense and Stem Cell Proliferation. Cell Host Microbe 2009, 5, 200–211. [Google Scholar] [CrossRef]
- Jones, R.M.; Luo, L.; Ardita, C.S.; Richardson, A.N.; Kwon, Y.M.; Mercante, J.W.; Alam, A.; Gates, C.L.; Wu, H.; Swanson, P.A.; et al. Symbiotic Lactobacilli Stimulate Gut Epithelial Proliferation via Nox-Mediated Generation of Reactive Oxygen Species. EMBO J. 2013, 32, 3017–3028. [Google Scholar] [CrossRef]
- Thomson, C.A.; Morgan, S.C.; Ohland, C.; McCoy, K.D. From Germ-Free to Wild: Modulating Microbiome Complexity to Understand Mucosal Immunology. Mucosal Immunol. 2022, 15, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Moysidou, C.-M.; Owens, R.M. Advances in Modelling the Human Microbiome–Gut–Brain Axis in Vitro. Biochem. Soc. Trans. 2021, 49, 187–201. [Google Scholar] [CrossRef]
- Pan, F.; Han, L.; Zhang, Y.; Yu, Y.; Liu, J. Optimization of Caco-2 and HT29 Co-Culture in Vitro Cell Models for Permeability Studies. Int. J. Food Sci. Nutr. 2015, 66, 680–685. [Google Scholar] [CrossRef]
- Jing, B.; Wang, Z.A.; Zhang, C.; Deng, Q.; Wei, J.; Luo, Y.; Zhang, X.; Li, J.; Du, Y. Establishment and Application of Peristaltic Human Gut-Vessel Microsystem for Studying Host–Microbial Interaction. Front. Bioeng. Biotechnol. 2020, 8, 272. [Google Scholar] [CrossRef]
- Lock, J.Y.; Caboni, M.; Strandwitz, P.; Morrissette, M.; DiBona, K.; Joughin, B.A.; Lewis, K.; Carrier, R.L. An in Vitro Intestinal Model Captures Immunomodulatory Properties of the Microbiota in Inflammation. Gut Microbes 2022, 14, 2039002. [Google Scholar] [CrossRef]
- Erttmann, S.F.; Swacha, P.; Aung, K.M.; Brindefalk, B.; Jiang, H.; Härtlova, A.; Uhlin, B.E.; Wai, S.N.; Gekara, N.O. The Gut Microbiota Prime Systemic Antiviral Immunity via the CGAS-STING-IFN-I Axis. Immunity 2022, 55, 847–861.e10. [Google Scholar] [CrossRef]
- Pearce, S.C.; Coia, H.G.; Karl, J.P.; Pantoja-Feliciano, I.G.; Zachos, N.C.; Racicot, K. Intestinal in Vitro and Ex Vivo Models to Study Host-Microbiome Interactions and Acute Stressors. Front. Physiol. 2018, 9, 1584. [Google Scholar] [CrossRef] [PubMed]
- Magryś, A.; Pawlik, M. Postbiotic Fractions of Probiotics Lactobacillus Plantarum 299v and Lactobacillus Rhamnosus GG Show Immune-Modulating Effects. Cells 2023, 12, 2538. [Google Scholar] [CrossRef] [PubMed]
- Zoumpopoulou, G.; Tsakalidou, E.; Dewulf, J.; Pot, B.; Grangette, C. Differential Crosstalk between Epithelial Cells, Dendritic Cells and Bacteria in a Co-Culture Model. Int. J. Food Microbiol. 2009, 131, 40–51. [Google Scholar] [CrossRef] [PubMed]
- Calvigioni, M.; Mazzantini, D.; Celandroni, F.; Ghelardi, E. Animal and In Vitro Models as Powerful Tools to Decipher the Effects of Enteric Pathogens on the Human Gut Microbiota. Microorganisms 2023, 12, 67. [Google Scholar] [CrossRef]
- Gościniak, A.; Eder, P.; Walkowiak, J.; Cielecka-Piontek, J. Artificial Gastrointestinal Models for Nutraceuticals Research-Achievements and Challenges: A Practical Review. Nutrients 2022, 14, 2560. [Google Scholar] [CrossRef]
- Alves, J.; Sargison, F.A.; Stawarz, H.; Fox, W.B.; Huete, S.G.; Hassan, A.; McTeir, B.; Pickering, A.C. A Case Report: Insights into Reducing Plastic Waste in a Microbiology Laboratory. Access Microbiol. 2021, 3, 000173. [Google Scholar] [CrossRef]
- Date, S.; Sato, T. Mini-Gut Organoids: Reconstitution of the Stem Cell Niche. Annu. Rev. Cell Dev. Biol. 2015, 31, 269–289. [Google Scholar] [CrossRef]
- Sakalem, M.E.; De Sibio, M.T.; da Costa, F.A.d.S.; de Oliveira, M. Historical Evolution of Spheroids and Organoids, and Possibilities of Use in Life Sciences and Medicine. Biotechnol. J. 2021, 16, 2000463. [Google Scholar] [CrossRef]
- Laschke, M.W.; Menger, M.D. Spheroids as Vascularization Units: From Angiogenesis Research to Tissue Engineering Applications. Biotechnol. Adv. 2017, 35, 782–791. [Google Scholar] [CrossRef]
- Lucafò, M.; Muzzo, A.; Marcuzzi, M.; Giorio, L.; Decorti, G.; Stocco, G. Patient-Derived Organoids for Therapy Personalization in Inflammatory Bowel Diseases. World J. Gastroenterol. 2022, 28, 2636–2653. [Google Scholar] [CrossRef]
- Fujii, M.; Matano, M.; Toshimitsu, K.; Takano, A.; Mikami, Y.; Nishikori, S.; Sugimoto, S.; Sato, T. Human Intestinal Organoids Maintain Self-Renewal Capacity and Cellular Diversity in Niche-Inspired Culture Condition. Cell Stem Cell 2018, 23, 787–793.e6. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Hong, S.-N.; Kim, E.-R.; Chang, D.-K.; Kim, Y.-H. Epithelial Regeneration Ability of Crohn’s Disease Assessed Using Patient-Derived Intestinal Organoids. Int. J. Mol. Sci. 2021, 22, 6013. [Google Scholar] [CrossRef] [PubMed]
- Dijkstra, K.K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; van de Haar, J.; Fanchi, L.F.; Slagter, M.; van der Velden, D.L.; Kaing, S.; Kelderman, S.; et al. Generation of Tumor-Reactive T Cells by Co-Culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 2018, 174, 1586–1598.e12. [Google Scholar] [CrossRef] [PubMed]
- Williamson, I.A.; Arnold, J.W.; Samsa, L.A.; Gaynor, L.; DiSalvo, M.; Cocchiaro, J.L.; Carroll, I.; Azcarate-Peril, M.A.; Rawls, J.F.; Allbritton, N.L.; et al. A High-Throughput Organoid Microinjection Platform to Study Gastrointestinal Microbiota and Luminal Physiology. Cell Mol. Gastroenterol. Hepatol. 2018, 6, 301–319. [Google Scholar] [CrossRef]
- Puschhof, J.; Pleguezuelos-Manzano, C.; Martinez-Silgado, A.; Akkerman, N.; Saftien, A.; Boot, C.; de Waal, A.; Beumer, J.; Dutta, D.; Heo, I.; et al. Intestinal Organoid Cocultures with Microbes. Nat. Protoc. 2021, 16, 4633–4649. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human Organoids: Model Systems for Human Biology and Medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, A.E.; Stoeger, V.; Owens, R.M. Lab-on-Chip Technologies for Exploring the Gut–Immune Axis in Metabolic Disease. Lab. Chip 2024, 24, 1266–1292. [Google Scholar] [CrossRef]
- Naumovska, E.; Aalderink, G.; Wong Valencia, C.; Kosim, K.; Nicolas, A.; Brown, S.; Vulto, P.; Erdmann, K.S.; Kurek, D. Direct On-Chip Differentiation of Intestinal Tubules from Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2020, 21, 4964. [Google Scholar] [CrossRef]
- Sontheimer-Phelps, A.; Chou, D.B.; Tovaglieri, A.; Ferrante, T.C.; Duckworth, T.; Fadel, C.; Frismantas, V.; Sutherland, A.D.; Jalili-Firoozinezhad, S.; Kasendra, M.; et al. Human Colon-on-a-Chip Enables Continuous In Vitro Analysis of Colon Mucus Layer Accumulation and Physiology. Cell Mol. Gastroenterol. Hepatol. 2020, 9, 507–526. [Google Scholar] [CrossRef]
- Whitesides, G.M. The Origins and the Future of Microfluidics. Nature 2006, 442, 368–373. [Google Scholar] [CrossRef]
- Mark, D.; Haeberle, S.; Roth, G.; von Stetten, F.; Zengerle, R. Microfluidic Lab-on-a-Chip Platforms: Requirements, Characteristics and Applications. Chem. Soc. Rev. 2010, 39, 1153–1182. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Yu, L.; Tian, F.; Zhao, J.; Zhai, Q. In Vitro Models to Study Human Gut-Microbiota Interactions: Applications, Advances, and Limitations. Microbiol. Res. 2023, 270, 127336. [Google Scholar] [CrossRef] [PubMed]
- Beaurivage, C.; Naumovska, E.; Chang, Y.X.; Elstak, E.D.; Nicolas, A.; Wouters, H.; van Moolenbroek, G.; Lanz, H.L.; Trietsch, S.J.; Joore, J.; et al. Development of a Gut-on-a-Chip Model for High Throughput Disease Modeling and Drug Discovery. Int. J. Mol. Sci. 2019, 20, 5661. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Li, H.; Collins, J.J.; Ingber, D.E. Contributions of Microbiome and Mechanical Deformation to Intestinal Bacterial Overgrowth and Inflammation in a Human Gut-on-a-Chip. Proc. Natl. Acad. Sci. USA 2016, 113, E7–E15. [Google Scholar] [CrossRef]
- Kasendra, M.; Tovaglieri, A.; Sontheimer-Phelps, A.; Jalili-Firoozinezhad, S.; Bein, A.; Chalkiadaki, A.; Scholl, W.; Zhang, C.; Rickner, H.; Richmond, C.A.; et al. Development of a Primary Human Small Intestine-on-a-Chip Using Biopsy-Derived Organoids. Sci. Rep. 2018, 8, 2871. [Google Scholar] [CrossRef]
- Boquet-Pujadas, A.; Feaugas, T.; Petracchini, A.; Grassart, A.; Mary, H.; Manich, M.; Gobaa, S.; Olivo-Marin, J.-C.; Sauvonnet, N.; Labruyère, E. 4D Live Imaging and Computational Modeling of a Functional Gut-on-a-Chip Evaluate How Peristalsis Facilitates Enteric Pathogen Invasion. Sci. Adv. 2024, 8, eabo5767. [Google Scholar] [CrossRef]
- Jeon, M.S.; Choi, Y.Y.; Mo, S.J.; Ha, J.H.; Lee, Y.S.; Lee, H.U.; Park, S.D.; Shim, J.-J.; Lee, J.-L.; Chung, B.G. Contributions of the Microbiome to Intestinal Inflammation in a Gut-on-a-Chip. Nano Converg. 2022, 9, 8. [Google Scholar] [CrossRef]
- Yuan, L.; de Haan, P.; Peterson, B.W.; de Jong, E.D.; Verpoorte, E.; van der Mei, H.C.; Busscher, H.J. Visualization of Bacterial Colonization and Cellular Layers in a Gut-on-a-Chip System Using Optical Coherence Tomography. Microsc. Microanal. 2020, 26, 1211–1219. [Google Scholar] [CrossRef]
- De Gregorio, V.; Sgambato, C.; Urciuolo, F.; Vecchione, R.; Netti, P.A.; Imparato, G. Immunoresponsive Microbiota-Gut-on-Chip Reproduces Barrier Dysfunction, Stromal Reshaping and Probiotics Translocation under Inflammation. Biomaterials 2022, 286, 121573. [Google Scholar] [CrossRef]
- Jalili-Firoozinezhad, S.; Gazzaniga, F.S.; Calamari, E.L.; Camacho, D.M.; Fadel, C.W.; Bein, A.; Swenor, B.; Nestor, B.; Cronce, M.J.; Tovaglieri, A.; et al. A Complex Human Gut Microbiome Cultured in an Anaerobic Intestine-on-a-Chip. Nat. Biomed. Eng. 2019, 3, 520–531. [Google Scholar] [CrossRef]
- Marzorati, M.; Vanhoecke, B.; De Ryck, T.; Sadaghian Sadabad, M.; Pinheiro, I.; Possemiers, S.; Van den Abbeele, P.; Derycke, L.; Bracke, M.; Pieters, J.; et al. The HMITM Module: A New Tool to Study the Host-Microbiota Interaction in the Human Gastrointestinal Tract in Vitro. BMC Microbiol. 2014, 14, 133. [Google Scholar] [CrossRef] [PubMed]
- Morelli, M.; Kurek, D.; Ng, C.P.; Queiroz, K. Gut-on-a-Chip Models: Current and Future Perspectives for Host–Microbial Interactions Research. Biomedicines 2023, 11, 619. [Google Scholar] [CrossRef] [PubMed]
- Beaurivage, C.; Kanapeckaite, A.; Loomans, C.; Erdmann, K.S.; Stallen, J.; Janssen, R.A.J. Development of a Human Primary Gut-on-a-Chip to Model Inflammatory Processes. Sci. Rep. 2020, 10, 21475. [Google Scholar] [CrossRef] [PubMed]
- Trujillo-de Santiago, G.; Lobo-Zegers, M.J.; Montes-Fonseca, S.L.; Zhang, Y.S.; Alvarez, M.M. Gut-Microbiota-on-a-Chip: An Enabling Field for Physiological Research. Microphysiol. Syst. 2018, 2, 7. [Google Scholar] [CrossRef]
- Kim, H.J.; Huh, D.; Hamilton, G.; Ingber, D.E. Human Gut-on-a-Chip Inhabited by Microbial Flora That Experiences Intestinal Peristalsis-like Motions and Flow. Lab. Chip 2012, 12, 2165–2174. [Google Scholar] [CrossRef]
- Van den Elsen, L.; Poyntz, H.C.; Weyrich, L.S.; Young, W.; E Forbes-Blom, E. Embracing the Gut Microbiota: The New Frontier for Inflammatory and Infectious Diseases. Clin. Transl. Immunol. 2017, 6, e125. [Google Scholar] [CrossRef]
- Bauer, E.; Thiele, I. From Metagenomic Data to Personalized in Silico Microbiotas: Predicting Dietary Supplements for Crohn’s Disease. NPJ Syst. Biol. Appl. 2018, 4, 27. [Google Scholar] [CrossRef]
- Park, S.-Y.; Ufondu, A.; Lee, K.; Jayaraman, A. Emerging Computational Tools and Models for Studying Gut Microbiota Composition and Function. Curr. Opin. Biotechnol. 2020, 66, 301–311. [Google Scholar] [CrossRef]
- Magnúsdóttir, S.; Heinken, A.; Kutt, L.; Ravcheev, D.A.; Bauer, E.; Noronha, A.; Greenhalgh, K.; Jäger, C.; Baginska, J.; Wilmes, P.; et al. Generation of Genome-Scale Metabolic Reconstructions for 773 Members of the Human Gut Microbiota. Nat. Biotechnol. 2017, 35, 81–89. [Google Scholar] [CrossRef]
- Heinken, A.; Sahoo, S.; Fleming, R.M.T.; Thiele, I. Systems-Level Characterization of a Host-Microbe Metabolic Symbiosis in the Mammalian Gut. Gut Microbes 2013, 4, 28–40. [Google Scholar] [CrossRef]
- Wendelsdorf, K.V.; Alam, M.; Bassaganya-Riera, J.; Bisset, K.; Eubank, S.; Hontecillas, R.; Hoops, S.; Marathe, M. Enteric Immunity Simulator: A Tool for in Silico Study of Gastroenteric Infections. IEEE Trans. Nanobiosci. 2012, 11, 273–288. [Google Scholar] [CrossRef] [PubMed]
- Volkova, A.; Ruggles, K.V. Predictive Metagenomic Analysis of Autoimmune Disease Identifies Robust Autoimmunity and Disease Specific Microbial Signatures. Front. Microbiol. 2021, 12, 621310. [Google Scholar] [CrossRef]
- Ezzamouri, B.; Shoaie, S.; Ledesma-Amaro, R. Synergies of Systems Biology and Synthetic Biology in Human Microbiome Studies. Front. Microbiol. 2021, 12, 681982. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Míguez, A.; Fdez-Riverola, F.; Sánchez, B.; Lourenço, A. Resources and Tools for the High-Throughput, Multi-Omic Study of Intestinal Microbiota. Brief. Bioinform. 2017, 20, 1032–1056. [Google Scholar] [CrossRef] [PubMed]
- Son, J.W.; Shoaie, S.; Lee, S. Systems Biology: A Multi-Omics Integration Approach to Metabolism and the Microbiome. Endocrinol. Metab. 2020, 35, 507–514. [Google Scholar] [CrossRef]
- Jing, Y.; Yuan, Y.; Monson, M.; Wang, P.; Mu, F.; Zhang, Q.; Na, W.; Zhang, K.; Wang, Y.; Leng, L.; et al. Multi-Omics Association Reveals the Effects of Intestinal Microbiome–Host Interactions on Fat Deposition in Broilers. Front. Microbiol. 2022, 12, 815538. [Google Scholar] [CrossRef]
- Wang, A.-J.; Song, D.; Hong, Y.-M.; Liu, N.-N. Multi-Omics Insights into the Interplay between Gut Microbiota and Colorectal Cancer in the “Microworld” Age. Mol. Omics 2023, 19, 283–296. [Google Scholar] [CrossRef]
- Heinken, A.; Thiele, I. Systems Biology of Host-Microbe Metabolomics. Wiley Interdiscip. Rev. Syst. Biol. Med. 2015, 7, 195–219. [Google Scholar] [CrossRef]
- Borenstein, E. Computational Systems Biology and in Silico Modeling of the Human Microbiome. Brief. Bioinform. 2012, 13, 769–780. [Google Scholar] [CrossRef]
- Borenstein, E. Metagenomic Systems Biology: Metabolic Modeling and Multi-Meta-Omic Analysis of the Human Microbiome. FASEB J. 2015, 29, 91.1. [Google Scholar] [CrossRef]
- Kumar, M.; Ji, B.; Babaei, P.; Das, P.; Lappa, D.; Ramakrishnan, G.; Fox, T.E.; Haque, R.; Petri, W.A.; Bäckhed, F.; et al. Gut Microbiota Dysbiosis Is Associated with Malnutrition and Reduced Plasma Amino Acid Levels: Lessons from Genome-Scale Metabolic Modeling. Metab. Eng. 2018, 49, 128–142. [Google Scholar] [CrossRef] [PubMed]
- Shoaie, S.; Karlsson, F.; Mardinoglu, A.; Nookaew, I.; Bordel, S.; Nielsen, J. Understanding the Interactions between Bacteria in the Human Gut through Metabolic Modeling. Sci. Rep. 2013, 3, 2532. [Google Scholar] [CrossRef] [PubMed]
- Shoaie, S.; Ghaffari, P.; Kovatcheva-Datchary, P.; Mardinoglu, A.; Sen, P.; Pujos-Guillot, E.; de Wouters, T.; Juste, C.; Rizkalla, S.; Chilloux, J.; et al. Quantifying Diet-Induced Metabolic Changes of the Human Gut Microbiome. Cell Metab. 2015, 22, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Thiele, I.; Heinken, A.; Fleming, R.M.T. A Systems Biology Approach to Studying the Role of Microbes in Human Health. Curr. Opin. Biotechnol. 2013, 24, 4–12. [Google Scholar] [CrossRef]
- Heirendt, L.; Arreckx, S.; Pfau, T.; Mendoza, S.N.; Richelle, A.; Heinken, A.; Haraldsdóttir, H.S.; Wachowiak, J.; Keating, S.M.; Vlasov, V.; et al. Creation and Analysis of Biochemical Constraint-Based Models Using the COBRA Toolbox v.3.0. Nat. Protoc. 2019, 14, 639–702. [Google Scholar] [CrossRef]
- Long, Y.; Luo, J. Open Access WMGHMDA: A Novel Weighted Meta-Graph-Based Model for Predicting Human Microbe-Disease Association on Heterogeneous Information Network. BMC Bioinform. 2019, 20, 541. [Google Scholar] [CrossRef]
- Luo, J.; Long, Y. NTSHMDA: Prediction of Human Microbe-Disease Association Based on Random Walk by Integrating Network Topological Similarity. IEEE/ACM Trans. Comput. Biol. Bioinform. 2020, 17, 1341–1351. [Google Scholar] [CrossRef]
- Niu, Y.-W.; Qu, C.-Q.; Wang, G.-H.; Yan, G.-Y. RWHMDA: Random Walk on Hypergraph for Microbe-Disease Association Prediction. Front. Microbiol. 2019, 10, 1578. [Google Scholar] [CrossRef]
- Huang, Y.A.; You, Z.H.; Chen, X.; Huang, Z.A.; Zhang, S.; Yan, G.Y. Prediction of Microbe-Disease Association from the Integration of Neighbor and Graph with Collaborative Recommendation Model. J. Transl. Med. 2017, 15, 209. [Google Scholar] [CrossRef]
- Li, L.; Wu, L.; Zhang, H.-S.; Wu, F.-X. A Fast Algorithm for Nonnegative Matrix Factorization and Its Convergence. IEEE Trans. Neural Netw. Learn. Syst. 2014, 25, 1855–1863. [Google Scholar] [CrossRef]
- Tian, L.-P.; Luo, P.; Wang, H.; Zheng, H.; Wu, F.-X. CASNMF: A Converged Algorithm for Symmetrical Nonnegative Matrix Factorization. Neurocomputing 2018, 275, 2031–2040. [Google Scholar] [CrossRef]
- Xu, D.; Xu, H.; Zhang, Y.; Wang, M.; Chen, W.; Gao, R. MDAKRLS: Predicting Human Microbe-Disease Association Based on Kronecker Regularized Least Squares and Similarities. J. Transl. Med. 2021, 19, 66. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.; Luo, J.; Zhang, Y.; Xia, Y. Predicting Human Microbe-Disease Associations via Graph Attention Networks with Inductive Matrix Completion. Brief. Bioinform. 2021, 22, bbaa146. [Google Scholar] [CrossRef]
- Shokri Garjan, H.; Omidi, Y.; Poursheikhali Asghari, M.; Ferdousi, R. In-Silico Computational Approaches to Study Microbiota Impacts on Diseases and Pharmacotherapy. Gut Pathog. 2023, 15, 10. [Google Scholar] [CrossRef] [PubMed]
- Baldini, F.; Heinken, A.; Heirendt, L.; Magnusdottir, S.; Fleming, R.M.T.; Thiele, I. The Microbiome Modeling Toolbox: From Microbial Interactions to Personalized Microbial Communities. Bioinformatics 2019, 35, 2332–2334. [Google Scholar] [CrossRef] [PubMed]
- Heirendt, L.; Thiele, I.; Fleming, R.M.T. DistributedFBA.Jl: High-Level, High-Performance Flux Balance Analysis in Julia. Bioinformatics 2017, 33, 1421–1423. [Google Scholar] [CrossRef]
- Stelling, J.; Klamt, S.; Bettenbrock, K.; Schuster, S.; Gilles, E.D. Metabolic Network Structure Determines Key Aspects of Functionality and Regulation. Nature 2002, 420, 190–193. [Google Scholar] [CrossRef]
- Reed, J.L.; Palsson, B. Thirteen Years of Building Constraint-Based in Silico Models of Escherichia Coli. J. Bacteriol. 2003, 185, 2692–2699. [Google Scholar] [CrossRef]
- Wu, J.; Singleton, S.S.; Bhuiyan, U.; Krammer, L.; Mazumder, R. Multi-Omics Approaches to Studying Gastrointestinal Microbiome in the Context of Precision Medicine and Machine Learning. Front. Mol. Biosci. 2023, 10, 1337373. [Google Scholar] [CrossRef]
- Abavisani, M.; Khoshrou, A.; Foroushan, S.K.; Ebadpour, N.; Sahebkar, A. Deciphering the Gut Microbiome: The Revolution of Artificial Intelligence in Microbiota Analysis and Intervention. Curr. Res. Biotechnol. 2024, 7, 100211. [Google Scholar] [CrossRef]
- Li, P.; Luo, H.; Ji, B.; Nielsen, J. Machine Learning for Data Integration in Human Gut Microbiome. Microb. Cell Fact. 2022, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Coelho, E.D.; Santiago, A.M.; Arrais, J.P.; Oliveira, J.L. Computational Methodology for Predicting the Landscape of the Human–Microbial Interactome Region Level Influence. J. Bioinform. Comput. Biol. 2015, 13, 1550023. [Google Scholar] [CrossRef]
- Menche, J.; Sharma, A.; Kitsak, M.; Ghiassian, S.D.; Vidal, M.; Loscalzo, J.; Barabási, A. Uncovering Disease-Disease Relationships Through the Incomplete Interactome. Science 2015, 347, 1257601. [Google Scholar] [CrossRef]
- Vidal, M.; Cusick, M.E.; Barabási, A. Interactome Networks and Human Disease. Cell 2011, 144, 986–998. [Google Scholar] [CrossRef]
- Heinken, A.; Basile, A.; Thiele, I. Advances in Constraint-Based Modelling of Microbial Communities. Curr. Opin. Syst. Biol. 2021, 27, 100346. [Google Scholar] [CrossRef]
- Naqvi, A.; Rangwala, H.; Keshavarzian, A.; Gillevet, P. Network-Based Modeling of the Human Gut Microbiome. Chem. Biodivers. 2010, 7, 1040–1050. [Google Scholar] [CrossRef]
- Oberhardt, M.A.; Puchałka, J.; Fryer, K.E.; Martins dos Santos, V.A.P.; Papin, J.A. Genome-Scale Metabolic Network Analysis of the Opportunistic Pathogen Pseudomonas Aeruginosa PAO1. J. Bacteriol. 2008, 190, 2790–2803. [Google Scholar] [CrossRef] [PubMed]
- Topçuoğlu, B.D.; Lesniak, N.A.; Ruffin, M.T.; Wiens, J.; Schloss, P.D. A Framework for Effective Application of Machine Learning to Microbiome-Based Classification Problems. mBio 2020, 11. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between Microbiota and Immunity in Health and Disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Muller, E.; Algavi, Y.M.; Borenstein, E. A Meta-Analysis Study of the Robustness and Universality of Gut Microbiome-Metabolome Associations. Microbiome 2021, 9, 203. [Google Scholar] [CrossRef]
- Li, Y.-N.; Su, J.-L.; Tan, S.-H.; Chen, X.-L.; Cheng, T.-L.; Jiang, Z.; Luo, Y.-Z.; Zhang, L.-M. Machine Learning Based on Metabolomics Unveils Neutrophil Extracellular Trap-Related Metabolic Signatures in Non-Small Cell Lung Cancer Patients Undergoing Chemoimmunotherapy. World J. Clin. Cases 2024, 12, 4091–4107. [Google Scholar] [CrossRef]
- Thompson, J.C.; Zavala, V.M.; Venturelli, O.S. Integrating a Tailored Recurrent Neural Network with Bayesian Experimental Design to Optimize Microbial Community Functions. PLoS Comput. Biol. 2023, 19, e1011436. [Google Scholar] [CrossRef]
- McCoubrey, L.E.; Elbadawi, M.; Orlu, M.; Gaisford, S.; Basit, A.W. Harnessing Machine Learning for Development of Microbiome Therapeutics. Gut Microbes 2021, 13, 1872323. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Jury, E.C.; Dönnes, P.; Ciurtin, C. Machine Learning Techniques for Personalised Medicine Approaches in Immune-Mediated Chronic in Fl Ammatory Diseases: Applications and Challenges. Front. Pharmacol. 2021, 12, 720694. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xu, Z.Z.; He, Y.; Yang, Y.; Liu, L.; Lin, Q.; Nie, Y.; Li, M.; Zhi, F.; Liu, S.; et al. Gut Microbiota Offers Universal Biomarkers across Ethnicity in Inflammatory Bowel Disease Diagnosis and Infliximab Response Prediction. mSystems 2018, 3, e00188-17. [Google Scholar] [CrossRef] [PubMed]
- Uehara, M.; Inoue, T.; Hase, S.; Sasaki, E.; Toyoda, A.; Sakakibara, Y. Decoding Host-Microbiome Interactions through Co-Expression Network Analysis within the Non-Human Primate Intestine. mSystems 2024, 9, e014052. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Shcherbin, E.; Mohimani, H. A Metabolome- and Metagenome-Wide Association Network Reveals Microbial Natural Products and Microbial Biotransformation Products from the Human Microbiota. mSystems 2019, 4, e00387-19. [Google Scholar] [CrossRef]
- Chiacchio, F.; Pennisi, M.; Russo, G.; Motta, S.; Pappalardo, F. Agent-Based Modeling of the Immune System: NetLogo, a Promising Framework. Biomed. Res. Int. 2014, 2014, 907171. [Google Scholar] [CrossRef]
- Vélez de Mendizábal, N.; Carneiro, J.; Solé, R.V.; Goñi, J.; Bragard, J.; Martinez-Forero, I.; Martinez-Pasamar, S.; Sepulcre, J.; Torrealdea, J.; Bagnato, F.; et al. Modeling the Effector—Regulatory T Cell Cross-Regulation Reveals the Intrinsic Character of Relapses in Multiple Sclerosis. BMC Syst. Biol. 2011, 5, 114. [Google Scholar] [CrossRef]
- Bianca, C.; Chiacchio, F.; Pappalardo, F.; Pennisi, M. Mathematical Modeling of the Immune System Recognition to Mammary Carcinoma Antigen. BMC Bioinform. 2012, 13, S21. [Google Scholar] [CrossRef]
- Pennisi, M. A Mathematical Model of Immune-System-Melanoma Competition. Comput. Math. Methods Med. 2012, 2012, 850754. [Google Scholar] [CrossRef]
- Pappalardo, F.; Brusic, V.; Fr, H. Immune System Modeling and Related Pathologies. Comput. Math. Methods Med. 2012, 2012, 274702. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Niu, X.; He, Q.; Chen, F.; Qi, R.-Q. Artificial Intelligence in Microbiomes Analysis: A Review of Applications in Dermatology. Front. Microbiol. 2023, 14, 1112010. [Google Scholar] [CrossRef]
- An, G.; Mi, Q.; Dutta-Moscato, J.; Vodovotz, Y. Agent-Based Models in Translational Systems Biology. Wiley Interdiscip. Rev. Syst. Biol. Med. 2009, 1, 159–171. [Google Scholar] [CrossRef] [PubMed]
- Bonabeau, E. Agent-Based Modeling: Methods and Techniques for Simulating Human Systems. Proc. Natl. Acad. Sci. USA 2002, 99 (Suppl. S3), 7280–7287. [Google Scholar] [CrossRef]
- Bauer, E.; Zimmermann, J.; Baldini, F.; Thiele, I.; Kaleta, C. BacArena: Individual-Based Metabolic Modeling of Heterogeneous Microbes in Complex Communities. PLoS Comput. Biol. 2017, 13, e1005544. [Google Scholar] [CrossRef] [PubMed]
- Railsback, S.; Lytinen, S.; Jackson, S. Agent-Based Simulation Platforms: Review and Development Recommendations. Simulation 2006, 82, 609–623. [Google Scholar] [CrossRef]
- Grimm, V.; Berger, U.; DeAngelis, D.L.; Polhill, J.G.; Giske, J.; Railsback, S.F. The ODD Protocol: A Review and First Update. Ecol. Model. 2010, 221, 2760–2768. [Google Scholar] [CrossRef]
- Thiele, J.C.; Kurth, W.; Grimm, V. Parameter Estimation and Sensitivity Analysis in Agent-Based Models. J. Artif. Soc. Soc. Simul. 2014, 17, 11. [Google Scholar] [CrossRef]
- MacAl, C.M.; North, M.J. Tutorial on Agent-Based Modelling and Simulation. J. Simul. 2010, 4, 151–162. [Google Scholar] [CrossRef]
- Perez, L.; Dragicevic, S. An Agent-Based Approach for Modeling Dynamics of Contagious Disease Spread. Int. J. Health Geogr. 2009, 8, 50. [Google Scholar] [CrossRef]
- Pennisi, M.; Rajput, A.-M.; Toldo, L.; Pappalardo, F. Agent Based Modeling of Treg-Teff Cross Regulation in Relapsing-Remitting Multiple Sclerosis. BMC Bioinform. 2013, 14 (Suppl. S16), S9. [Google Scholar] [CrossRef]
- Kitano, H. Systems Biology: A Brief Overview. Science 2002, 295, 1662–1664. [Google Scholar] [CrossRef] [PubMed]
- Kerepesi, C.; Bakács, T.; Szabados, T. MiStImm: An Agent-Based Simulation Tool to Study the Self-Nonself Discrimination of the Adaptive Immune Response. Theor. Biol. Med. Model. 2019, 16, 9. [Google Scholar] [CrossRef] [PubMed]
- Castiglione, F.; Bernaschi, M. C-ImmSim: Playing with the Immune Response; Katholieke Universiteit Leuven: Leuven, Belgium, 2004. [Google Scholar]
- Baldazzi, V.; Castiglione, F.; Bernaschi, M. An Enhanced Agent Based Model of the Immune System Response. Cell Immunol. 2006, 244, 77–79. [Google Scholar] [CrossRef]
- Folcik, V.A.; An, G.C.; Orosz, C.G. The Basic Immune Simulator: An Agent-Based Model to Study the Interactions between Innate and Adaptive Immunity. Theor. Biol. Med. Model. 2007, 4, 39. [Google Scholar] [CrossRef]
- Mack, G. SIMMUNE, A Tool for Simulating and Analyzing Immune System Behavior. arXiv 2018. [Google Scholar] [CrossRef]
- Efroni, S.; Harel, D.; Cohen, I.R. Toward Rigorous Comprehension of Biological Complexity: Modeling, Execution, and Visualization of Thymic T-Cell Maturation. Genome Res. 2003, 13, 2485–2497. [Google Scholar] [CrossRef]
- Efroni, S.; Harel, D.; Cohen, I. Reactive Animation: Realistic Modeling of Complex Dynamic. Computer 2005, 38, 38–47. [Google Scholar] [CrossRef]
- Mata, J.; Cohn, M. Cellular Automata-Based Modeling Program: Synthetic Immune System. Immunol. Rev. 2007, 216, 198–212. [Google Scholar] [CrossRef]
- Kohler, B.; Puzone, R.; Seiden, P.E.; Celada, F. A Systematic Approach to Vaccine Complexity Using an Automaton Model of the Cellular and Humoral Immune System. I. Viral Characteristics and Polarized Responses. Vaccine 2000, 19, 862–876. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.J.; Agosto, H.; Litwin, S.; Welsh, J.D.; Shlomchik, M.; Weigert, M.; Seiden, P.E. A Solution to the Rheumatoid Factor Paradox: Pathologic Rheumatoid Factors Can Be Tolerized by Competition with Natural Rheumatoid Factors. J. Immunol. 2009, 159, 1728–1738. [Google Scholar] [CrossRef]
- Bauer, A.L.; Beauchemin, C.A.A.; Perelson, A.S. Agent-Based Modeling of Host-Pathogen Systems: The Successes and Challenges. Inf. Sci. 2009, 179, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- Polys, N.F.; Bowman, D.A.; Laubenbacher, R.; Duca, K. PathSim Visualizer: An Information-Rich Virtual Environment Framework for Systems Biology. In Proceedings of the Ninth International Conference on 3D Web Technology (Web3D ‘04), Association for Computing Machinery, Monterey, CA, USA, 5–8 April 2004; pp. 7–14. [Google Scholar]
- Mei, Y.; Abedi, V.; Carbo, A.; Zhang, X.; Lu, P.; Philipson, C.; Hontecillas, R.; Hoops, S.; Liles, N.; Bassaganya-Riera, J. Multiscale Modeling of Mucosal Immune Responses. BMC Bioinform. 2015, 16, S2. [Google Scholar] [CrossRef]
- Wendelsdorf, K.; Bassaganya-Riera, J.; Bisset, K.; Eubank, S.; Hontecillas, R.; Marathe, M. ENteric Immunity SImulator: A Tool for in Silico Study of Gut Immunopathologies (166.15). J. Immunol. 2011, 186, 166.15. [Google Scholar] [CrossRef]
| Models Used for Studying the Gut Microbiota-Immune System Interaction | Advantages | Disadvantages |
|---|---|---|
| In Vivo | Comprehensive biological context; manipulates microbial compositions; insights into dynamic interactions. | Ethical concerns; high cost; labor-intensive; variability; limited human applicability. |
| In Vitro | Ethical; high experimental control; cost-effective; detailed mechanistic studies. | Lacks full biological complexity; requires advanced technology; some models don’t replicate gut environment. |
| In Silico | Cost-effective; large-scale simulations; models complex interactions; aids experimental design. | Requires robust data for accuracy; potential oversimplification of biological processes. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bertorello, S.; Cei, F.; Fink, D.; Niccolai, E.; Amedei, A. The Future Exploring of Gut Microbiome-Immunity Interactions: From In Vivo/Vitro Models to In Silico Innovations. Microorganisms 2024, 12, 1828. https://doi.org/10.3390/microorganisms12091828
Bertorello S, Cei F, Fink D, Niccolai E, Amedei A. The Future Exploring of Gut Microbiome-Immunity Interactions: From In Vivo/Vitro Models to In Silico Innovations. Microorganisms. 2024; 12(9):1828. https://doi.org/10.3390/microorganisms12091828
Chicago/Turabian StyleBertorello, Sara, Francesco Cei, Dorian Fink, Elena Niccolai, and Amedeo Amedei. 2024. "The Future Exploring of Gut Microbiome-Immunity Interactions: From In Vivo/Vitro Models to In Silico Innovations" Microorganisms 12, no. 9: 1828. https://doi.org/10.3390/microorganisms12091828
APA StyleBertorello, S., Cei, F., Fink, D., Niccolai, E., & Amedei, A. (2024). The Future Exploring of Gut Microbiome-Immunity Interactions: From In Vivo/Vitro Models to In Silico Innovations. Microorganisms, 12(9), 1828. https://doi.org/10.3390/microorganisms12091828








