Diversity of Anaplasmataceae Transmitted by Ticks (Ixodidae) and the First Molecular Evidence of Anaplasma phagocytophilum and Candidatus Anaplasma boleense in Paraguay
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Areas and Sample Collection
2.2. Classical and Molecular Identification of Tick Species
2.3. DNA Extraction from Ticks and Host Blood
| Target Gene | Primers Code | Primers (5′→3′) | Amplicon Size | Reference |
|---|---|---|---|---|
| Ticks DNA 16S rRNA (PCR) | 16S +1 | AACGAACGCTGGCGGCAAGC | 460 bp | [33] |
| 16S −1 | AGTAYCGRACCAGATAGCCGC | |||
| Vertebrate cytb (PCR) | Cyb1 | GAAGATGCWGTWGGWTGTACKGC | 358 bp | [35] |
| Cyb2 | AGMGCTTCWCCTTCWACRTCYTC | |||
| Anaplasmataceae 16S rRNA (PCR-HRM) | EHR16SD | GGTACCYACAGAAGAAGTCC | 345 bp | [36] |
| EHR16RD | TAGCACTCATCGTTTACAGC | |||
| Anaplasmataceae groEL (Nested-PCR) | gro607 F a | GAAGATGCWGTWGGWTGTACKGC | 664 bp | [37] |
| gro1294 R a | AGMGCTTCWCCTTCWACRTCYTC | |||
| gro677 F b | ATTACTCAGAGTGCTTCTCARTG | 315 bp | ||
| gro1121 R b | TGCATACCRTCAGTYTTTTCAAC |
2.4. Blood Feeding Source in Free-Living Ticks
2.5. Optimization of PCR-HRM for Detecting and Differentiating Genera in the Family Anaplasmataceae
2.6. Molecular Identification of Anaplasmataceae Based on the 16S rRNA and groEL Gene
2.7. Purification, Sequencing of Amplified Products, and Phylogenetic Analysis
2.8. Data Analysis and Mapping
2.9. Ethical Approval
3. Results
3.1. Taxonomic Identification of Ticks Collected from Hosts and Free-Living Ticks
3.2. Blood Feeding Sources of Free-Living Ticks
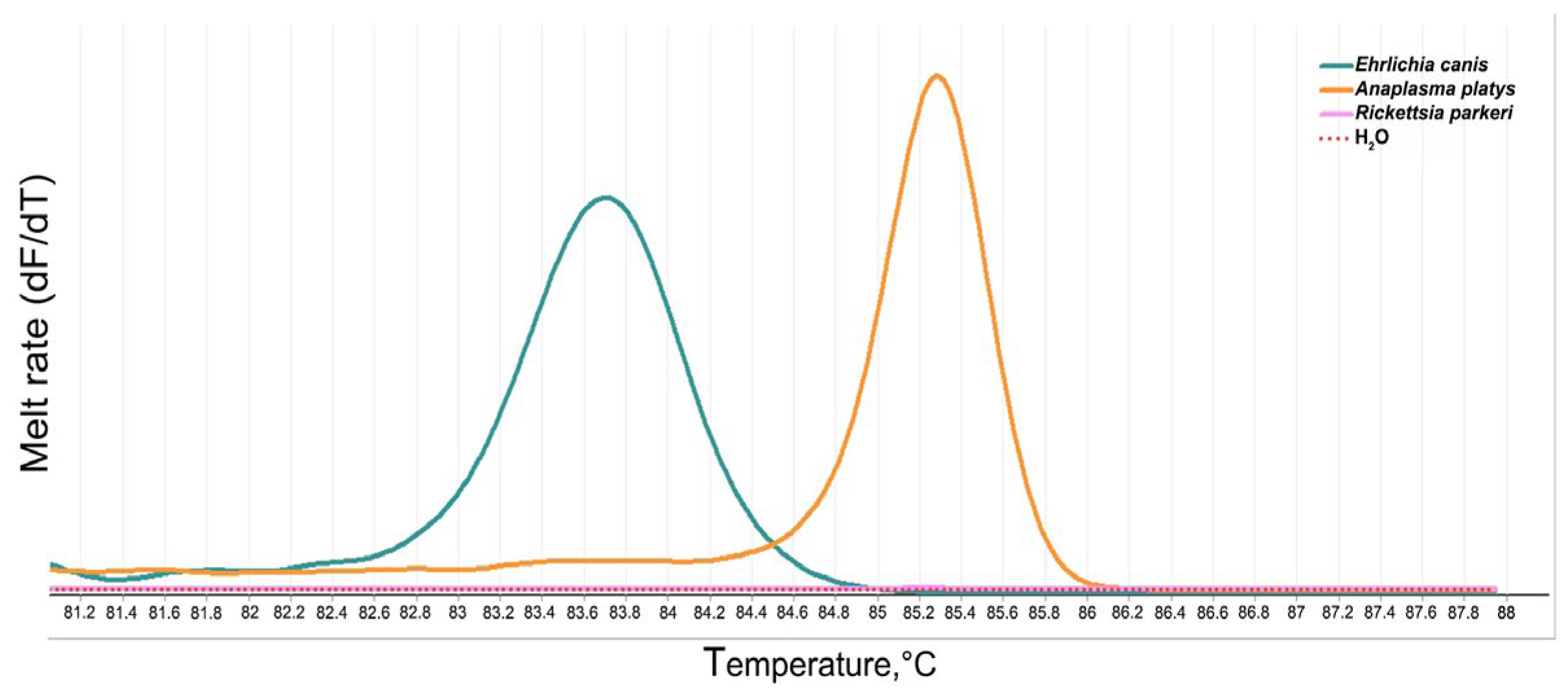
3.3. Optimization of PCR-HRM for the Detection and Differentiation of the Genus of the Anaplasmataceae Family
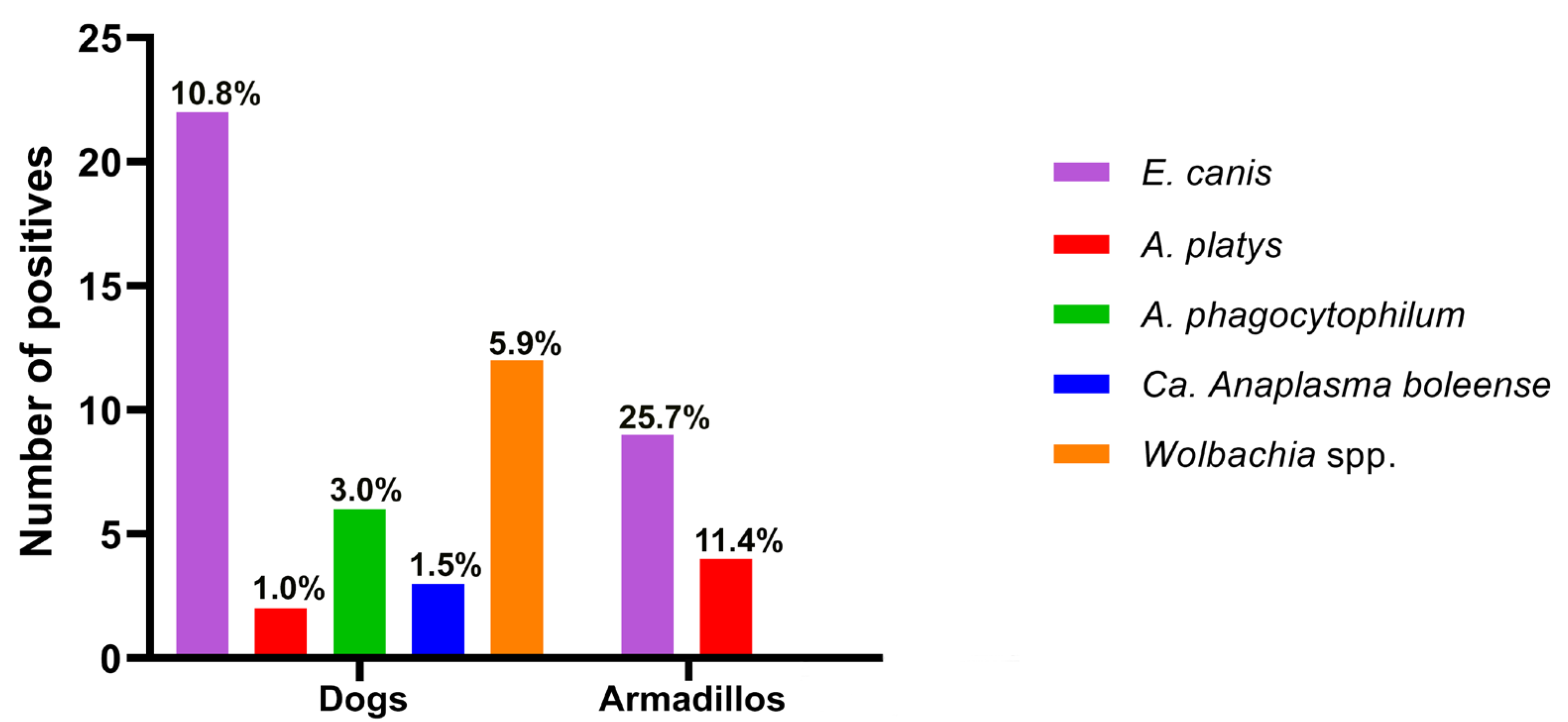
3.4. Detection and Differentiation of the Anaplasmataceae Family Using HRM in Host Blood and Ticks
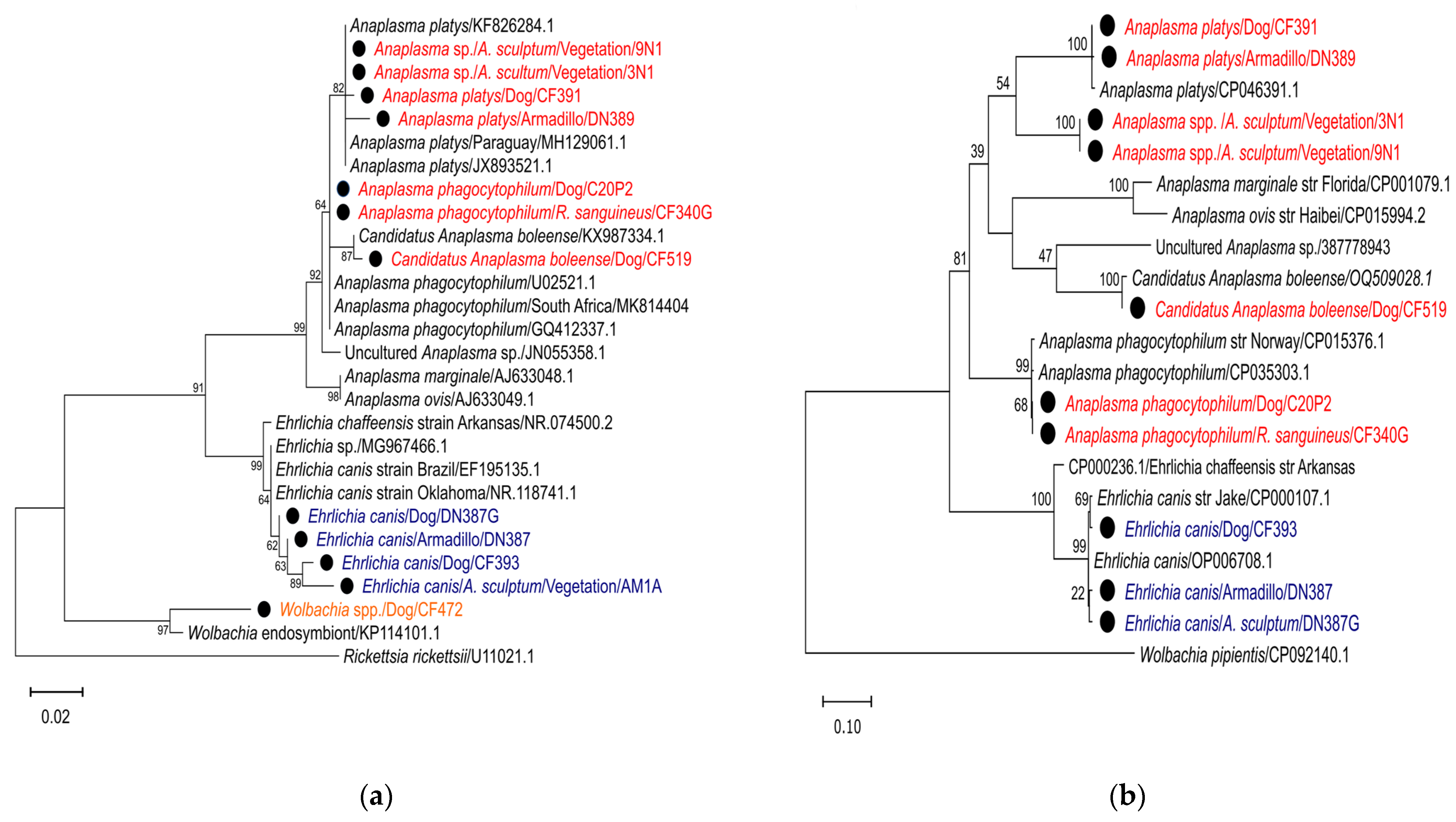
3.5. Sequence Analysis and Phylogeny of Anaplasmataceae Family
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization (WHO). Vector-Borne Diseases. 2020. Available online: https://www.who.int/news-room/fact-sheets/detail/vector-borne-diseases (accessed on 30 April 2024).
- Müller, R.; Reuss, F.; Kendrovski, V.; Montag, D. Vector-Borne Diseases. In Biodiversity and Health in the Face of Climate Change; Marselle, M., Stadler, J., Korn, H., Irvine, K., Bonn, A., Eds.; Springer: Cham, Switzerland, 2019; pp. 67–90. [Google Scholar] [CrossRef]
- Wilson, A.L.; Courtenay, O.; Kelly-Hope, L.A.; Scott, T.W.; Takken, W.; Torr, S.J.; Lindsay, S.W. The importance of vector control for the control and elimination of vector-borne diseases. PLoS Negl. Trop. Dis. 2020, 14, e0007831. [Google Scholar] [CrossRef] [PubMed]
- Marcos-Marcos, J.; de Labry-Lima, A.O.; Toro-Cardenas, S.; Lacasaña, M.; Degroote, S.; Ridde, V.; Bermudez-Tamayo, C. Impact, economic evaluation, and sustainability of integrated vector management in urban settings to prevent vector-borne diseases: A scoping review. Infect. Dis. Poverty 2018, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- De La Fuente, J.; Antunes, S.; Bonnet, S.; Cabezas-Cruz, A.; Domingos, A.G.; Estrada-Peña, A.; Johnson, N.; Kocan, K.M.; Mansfield, K.L.; Nijhof, A.M.; et al. Tick-Pathogen Interactions and Vector Competence: Identification of Molecular Drivers for Tick-Borne Diseases. Front. Cell. Infect. Microbiol. 2017, 7, 114. [Google Scholar] [CrossRef]
- Rochlin, I.; Toledo, A. Emerging tick-borne pathogens of public health importance: A mini-review. J. Med. Microbiol. 2020, 69, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Charles, R.A.; Bermúdez, S.; Banović, P.; Alvarez, D.O.; Díaz-Sánchez, A.A.; Corona-González, B.; Etter, E.M.C.; González, I.R.; Ghafar, A.; Jabbar, A.; et al. Ticks and tick-borne diseases in central america and the caribbean: A one health perspective. Pathogens 2021, 10, 1273. [Google Scholar] [CrossRef] [PubMed]
- Rar, V.; Golovljova, I. Anaplasma, Ehrlichia, and “Candidatus Neoehrlichia” bacteria: Pathogenicity, biodiversity, and molecular genetic characteristics, a review. Infect. Genet. Evol. 2011, 11, 1842–1861. [Google Scholar] [CrossRef] [PubMed]
- Cicculli, V.; DeCarreaux, D.; Ayhan, N.; Casabianca, F.; de Lamballerie, X.; Charrel, R.; Falchi, A. Molecular screening of Anaplasmataceae in ticks collected from cattle in Corsica, France. Exp. Appl. Acarol. 2020, 81, 561–574. [Google Scholar] [CrossRef] [PubMed]
- Di Cataldo, S.; Cevidanes, A.; Ulloa-Contreras, C.; Hidalgo-Hermoso, E.; Gargano, V.; Sacristán, I.; Sallaberry-Pincheira, N.; Penaloza-Madrid, D.; González-Acuña, D.; Napolitano, C.; et al. Mapping the distribution and risk factors of Anaplasmataceae in wild and domestic canines in Chile and their association with Rhipicephalus sanguineus species complex lineages. Ticks Tick Borne Dis. 2021, 12, 101752. [Google Scholar]
- Sun, J.; Liu, H.; Yao, X.Y.; Zhang, Y.Q.; Lv, Z.H.; Shao, J.W. Circulation of four species of Anaplasmataceae bacteria in ticks in Harbin, northeastern China. Ticks Tick Borne Dis. 2023, 14, 102136. [Google Scholar] [CrossRef] [PubMed]
- Muraro, L.S.; Souza, A.d.O.; Leite, T.N.S.; Cândido, S.L.; Melo, A.L.T.; Toma, H.S.; Carvalho, M.B.; Dutra, V.; Nakazato, L.; Cabezas-Cruz, A.; et al. First evidence of Ehrlichia minasensis infection in horses from Brazil. Pathogens 2021, 10, 265. [Google Scholar] [CrossRef]
- Rar, V.; Tkachev, S.; Tikunova, N. Genetic diversity of Anaplasma bacteria: Twenty years later. Infect. Genet. Evol. 2021, 91, 104833. [Google Scholar] [CrossRef] [PubMed]
- Madison-Antenucci, S.; Kramer, L.D.; Gebhardt, L.L.; Kauffman, E. Emerging Tick-Borne Diseases. Clin. Microbiol. Rev. 2020, 33, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Rodino, K.G.; Theel, E.S.; Pritt, B.S. Tick-Borne Diseases in the United States. Clin. Chem. 2020, 66, 537–548. [Google Scholar] [CrossRef] [PubMed]
- Buczek, A.; Buczek, W. Importation of Ticks on Companion Animals and the Risk of Spread of Tick-Borne Diseases to Non-Endemic Regions in Europe. Animals 2020, 11, 6. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, L. The Impacts of Climate Change on Ticks and Tick-Borne Disease Risk. Annu. Rev. Entomol. 2021, 66, 273–288. [Google Scholar] [CrossRef]
- Sanchez-Vicente, S.; Tagliafierro, T.; Coleman, J.L.; Benach, J.L.; Tokarz, R. Polymicrobial nature of tick-borne diseases. mBio 2019, 10, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Dahmani, M.; Davoust, B.; Sambou, M.; Bassene, H.; Scandola, P.; Ameur, T.; Raoult, D.; Fenollar, F.; Mediannikov, O. Molecular investigation and phylogeny of species of the Anaplasmataceae infecting animals and ticks in Senegal. Parasite Vectors 2019, 12, 495. [Google Scholar] [CrossRef] [PubMed]
- Romer, Y.; Borrás, P.; Govedic, F.; Nava, S.; Carranza, J.I.; Santini, S.; Armitano, R.; Lloveras, S. Clinical and epidemiological comparison of Rickettsia parkeri rickettsiosis, related to Amblyomma triste and Amblyomma tigrinum, in Argentina. Ticks Tick Borne Dis. 2020, 11, 101436. [Google Scholar] [CrossRef] [PubMed]
- Ogata, S.; Pereira, J.A.C.; Jhonny, L.V.A.; Carolina, H.P.G.; Matsuno, K.; Orba, Y.; Sawa, H.; Kawamori, F.; Nonaka, N.; Nakao, R. Molecular Survey of Babesia and Anaplasma Infection in Cattle in Bolivia. Vet. Sci. 2021, 8, 188. [Google Scholar] [CrossRef]
- Alcon-Chino, M.E.T.; De-Simone, S.G. Recent Advances in the Immunologic Method Applied to Tick-Borne Diseases in Brazil. Pathogens 2022, 11, 870. [Google Scholar] [CrossRef]
- Ogrzewalska, M.; Literak, I.; Martins, T.F.; Labruna, M.B. Rickettsial infections in ticks from wild birds in Paraguay. Ticks Tick Borne Dis. 2014, 5, 83–89. [Google Scholar] [CrossRef]
- Tintel, A.; José, M.; Paola, A.S. Ehrlichiosis, enfermedad transmitida por garrapatas y potencial zoonosis en Paraguay (Ehrlichiosis, tick-borne disease: A potential zoonosis in Paraguay). REDVET Rev. Electrón. Vet. 2016, 17, 1–9. [Google Scholar]
- Pérez-Macchi, S.; Pedrozo, R.; Bittencourt, P.; Müller, A. Prevalence, molecular characterization and risk factor analysis of Ehrlichia canis and Anaplasma platys in domestic dogs from Paraguay. Comp. Immunol. Microbiol. Infect. Dis. 2019, 62, 31–39. [Google Scholar] [CrossRef]
- Tintel, A.; Selich, P.A.; Rolón, M.S.; Vega Gómez, C. Detección canina de Anaplasma platys mediante PCR en tiempo real. Cienc. Lat. Rev. Científica Multidiscip. 2023, 7, 1674–1680. [Google Scholar] [CrossRef]
- Owen, R.D.; Sánchez, H.; Rodríguez, L.; Jonsson, C.B. Composition and Characteristics of a Diverse Didelphid Community (Mammalia: Didelphi-morphia) in Sub-tropical South America. Occas. Pap. Tex. Tech Univ. Mus. 2018, 2018, 358. [Google Scholar] [PubMed]
- Weaver, G.V.; Anderson, N.; Garrett, K.; Thompson, A.T.; Yabsley, M.J. Ticks and Tick-Borne Pathogens in Domestic Animals, Wild Pigs, and Off-Host Environmental Sampling in Guam, USA. Front. Vet. Sci. 2022, 8, 803424. [Google Scholar] [CrossRef] [PubMed]
- Barros-Battesti, D.M.; Arzua, M.; Bechara, G.H. Carrapatos de Importância Médico-Veterinária da Região Neotropical: Um Guia Ilustrado para Identificação de Espécies; Vox/ICTTD-3/Butantan: São Paulo, Brazil, 2006; pp. 1–223. [Google Scholar]
- Dantas-Torres, F.; Fernandes Martins, T.; Muñoz-Leal, S.; Onofrio, V.C.; Barros-Battesti, D.M. Ticks (Ixodida: Argasidae, Ixodidae) of Brazil: Updated species checklist and taxonomic keys. Ticks Tick Borne Dis. 2019, 10, 101252. [Google Scholar] [CrossRef]
- Martins, T.F.; Onofrio, V.C.; Barros-Battesti, D.M.; Labruna, M.B. Nymphs of the genus Amblyomma (Acari: Ixodidae) of Brazil: Descriptions, redescriptions, and identification key. Ticks Tick Borne Dis. 2010, 1, 75–99. [Google Scholar] [CrossRef]
- Nava, S.; Venzal, J.M.; Gonzazález-Acuña, D.; Martins, T.; Guglielmone, A. Ticks of the Southern Cone of America: Diagnosis, Distribution, and Hosts with Taxonomy, Ecology and Sanitary Importance; Academic Press: Cambridge, MA, USA, 2017; 375p, Available online: https://www.sciencedirect.com/book/9780128110751/ticks-of-the-southern-cone-of-america (accessed on 6 January 2024).
- Mangold, A.J.; Bargues, M.D.; Mas-Coma, S. Mitochondrial 16S rDNA sequences and phylogenetic relationships of species of Rhipicephalus and other tick genera among Metastriata (Acari: Ixodidae). Parasitol. Res. 1998, 84, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Adegoke, A.; Kumar, D.; Budachetri, K.; Karim, S. Hematophagy and tick-borne Rickettsial pathogen shape the microbial community structure and predicted functions within the tick vector, Amblyomma maculatum. Front. Cell. Infect. Microbiol. 2022, 12, 1037387. [Google Scholar] [CrossRef]
- Boakye, D.A.; Tang, J.; Truc, P.; Merriweather, A.; Unnasch, T.R. Identification of bloodmeals in haematophagous Diptera by cytochrome B heteroduplex analysis. Med. Vet. Entomol. 1999, 13, 282–287. [Google Scholar] [CrossRef] [PubMed]
- Parola, P.; Roux, V.; Camicas, J.L.; Baradji, I.; Brouqui, P.; Raoult, D. Detection of Ehrlichiae in African ticks by polymerase chain reaction. Trans. R. Soc. Trop. Med. Hyg. 2000, 94, 707–708. [Google Scholar] [CrossRef] [PubMed]
- Tabara, K.; Arai, S.; Kawabuchi, T.; Itagaki, A.; Ishihara, C.; Satoh, H.; Okabe, N.; Tsuji, M. Molecular survey of Babesia microti, Ehrlichia species and Candidatus neoehrlichia mikurensis in wild rodents from Shimane Prefecture, Japan. Microbiol. Immunol. 2007, 51, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Ruijter, J.M.; Benes, V.; van den Hoff, M.J.B. Web-based LinRegPCR: Application for the visualization and analysis of (RT)-qPCR amplification and melting data. BMC Bioinform. 2021, 22, 398. [Google Scholar] [CrossRef] [PubMed]
- Andreassen, A.; Jore, S.; Cuber, P.; Dudman, S.; Tengs, T.; Isaksen, K.; Hygen, H.O.; Viljugrein, H.; Ånestad, G.; Ottesen, P.; et al. Prevalence of tick borne encephalitis virus in tick nymphs in relation to climatic factors on the southern coast of Norway. Parasites Vectors 2012, 5, 177. [Google Scholar] [CrossRef]
- Sergeant, E.S.G. AusVet Animal Health Services and Australian Biosecurity Cooperative Research Centre for Emerging Infectious Disease. 2009. Available online: https://epitools.ausvet.com.au/ (accessed on 5 December 2023).
- Maggi, R.G.; Krämer, F. A review on the occurrence of companion vector-borne diseases in pet animals in Latin America. Parasites Vectors 2019, 12, 145. [Google Scholar] [CrossRef]
- Noguera Zayas, L.P.; Rüegg, S.; Torgerson, P. The burden of zoonoses in Paraguay: A systematic review. PLoS Negl. Trop. Dis. 2021, 15, e0009909. [Google Scholar] [CrossRef]
- Nava, S.; Lareschi, M.; Rebollo, C.; Usher, C.B.; Beati, L.; Robbins, R.G.; Durden, L.A.; Mangold, A.J.; Guglielmone, A.A. The ticks (Acari: Ixodida: Argasidae, Ixodidae) of Paraguay. Ann. Trop. Med. Parasitol. 2007, 101, 255–270. [Google Scholar] [CrossRef]
- Kirstein, F.; Gray, J.S. A molecular marker for the identification of the zoonotic reservoirs of Lyme borreliosis by analysis of the blood meal in its European vector Ixodes ricinus. Appl. Environ. Microbiol. 1996, 62, 4060–4065. [Google Scholar] [CrossRef]
- Pierce, K.A.; Paddock, C.D.; Sumner, J.W.; Nicholson, W.L. Pathogen prevalence and blood meal identification in Amblyomma ticks as a means of reservoir host determination for Ehrlichial pathogens. Clin. Microbiol. Infect. 2009, 15, 37–38. [Google Scholar] [CrossRef] [PubMed]
- Ngo, K.A.; Kramer, L.D. Identification of Mosquito Bloodmeals Using Polymerase Chain Reaction (PCR) with Order-Specific Primers. J. Med. Entomol. 2003, 40, 215–222. [Google Scholar] [CrossRef]
- Maleki-Ravasan, N.; Oshaghi, M.; Javadian, E.; Rassi, Y.; Sadraei, J.; Mohtarami, F. Blood Meal Identification in Field-Captured Sand flies: Comparison of PCR-RFLP and ELISA Assays. Iran. J. Arthropod. Borne Dis. 2009, 3, 8–18. [Google Scholar] [PubMed]
- Oundo, J.W.; Villinger, J.; Jeneby, M.; Ong’amo, G.; Otiende, M.Y.; Makhulu, E.E.; Musa, A.A.; Ouso, D.O.; Wambua, L. Pathogens, endosymbionts, and blood-meal sources of host-seeking ticks in the fast-changing Maasai Mara wildlife ecosystem. PLoS ONE 2020, 15, e0228366. [Google Scholar] [CrossRef]
- Hoffmann, A.; Fingerle, V.; Noll, M. Analysis of Tick Surface Decontamination Methods. Microorganisms 2020, 8, 987. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Vivas, R.I.; Apanaskevich, D.A.; Ojeda-Chi, M.M.; Trinidad-Martínez, I.; Reyes-Novelo, E.; Esteve-Gassent, M.D.; De León, A.P. Veterinary Parasitology Ticks collected from humans, domestic animals, and wildlife in Yucatan, Mexico. Vet. Parasitol. 2016, 215, 106–113. [Google Scholar] [CrossRef]
- Buddhachat, K.; Meerod, T.; Pradit, W.; Siengdee, P.; Chomdej, S.; Nganvongpanit, K. Simultaneous differential detection of canine blood parasites: Multiplex high-resolution melting analysis (mHRM). Ticks Tick Borne Dis. 2020, 11, 101370. [Google Scholar] [CrossRef]
- Rodino, K.G.; Wolf, M.J.; Sheldon, S.; Kingry, L.C.; Petersen, J.M.; Patel, R.; Pritt, B.S. Detection of Tick-Borne Bacteria from Whole Blood Using 16S. J. Clin. Microbiol. 2021, 59, 10–1128. [Google Scholar] [CrossRef]
- Reed, G.H.; Kent, J.O.; Wittwer, C.T. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics 2007, 8, 597–608. [Google Scholar] [CrossRef]
- Higuera, S.L.; Guhl, F.; Ramírez, J.D. Identification of Trypanosoma cruzi Discrete Typing Units (DTUs) through the implementation of a High-Resolution Melting (HRM) genotyping assay. Parasites Vectors 2013, 6, 112. [Google Scholar] [CrossRef]
- Zampieri, R.A.; Laranjeira-Silva, M.F.; Muxel, S.M.; Stocco de Lima, A.C.; Shaw, J.J.; Floeter-Winter, L.M. High Resolution Melting Analysis Targeting hsp70 as a Fast and Efficient Method for the Discrimination of Leishmania Species. PLoS Negl. Trop. Dis. 2016, 10, e0004485. [Google Scholar] [CrossRef]
- Kagkli, D.M.; Folloni, S.; Barbau-Piednoir, E.; van den Eede, G.; van den Bulcke, M. Towards a pathogenic Escherichia coli detection platform using multiplex SYBR®green real-time PCR methods and high-resolution melting analysis. PLoS ONE 2012, 7, e39287. [Google Scholar] [CrossRef]
- Goldschmidt, P.; Degorge, S.; Che Sarria, P.; Benallaoua, D.; Semoun, O.; Borderie, V.; Laroche, L.; Chaumeil, C. New strategy for rapid diagnosis and characterization of fungal infections: The example of corneal scrapings. PLoS ONE 2012, 7, e37660. [Google Scholar] [CrossRef] [PubMed]
- Barrera-Illanes, A.N.; Micieli, M.V.; Ibáñez-Shimabukuro, M.; Santini, M.S.; Martins, A.J.; Ons, S. First report on knockdown resistance mutations in wild populations of Aedes aegypti from Argentina determined by a novel multiplex high-resolution melting polymerase chain reaction method. Parasites Vectors 2023, 16, 222. [Google Scholar] [CrossRef]
- Harrus, S.; Waner, T. Diagnosis of canine monocytotropic ehrlichiosis (Ehrlichia canis): An overview. Vet. J. 2011, 187, 292–296. [Google Scholar] [CrossRef]
- Nava, S.; Mastropaolo, M.; Venzal, J.M. Veterinary Parasitology Mitochondrial DNA analysis of Rhipicephalus sanguineus sensu lato (Acari: Ixodidae) in the Southern Cone of South America. Vet. Parasitol. 2012, 190, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Sykes, J.E. Canine and Feline Infectious Diseases; Sykes, J.E., Ed.; W.B. Saunders: Saint Louis, MO, USA, 2014; Chapter 28—Ehrlichiosis; p. 278. [Google Scholar]
- Shilton, C.; Foster, A.; Reid, T.; Keyburn, A.; Besier, S. Detection of Ehrlichia canis in WA and the NT. Scope Off. Newsletter Aust. Soc. Vet. Pathol. 2020, 1, 12–16. [Google Scholar]
- André, M.R. Diversity of Anaplasma and Ehrlichia/Neoehrlichia Agents in terrestrial wild carnivores worldwide: Implications for human and domestic animal health and wildlife conservation. Front. Vet. Sci. 2018, 5, 293. [Google Scholar] [CrossRef] [PubMed]
- Perez, M.; Rikihisa, Y.; Wen, B. Ehrlichia canis-like agent isolated from a man in Venezuela: Antigenic and genetic characterization. J. Clin. Microbiol. 1996, 34, 2133–2139. [Google Scholar] [CrossRef]
- Perez, M.; Bodor, M.; Zhang, C.; Xiong, Q.; Rikihisa, Y. Human Infection with Ehrlichia canis Accompanied by Clinical Signs in Venezuela. Ann. N. Y. Acad. Sci. 2006, 1078, 110–117. [Google Scholar] [CrossRef]
- Bouza-Mora, L.; Dolz, G.; Solórzano-Morales, A.; Romero-Zuñiga, J.J.; Salazar-Sánchez, L.; Labruna, M.B.; Aguiar, D.M. Novel genotype of Ehrlichia canis detected in samples of human blood bank donors in Costa Rica. Ticks Tick Borne Dis. 2017, 8, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Snellgrove, A.N.; Krapiunaya, I.; Ford, S.L.; Stanley, H.M.; Wickson, A.G.; Hartzer, K.L.; Levin, M.L. Vector competence of Rhipicephalus sanguineus sensu stricto for Anaplasma platys. Ticks Tick Borne Dis. 2020, 11, 101517. [Google Scholar] [CrossRef] [PubMed]
- Dantas-Torres, F. Canine vector-borne diseases in Brazil. Parasites Vectors 2008, 1, 25. [Google Scholar] [CrossRef] [PubMed]
- Dumic, I.; Jevtic, D.; Veselinovic, M.; Nordstrom, C.W.; Jovanovic, M.; Mogulla, V.; Veselinovic, E.M.; Hudson, A.; Simeunovic, G.; Petcu, E.; et al. Human Granulocytic Anaplasmosis—A Systematic Review of Published Cases. Microorganisms 2022, 10, 1433. [Google Scholar] [CrossRef] [PubMed]
- Stuen, S.; Granquist, E.G.; Silaghi, C. Anaplasma phagocytophilum—A widespread multi-host pathogen with highly adaptive strategies. Front. Cell. Infect. Microbiol. 2013, 3, 31. [Google Scholar] [CrossRef] [PubMed]
- El Hamiani Khatat, S.; Daminet, S.; Duchateau, L.; Elhachimi, L.; Kachani, M.; Sahibi, H. Epidemiological and Clinicopathological Features of Anaplasma phagocytophilum Infection in Dogs: A Systematic Review. Front. Vet. Sci. 2021, 8, 686644. [Google Scholar] [CrossRef]
- Kowalewski, M.; Pontón, F.; Ramírez Pinto, F.; Velázquez, M.C. Estado sanitario de perros en áreas de interfase entre animales domésticos y silvestres de la Reserva Natural del Bosque Mbaracayú, Paraguay. Rev. Soc. Cient. Parag. 2019, 24, 114–125. [Google Scholar] [CrossRef]
- Kang, Y.-J.; Diao, X.-N.; Zhao, G.-Y.; Chen, M.-H.; Xiong, Y.; Shi, M.; Fu, W.-M.; Guo, Y.-J.; Pan, B.; Chen, X.-P.; et al. Extensive diversity of Rickettsiales bacteria in two species of ticks from China and the evolution of the Rickettsiales. BMC Evol. Biol. 2014, 14, 167. [Google Scholar] [CrossRef]
- Guo, W.-P.; Tian, J.-H.; Lin, X.-D.; Ni, X.-B.; Chen, X.-P.; Liao, Y.; Yang, S.-Y.; Dumler, J.S.; Holmes, E.C.; Zhang, Y.-Z. Extensive genetic diversity of Rickettsiales bacteria in multiple mosquito species. Sci. Rep. 2016, 6, 38770. [Google Scholar] [CrossRef]
- Koh, F.X.; Panchadcharam, C.; Sitam, F.T.; Tay, S.T. Molecular investigation of Anaplasma spp. in domestic and wildlife animals in Peninsular Malaysia. Vet. Parasitol. Reg. Stud. Rep. 2018, 13, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Hailemariam, Z.; Krücken, J.; Baumann, M.; Ahmed, J.S.; Clausen, P.H.; Nijhof, A.M. Molecular detection of tick-borne pathogens in cattle from Southwestern Ethiopia. PLoS ONE 2017, 12, e0188248. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, S.d.J.; Matos, C.A.; Freschi, C.R.; de Souza Ramos, I.A.; Machado, R.Z.; André, M.R. Diversity of Anaplasma species in cattle in Mozambique. Ticks Tick Borne Dis. 2019, 10, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Kolo, A.O.; Collins, N.E.; Brayton, K.A.; Chaisi, M.; Blumberg, L.; Frean, J.; Gall, C.A.; MWentzel, J.; Wills-Berriman, S.; Boni, L.D.; et al. Anaplasma phagocytophilum and Other Anaplasma spp. in Various Hosts in the Mnisi Community, Mpumalanga Province, South Africa. Microorganisms 2020, 8, 1812. [Google Scholar] [CrossRef] [PubMed]
- Orozco, M.M.; Argibay, H.D.; Minatel, L.; Guillemi, E.C.; Berra, Y.; Schapira, A.; Di Nucci, D.; Marcos, A.; Lois, F.; Falzone, M.; et al. A participatory surveillance of marsh deer (Blastocerus dichotomus) morbidity and mortality in Argentina: First results. BMC Vet. Res. 2020, 16, 321. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, P.S.; Panizza, M.N.M.; Ríos, I.J.M.G.; Tarragona, E.L.; Trova, G.B.; Negrette, O.S.; Primo, M.E.; Nava, S. Molecular detection and phylogenetic analysis of Anaplasma platys-like and Candidatus Anaplasma boleense strains from Argentina. Comp. Immunol. Microbiol. Infect. Dis. 2023, 96, 101980. [Google Scholar] [CrossRef] [PubMed]
- Winter, M.; Sebastian, P.S.; Tarragona, E.L.; Flores, F.S.; Abate, S.D.; Nava, S. Tick-borne microorganisms in Amblyomma tigrinum (Acari: Ixodidae) from the Patagonian region of Argentina. Exp. Appl. Acarol. 2024, 92, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Silva, E.M.C.; Marques, I.C.L.; de Mello, V.V.C.; do Amaral, R.B.; Gonçalves, L.R.; Braga, M.D.S.C.O.; dos Santos Ribeiro, L.S.; Machado, R.Z.; André, M.R.; de Carvalho Neta, A.V. Molecular and serological detection of Anaplasma spp. in small ruminants in an area of Cerrado Biome in northeastern Brazil. Ticks Tick Borne Dis. 2024, 15, 102254. [Google Scholar] [CrossRef] [PubMed]
- Thinnabut, K.; Rodpai, R.; Sanpool, O.; Maleewong, W.; Tangkawanit, U. Genetic diversity of tick (Acari: Ixodidae) populations and molecular detection of Anaplasma and Ehrlichia infesting beef cattle from upper-northeastern Thailand. Infect. Genet. Evol. 2023, 107, 105394. [Google Scholar] [CrossRef]
- Laidoudi, Y.; Marié, J.L.; Tahir, D.; Watier-Grillot, S.; Mediannikov, O.; Davoust, B. Detection of Canine Vector-Borne Filariasis and Their Wolbachia Endosymbionts in French Guiana. Microorganisms 2020, 8, 770. [Google Scholar] [CrossRef]


| Type of Habitat (n) | Hosts n (%) | Ticks (n) | Life Cycle | GenBank Accession Number | |||
|---|---|---|---|---|---|---|---|
| F | M | N | L | ||||
| Dog (203) | 23 (11.3) | R. sanguineus (24) | 10 | 6 | 8 | - | OR976129, OR976130 (1F, 1N) |
| A. ovale (2) | 2 | - | - | - | OR976131 (1F) | ||
| Armadillo (35) | 13 (37.1) | A. sculptum (39) | 21 | - | 18 | - | OR976133, OR976134 (1F, 1N) |
| A. coelebs (4) | - | - | 4 | - | OR976132 (1N) | ||
| Free-living (1855) | A. sculptum (808) | 46 | 24 | 738 | - | OR976135, OR976136, OR976137 (1F, 2N) | |
| Amblyomma spp. (1047) | 0 | 0 | 0 | 1047 | |||
| n: 1924 | 79 | 30 | 768 | 1047 | |||
| Ticks (n) | Life Cycle (n) | Total Detected | Blood Meal Source | |||
|---|---|---|---|---|---|---|
| Rodent | Human | Armadillo | Dog | |||
| A.sculptum (57) | Female (32) | 12 | 10 | - | 1 | 1 |
| Nymph a (25) | 10 | - | 5 | 3 | 2 | |
| Amblyomma spp. (7) | Larvae b (7) | - | - | - | - | - |
| n: 64 | Total (n/%) | 22 (34.4) | 10 (45.5) | 5 (22.7) | 4 (18.2) | 3 (13.6) |
| Host/Habitat (n) | Ticks (n) | Life Cycle (n) | No. of Ticks Infected with Bacteria Anaplasmataceae | Infected/ Collected (%) | ||||
|---|---|---|---|---|---|---|---|---|
| Ehrlichia canis | Anaplasma platys | Anaplasma phago. | Ca. Anaplasma boleense | Anaplasma spp. | ||||
| Dogs (26) | R. sanguineus (24) | Female (10) | - | 2 | 2 | 1 | - | 9/26 (34.6) |
| Male (6) | 4 | - | - | - | - | |||
| Nymph (8) | - | - | - | - | - | |||
| A. ovale (2) | Female (2) | - | - | - | - | - | ||
| Armadillo (43) | A.coelebs (4) | Nymph (4) | - | - | - | - | - | 10/43 (23.3) |
| A.sculptum (39) | Female (21) | 6 | - | - | - | - | ||
| Nymph (18) | 4 | - | - | - | - | |||
| Free-living (70) | A.sculptum (70) | Female (46) | 5 | - | - | - | 8 | 15/70 (21.4) |
| Male (24) | - | - | - | - | 2 | |||
| n: 139 | Total (n/%) | 19 (13.7) | 2 (1.4) | 2 (1.4) | 1 (0.7) | 10 (7.2) | 34 (24.5) | |
| Free-Living Ticks (Life Cycle Stage) | No. of Pools | Anaplasmataceae Positive (Minimum Infection Rate %) | ||||
|---|---|---|---|---|---|---|
| Ehrlichia canis | Anaplasma platys | Anaplasma phago | Ca. Anaplasma boleense | Anaplasma spp. | ||
| A.sculptum (nymphs a) | 4 | 1 (1.25) | - | - | - | - |
| 10 | - | - | - | - | 2 (1.0) | |
| 21 | - | - | - | - | - | |
| Amblyomma spp. (larvae b) | 2 | - | - | - | - | - |
| 7 | - | - | - | - | - | |
| Sample Code | Sources | Gene | GenBank Accession Number | Species | Closest Match Accession Number, Similarity (%) |
|---|---|---|---|---|---|
| CF393 | Canis familiaris | 16S rRNA | OR976118 | Ehrlichia canis | MN922610.1, (99.13) |
| groEL | PP928796 | OP006713.1, (99.72) | |||
| DN387 | Armadillo | 16S rRNA | OR976119 | Ehrlichia canis | MN922610.1, (99.71) |
| groEL | PP928797 | OP006708.1, (99.15) | |||
| DN387G | A. sculptum a | 16S rRNA | OR976120 | Ehrlichia canis | MN922610.1, (100.0) |
| groEL | PP928798 | OP006708.1, (99.15) | |||
| CF391 | Canis familiaris | 16S rRNA | OR976123 | Anaplasma platys | MN630836.1, (99.13) |
| groEL | PP928804 | CP046391.1, (99.43) | |||
| DN389 | Armadillo | 16S rRNA | OR976122 | Anaplasma platys | MN630836.1, (99.10) |
| groEL | PP928805 | CP046391.1, (99.14) | |||
| 3N1 | A. sculptum/nymph b | 16S rRNA | PP930511 | Anaplasma spp. | MT019534.1, (100.0) |
| groEL | PP928799 | LC381241.1, (85.01) | |||
| 9N1 | A. sculptum/nymph b | 16S rRNA | PP930512 | Anaplasma spp. | MT019534.1, (100.0) |
| groEL | PP928799 | LC381241.1, (85.01) | |||
| C20P2 | Canis familiaris | 16S rRNA | OR976125 | Anaplasma phagocytophilum | CP015376.1, (99.42) |
| groEL | PP928801 | OQ453812.1, (100.0) | |||
| C20P2G | R. sanguineus c | 16S rRNA | OR976126 | Anaplasma phagocytophilum | CP015376.1, (99.42) |
| groEL | PP928802 | OQ453812.1, (100.0) | |||
| CF519 | Canis familiaris | 16S rRNA | OR976128 | Ca. A. boleense | KX987335.1, (99.71) |
| groEL | PP928803 | OQ509028.1, (99.14) | |||
| CF472 | Canis familiaris | 16S rRNA | OR976117 | Wolbachia spp. | MT792375.1, (99.13) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salvioni Recalde, O.D.; Rolón, M.S.; Velázquez, M.C.; Kowalewski, M.M.; Alfonso Ruiz Diaz, J.J.; Rojas de Arias, A.; Moraes, M.O.; Magdinier Gomes, H.; de Azevedo Baêta, B.; Dias Cordeiro, M.; et al. Diversity of Anaplasmataceae Transmitted by Ticks (Ixodidae) and the First Molecular Evidence of Anaplasma phagocytophilum and Candidatus Anaplasma boleense in Paraguay. Microorganisms 2024, 12, 1893. https://doi.org/10.3390/microorganisms12091893
Salvioni Recalde OD, Rolón MS, Velázquez MC, Kowalewski MM, Alfonso Ruiz Diaz JJ, Rojas de Arias A, Moraes MO, Magdinier Gomes H, de Azevedo Baêta B, Dias Cordeiro M, et al. Diversity of Anaplasmataceae Transmitted by Ticks (Ixodidae) and the First Molecular Evidence of Anaplasma phagocytophilum and Candidatus Anaplasma boleense in Paraguay. Microorganisms. 2024; 12(9):1893. https://doi.org/10.3390/microorganisms12091893
Chicago/Turabian StyleSalvioni Recalde, Oscar Daniel, Miriam Soledad Rolón, Myriam Celeste Velázquez, Martin M. Kowalewski, Jorge Javier Alfonso Ruiz Diaz, Antonieta Rojas de Arias, Milton Ozório Moraes, Harrison Magdinier Gomes, Bruna de Azevedo Baêta, Matheus Dias Cordeiro, and et al. 2024. "Diversity of Anaplasmataceae Transmitted by Ticks (Ixodidae) and the First Molecular Evidence of Anaplasma phagocytophilum and Candidatus Anaplasma boleense in Paraguay" Microorganisms 12, no. 9: 1893. https://doi.org/10.3390/microorganisms12091893






