Changes in Cyanobacterial Phytoplankton Communities in Lake-Water Mesocosms Treated with Either Glucose or Hydrogen Peroxide
Abstract
1. Introduction
2. Material and Methods
2.1. Muskegon Lake and the AWRI Research Facility
2.2. Experimental Design
2.3. Mesocosm Sampling and Amplicon Sequencing
2.4. Amplicon Processing
2.5. Sequence Data Analysis
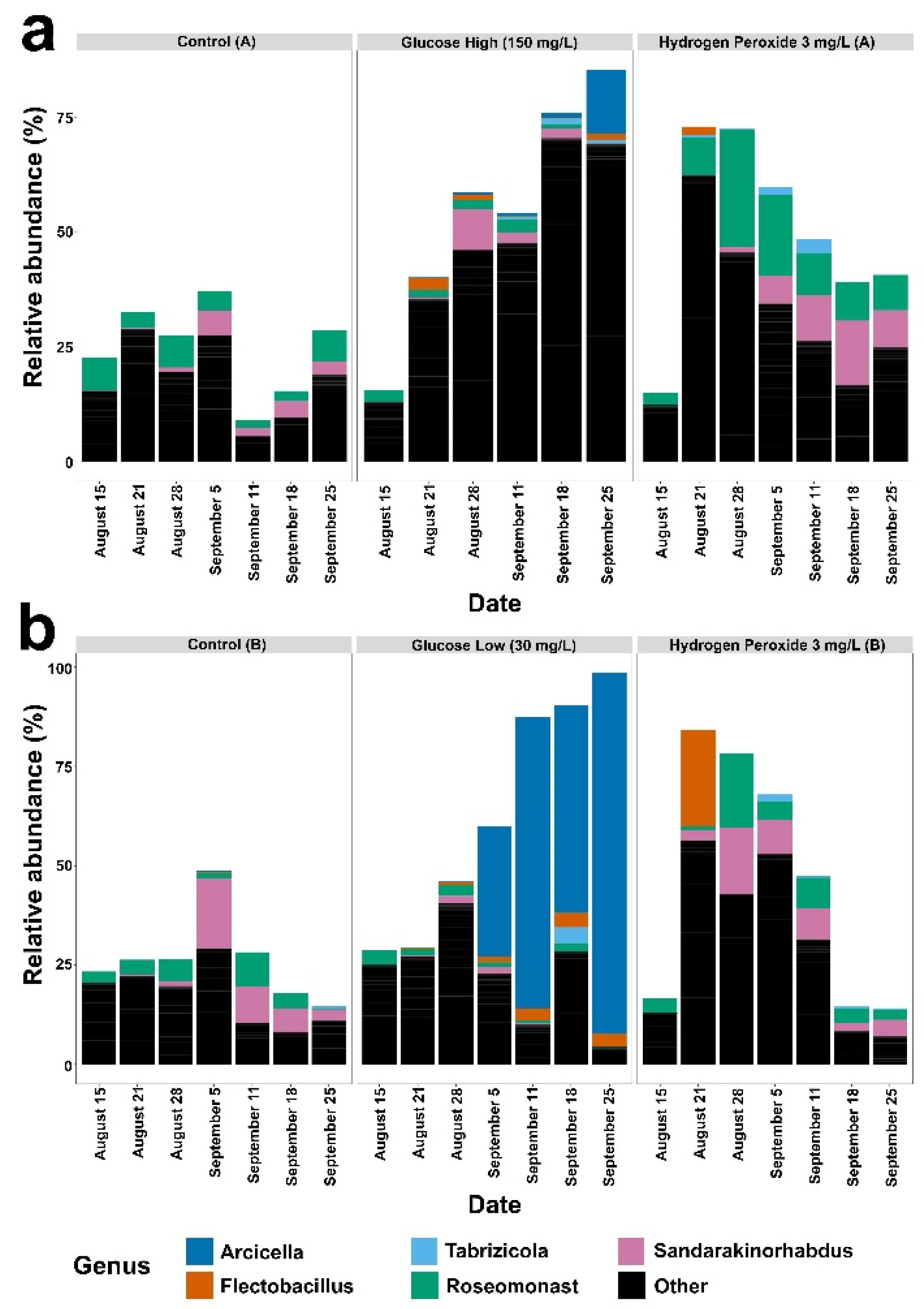
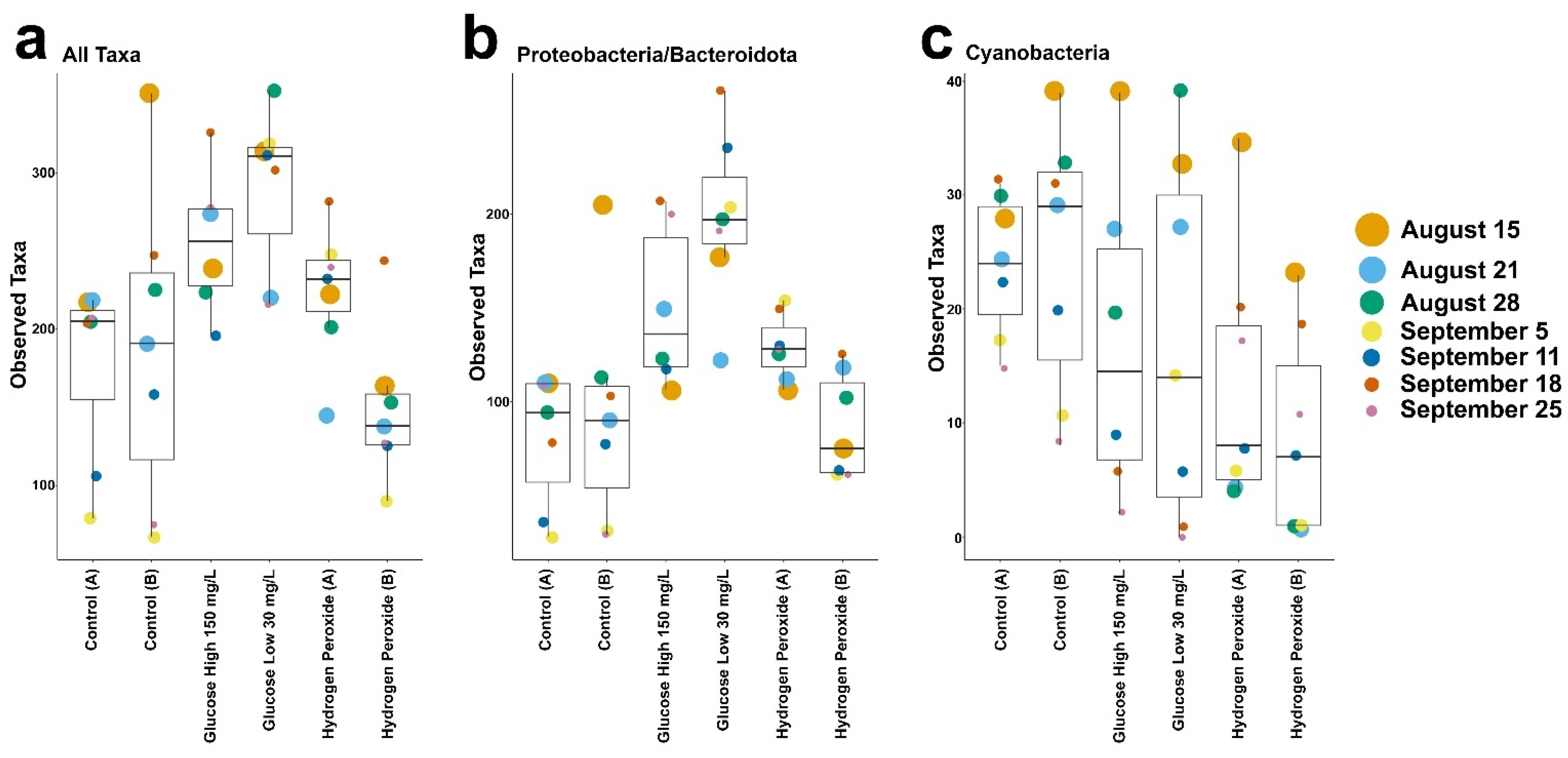
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Elser, J.J.; Bracken, M.E.; Cleland, E.E.; Gruner, D.S.; Harpole, W.S.; Hillebrand, H.; Ngai, J.T.; Seabloom, E.W.; Shurin, J.B.; Smith, J.E. Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol. Lett. 2007, 10, 1135–1142. [Google Scholar] [CrossRef]
- Fu, H.; Uchimiya, M.; Gore, J.; Moran, M.A. Ecological drivers of bacterial community assembly in synthetic phycospheres. Proc. Natl. Acad. Sci. USA 2020, 117, 3656–3662. [Google Scholar] [CrossRef] [PubMed]
- Wurtsbaugh, W.A.; Paerl, H.W.; Dodds, W.K. Nutrients, eutrophication and harmful algal blooms along the freshwater to marine continuum. Wiley Interdiscipl. Rev. Water 2019, 6, e1373. [Google Scholar] [CrossRef]
- Canfield, D.E., Jr.; Bachmann, R.W.; Hoyer, M.V. Restoration of Lake Okeechobee, Florida: Mission impossible? Lake Reserv. Manag. 2020, 37, 95–111. [Google Scholar] [CrossRef]
- Phlips, E.J.; Badylak, S.; Mathews, A.L.; Milbrandt, E.C.; Montefiore, L.R.; Morrison, E.S.; Stelling, B. Algal blooms in a river-dominated estuary and nearshore region of Florida, USA: The influence of regulated discharges from water control structures on hydrologic and nutrient conditions. Hydrobiologia 2023, 850, 4385–4411. [Google Scholar] [CrossRef]
- Summers, E.J.; Ryder, J.L. A critical review of operational strategies for the management of harmful algal blooms (HABs) in inland reservoirs. J. Environ. Manag. 2023, 330, 117141. [Google Scholar] [CrossRef]
- Merder, J.; Harris, T.; Zhao, G.; Stasinopoulos, D.M.; Ribby, R.A.; Michalak, A.M. Geographic redistribution of microcystin hotspots in response to climate warming. Nat. Water 2023, 1, 844–854. [Google Scholar] [CrossRef]
- Greenfield, D.I.; Duquette, A.; Goodson, A.; Keppler, C.J.; Williams, S.H.; Brock, L.M.; Stackley, K.D.; White, D.; Wilde, S.B. The effects of three chemical algaecides on cell numbers and toxin content of the cyanobacteria Microcystis aeruginosa and Anabaenopsis sp. Environ. Manag. 2014, 54, 1110–1120. [Google Scholar] [CrossRef]
- Moreno-Andrés, J.; Rivas-Zaballos, I.; Acevedo-Merino, A.; Nebot, E. On the efficacy of H2O2 or S2O82- at promoting the inactivation of a consortium of cyanobacteria and bacteria in algae-laden water. Microorganisms 2022, 10, 735. [Google Scholar] [CrossRef]
- Vesper, S.; Sienkiewicz, N.; Struewing, I.; Linz, D.; Lu, J. Prophylactic addition of glucose suppresses cyanobacterial abundance in lake water. Life 2022, 12, 385. [Google Scholar] [CrossRef]
- Linz, D.; Struewing, I.; Sienkiewicz Steinman, A.D.; Partridge, C.G.; McIntosh, K.; Allen, J.; Lu, J.; Vesper, S. Periodic addition of glucose suppressed cyanobacterial abundance in additive lake water samples during the entire bloom season. J. Water Resour. Prot. 2024, 16, 140–155. [Google Scholar] [CrossRef] [PubMed]
- Steinman, A.D.; Ogdahl, M.; Rediske, R.; Ruetz, C.R., III; Biddanda, B.A.; Nemeth, L. Current status and trends in Muskegon Lake, Michigan. J. Great Lakes Res. 2008, 34, 169–188. [Google Scholar] [CrossRef]
- Marko, K.M.; Rutherford, E.S.; Eadie, B.J.; Johengen, T.H.; Lansing, M.B. Delivery of nutrients and seston from the Muskegon River Watershed to near shore Lake Michigan. J. Great Lakes Res. 2013, 39, 672–681. [Google Scholar] [CrossRef]
- Mader, M.M.; Ruetz, C.R.; Woznicki, S.A.; Steinman, A.D. Land cover and water quality of drowned river mouths: Evidence of an environmental gradient along the eastern Lake Michigan shoreline. J. Great Lakes Res. 2023, 49, 102237. [Google Scholar] [CrossRef]
- Bagley, M.; Pilgrim, E.; Knapp, M.; Yoder, C.; Domingo, J.S.; Banerji, A. High-throughput environmental DNA analysis informs a biological assessment of an urban stream. Ecol. Indic. 2019, 104, 378–389. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotech. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web- based tools. Nucleic Acids Res 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Yilmaz, P.; Parfrey, L.W.; Yarza, P.; Gerken, J.; Pruesse, E.; Quast, C.; Schweer, T.; Peplies, J.; Ludwig, W.; Glöckner, F.O. The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res 2014, 42, D643–D648. [Google Scholar] [CrossRef]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2021. Available online: https://www.R-project.org/ (accessed on 6 March 2023).
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Stevens, M.H.H. Vegan: Community Ecology Package. R package, version 2.5–7; Scientific Research, Inc.: Glendale, CA, USA, 2018.
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. Available online: https://ggplot2.tidyverse.org (accessed on 7 July 2023).
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Aus. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Harke, M.J.; Steffen, M.M.; Gobler, C.J.; Otten, T.G.; Wilhelm, S.W.; Wood, S.A.; Paerl, H.W. A review of the global ecology, genomics, and biogeography of the toxic cyanobacterium, microcystis spp. Harmful Algae 2016, 54, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Urakawa, H.; Steele, J.H.; Hancock, T.L.; Dahedl, E.K.; Schroeder, E.R.; Sereda, J.V.; Kratz, M.A.; García, P.E.; Armstrong, R.A. Interaction among spring phytoplankton succession, water discharge patterns, and hydrogen peroxide dynamics in the Caloosahatchee River in southwest Florida. Harmful Algae 2023, 126, 102434. [Google Scholar] [CrossRef] [PubMed]
- Gillett, N.D.; Steinman, A.D. An analysis of long-term phytoplankton dynamics in Muskegon Lake, a Great Lakes Area of Concern. J. Great Lakes Res. 2011, 37, 335–342. [Google Scholar] [CrossRef]
- Alegría-Gómez, J.; Castañón-González, J.H.; Hernández-García, J.A.; González-Terreros, E.; Velázquez-Ríos, I.O.; Ruíz-Valdiviezo, V.M. Changes in the abundance and diversity of bacterial and archaeal communities at different depths in a eutrophic freshwater lake in southwestern Mexico. Environ. Sci. Pollut. Res. Int. 2023, 30, 98362–98376. [Google Scholar] [CrossRef]
- Sanseverino, I.; Pretto, P.; António, D.C.; Lahm, A.; Facca, C.; Loos, R.; Skejo, H.; Beghi, A.; Pandolfi, F.; Genoni, P.; et al. Metagenomics analysis to investigate the microbial communities and their functional profile during cyanobacterial blooms in Lake Varese. Microb. Ecol. 2022, 83, 850–868. [Google Scholar] [CrossRef]
- Struewing, I.; Sienkiewicz, N.; Zhang, C.; Dugan, N.; Lu, J. Effective early treatment of Microcystis exponential growth and microcystin production with hydrogen peroxide and hydroxyapatite. Toxins 2022, 15, 3. [Google Scholar] [CrossRef]
- Piel, T.; Sandrini, G.; Weenink, E.F.J.; Qin, H.; Herk, M.J.V.; Morales-Grooters, M.L.; Schuurmans, J.M.; Slot, P.C.; Wijn, G.; Arntz, J.; et al. Shifts in phytoplankton and zooplankton communities in three cyanobacteria-dominated lakes after treatment with hydrogen peroxide. Harmful Algae 2024, 133, 102585. [Google Scholar] [CrossRef]
- Chen, C.; Wang, Y.; Chen, K.; Shi, X.; Yang, G. Using hydrogen peroxide to control cyanobacterial blooms: A mesocosm study focused on the effects of algal density in Lake Chaohu, China. Environ. Pollut. 2021, 272, 115923. [Google Scholar] [CrossRef]
- Huang, J.; Ghaly, M.; Hobson, P.; Chow, C.W.K. Innovative method of utilising hydrogen peroxide for source water management of cyanobacteria. Environ. Sci. Pollut. Res. Int. 2022, 29, 22651–22660. [Google Scholar] [CrossRef] [PubMed]
- Lusty, M.W.; Gobler, C.J. The efficacy of hydrogen peroxide in mitigating cyanobacterial blooms and altering microbial communities across four lakes in NY, USA. Toxins 2020, 12, 428. [Google Scholar] [CrossRef] [PubMed]
- Anantapantula, S.S.; Wilson, A.E. Most treatments to control freshwater algal blooms are not effective: Meta-analysis of field experiments. Water Res. 2023, 243, 120342. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Linz, D.; Partridge, C.G.; Hassett, M.C.; Sienkiewicz, N.; Tyrrell, K.; Henderson, A.; Tardani, R.; Lu, J.; Steinman, A.D.; Vesper, S. Changes in Cyanobacterial Phytoplankton Communities in Lake-Water Mesocosms Treated with Either Glucose or Hydrogen Peroxide. Microorganisms 2024, 12, 1925. https://doi.org/10.3390/microorganisms12091925
Linz D, Partridge CG, Hassett MC, Sienkiewicz N, Tyrrell K, Henderson A, Tardani R, Lu J, Steinman AD, Vesper S. Changes in Cyanobacterial Phytoplankton Communities in Lake-Water Mesocosms Treated with Either Glucose or Hydrogen Peroxide. Microorganisms. 2024; 12(9):1925. https://doi.org/10.3390/microorganisms12091925
Chicago/Turabian StyleLinz, David, Charlyn G. Partridge, Michael C. Hassett, Nathan Sienkiewicz, Katie Tyrrell, Aimèe Henderson, Renee Tardani, Jingrang Lu, Alan D. Steinman, and Stephen Vesper. 2024. "Changes in Cyanobacterial Phytoplankton Communities in Lake-Water Mesocosms Treated with Either Glucose or Hydrogen Peroxide" Microorganisms 12, no. 9: 1925. https://doi.org/10.3390/microorganisms12091925
APA StyleLinz, D., Partridge, C. G., Hassett, M. C., Sienkiewicz, N., Tyrrell, K., Henderson, A., Tardani, R., Lu, J., Steinman, A. D., & Vesper, S. (2024). Changes in Cyanobacterial Phytoplankton Communities in Lake-Water Mesocosms Treated with Either Glucose or Hydrogen Peroxide. Microorganisms, 12(9), 1925. https://doi.org/10.3390/microorganisms12091925




