Root Endophytic Microorganisms Contribute to the Attribute of Full-Year Shooting in Woody Bamboo Cephalostachyum pingbianense
Abstract
1. Introduction
2. Materials and Methods
2.1. Root Sample Collection and Surface Sterilization
2.2. DNA Extraction and Illumina Sequencing
2.3. Data Processing and Analysis
3. Results
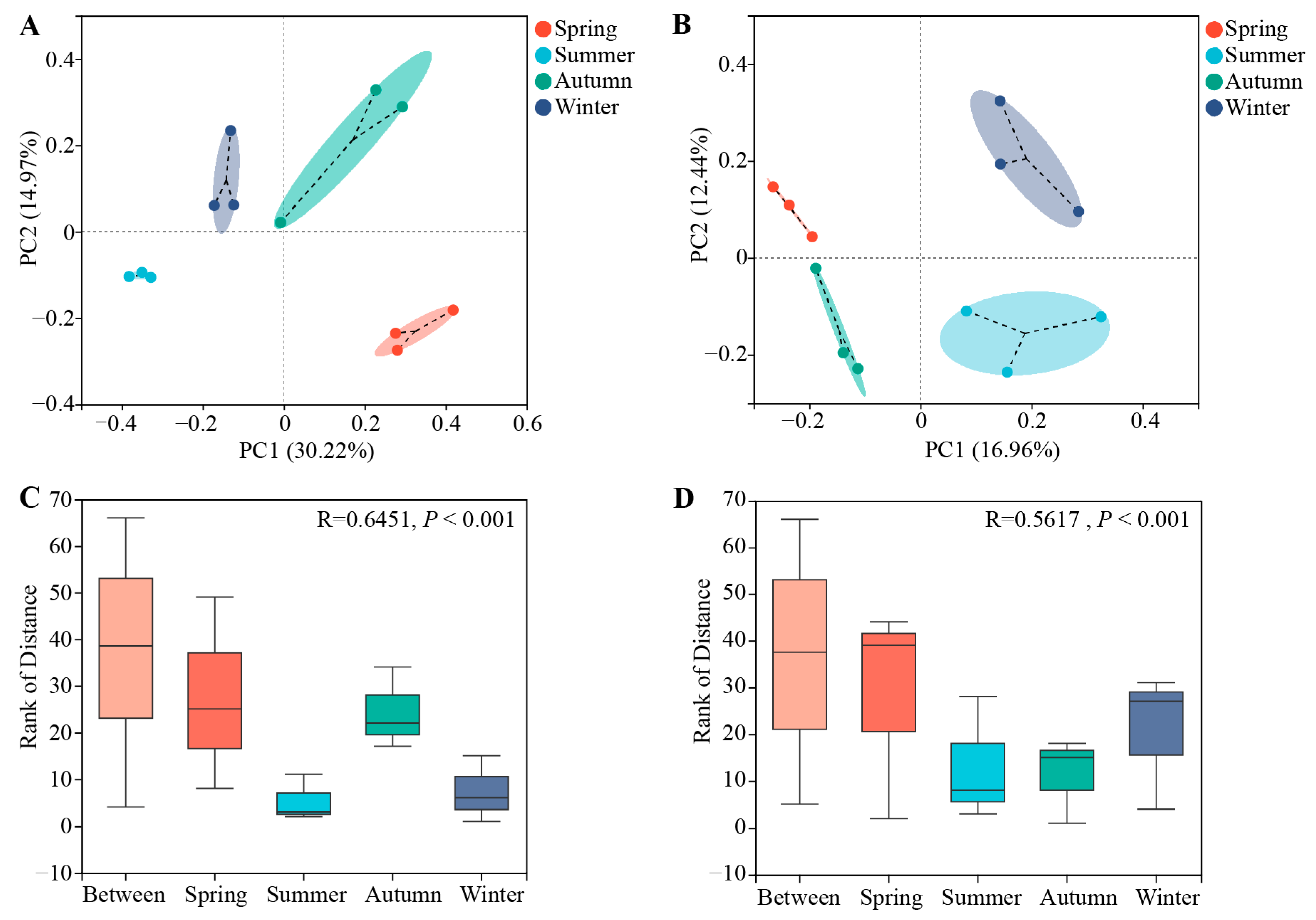
3.1. Community Diversities of Bacterial and Fungal Root Endophytes in C. pingbianense across Different Seasons
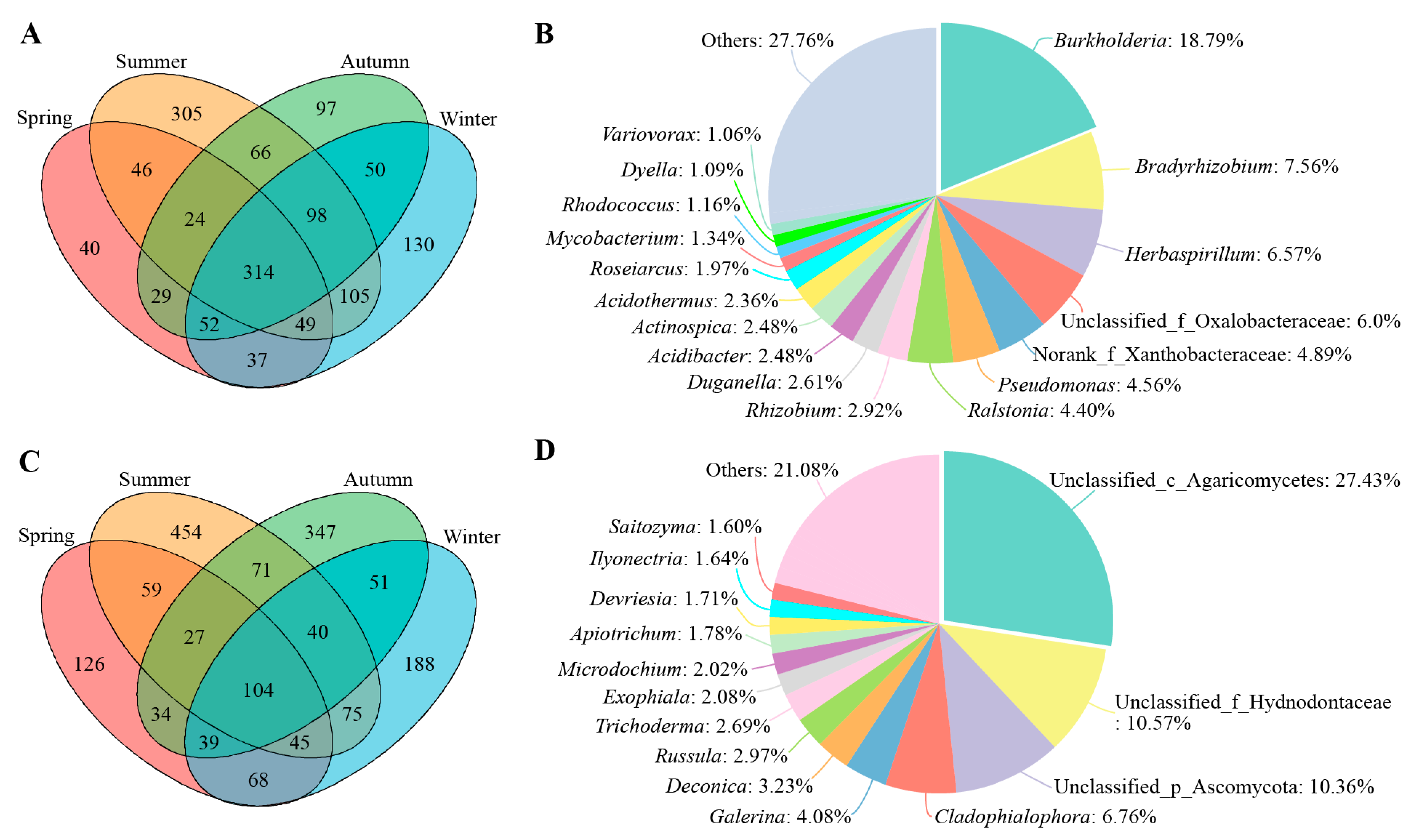
3.2. Community Composition of Bacterial and Fungal Root Endophytes in C. pingbianense across Different Seasons
3.3. Taxa Characterizing Seasonal Differences within the Root Endophytic Microbiome
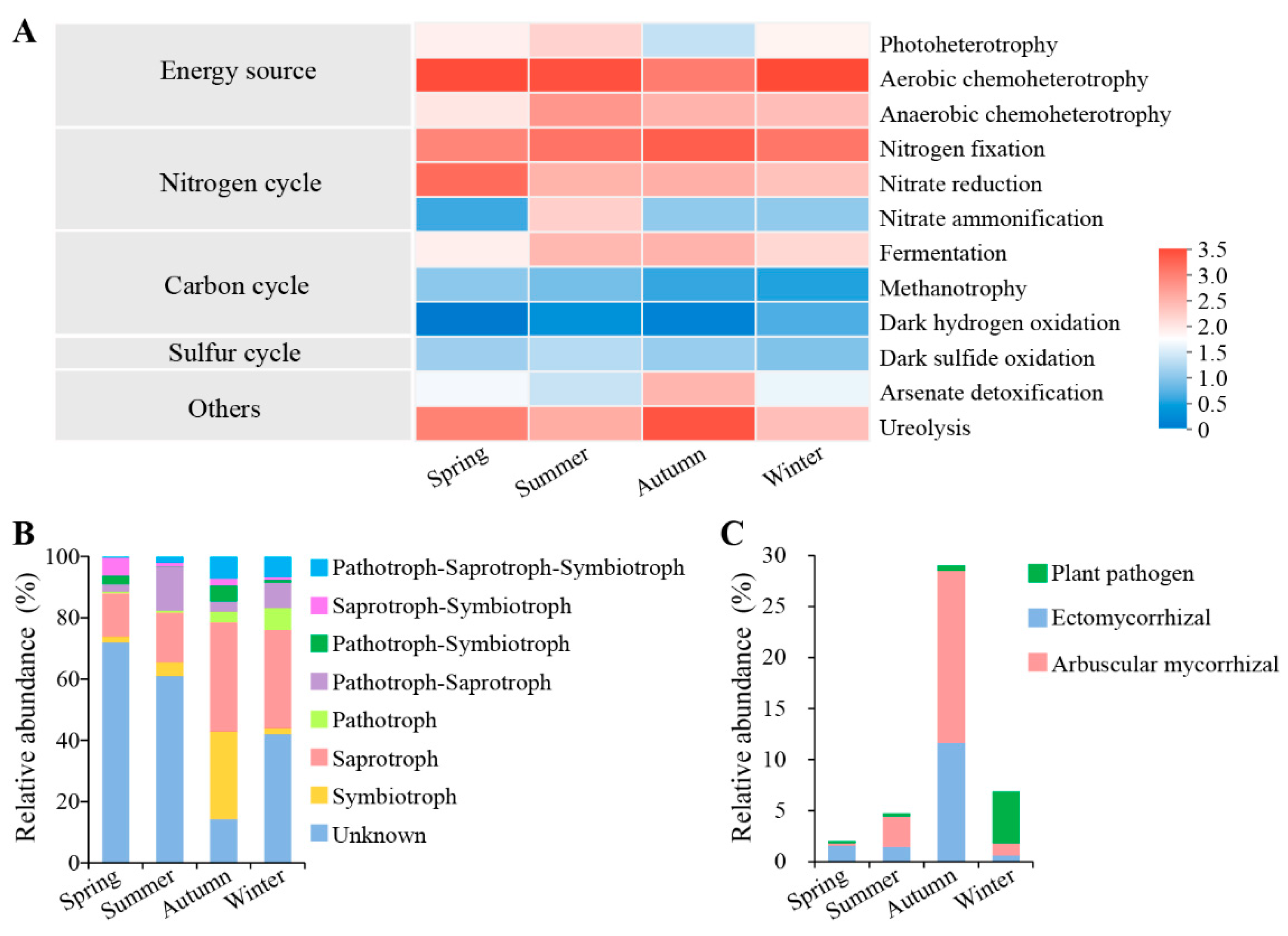
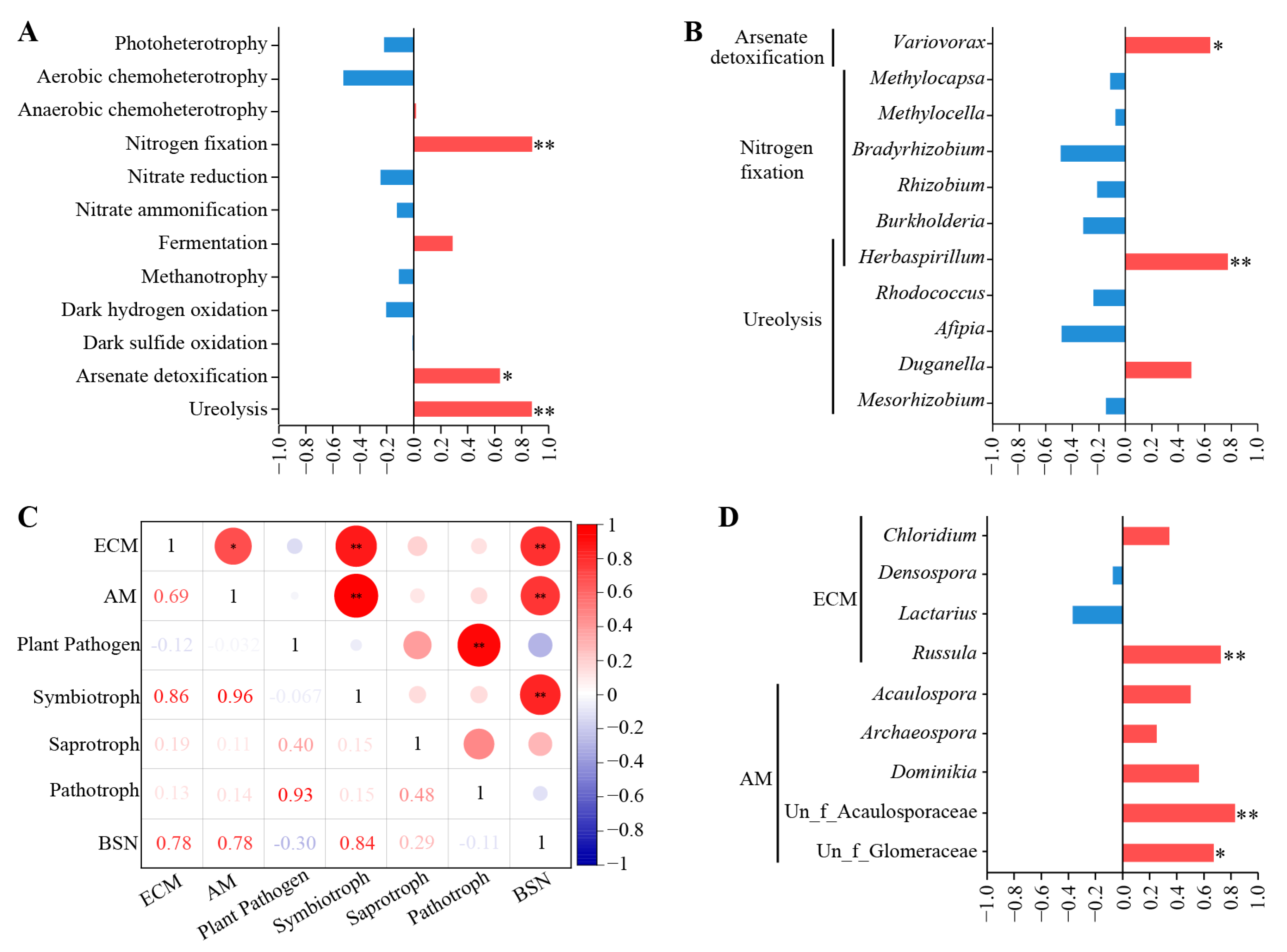
3.4. Root Endophytic Microbial Function Prediction
3.5. Correlation between Bamboo Shoot Number and Root Endophytic Microorganism
4. Discussion
4.1. The Seasonal Variations in the Bacterial and Fungal Communities in C. pingbianense Roots May Be Influenced by Temperature and Precipitation
4.2. The Abundant Nitrogen-Fixing Bacteria and Mycorrhizal Fungi in C. pingbianense Roots May Enhance Nutrient Absorption and Utilization
4.3. Root Endophytic Microorganisms Have a Positive Impact on the Year-Round Bamboo Shooting Characteristic of C. pingbianense
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Verma, S.K.; Sahu, P.K.; Kumar, K.; Pal, G.; Gond, S.K.; Kharwar, R.N.; White, J.F. Endophyte roles in nutrient acquisition, root system architecture development and oxidative stress tolerance. J. Appl. Microbiol. 2021, 131, 2161–2177. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.S.; Jain, R.; Bhardwaj, P.; Thakur, A.; Kumari, M.; Bhushan, S.; Kumar, S. Plant probiotics—Endophytes pivotal to plant health. Microbiol. Res. 2022, 263, 127148. [Google Scholar] [CrossRef]
- Guo, X.Y.; Peng, W.R.; Xu, X.Y.; Xie, K.W.; Yang, X.Y. The potential of endophytes in improving salt-alkali tolerance and salinity resistance in plants. Int. J. Mol. Sci. 2023, 24, 16917. [Google Scholar] [CrossRef] [PubMed]
- Bastías, D.A.; Gianoli, E.; Gundel, P.E. Fungal endophytes can eliminate the plant growth-defence trade-off. New Phytol. 2021, 230, 2105–2113. [Google Scholar] [CrossRef]
- Pang, F.H.; Tao, A.L.; Ayra-Pardo, C.; Wang, T.; Yu, Z.W.; Huang, S.L. Plant organ- and growth stage-diversity of endophytic bacteria with potential as biofertilisers isolated from wheat (Triticum aestivum L.). BMC Plant Biol. 2022, 22, 276. [Google Scholar] [CrossRef]
- Kumar, K.K.; Dara, S.K. Fungal and bacterial endophytes as microbial control agents for plant-parasitic nematodes. Int. J. Environ. Res. Public Health 2021, 18, 4269. [Google Scholar] [CrossRef]
- Berg, G. Plant-microbe interactions promoting plant growth and health: Perspectives for controlled use of microorganisms in agriculture. Appl. Microbiol. Biotechnol. 2009, 84, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Yan, K.; Zhang, J.; Cai, Y.; Cao, G.; Meng, L.; Soaud, S.A.; Heakel, R.M.Y.; Ihtisham, M.; Zhao, X.; Wei, Q.; et al. Comparative analysis of endophytic fungal communities in bamboo species Phyllostachys edulis, Bambusa rigida, and Pleioblastus amarus. Sci. Rep. 2023, 13, 20910. [Google Scholar] [CrossRef]
- Chen, M.; Chen, J.Z.; Liu, J.M.; Wu, M.Y.; Yan, Q.; Li, P.; Huang, L.T.; Xiao, X.F. Diversity analysis of rhizosphere soil fungi and endophytic fungi in Ampelocalamus luodianensis. Acta Ecol. Sin. 2021, 41, 4120–4130. [Google Scholar]
- Zhang, X.P.; Zhong, Z.K.; Gai, X.; Du, X.H.; Bian, F.Y.; Yang, C.B.; Gao, G.B.; Wen, X. Changes of root endophytic bacterial community along a chronosequence of intensively managed Lei bamboo (Phyllostachys praecox) forests in subtropical China. Microorganisms 2019, 7, 616. [Google Scholar] [CrossRef]
- Song, X. The Diversity of Endophyte Isolated from Bambusa Chungii and Biological Function. Master’s Thesis, South China Agricultural University, Guangzhou, China, 2020. [Google Scholar]
- Yuan, Z.S. Study on the growth-promoting effects of entophytic bacteria on Moso bamboo. J. Anhui Agric. Sci. 2019, 47, 120–122. [Google Scholar]
- Yuan, Z.S.; Liu, F.; Zhang, G.F. Characteristics and biodiversity of endophytic phosphorus- and potassium-solubilizing bacteria in Moso bamboo (Phyllostachys edulis). Acta Biol. Hung. 2015, 66, 449–459. [Google Scholar] [CrossRef]
- Wang, K.; Wu, Y.; Ye, M.Y.; Yang, Y.F.; Asiegbu, F.O.; Overmyer, K.; Liu, S.K.; Cui, F.Q. Comparative genomics reveals potential mechanisms of plant beneficial effects of a novel bamboo-endophytic bacterial isolate Paraburkholderia sacchari Suichang 626. Front. Microbiol. 2021, 12, 686998. [Google Scholar]
- Liu, F.; Yuan, Z.S.; Zhang, X.T.; Zhang, G.F.; Xie, B.G. Characteristics and diversity of endophytic bacteria in moso bamboo (Phyllostachys edulis) based on 16S rDNA sequencing. Arch. Microbiol. 2017, 199, 1259–1266. [Google Scholar] [CrossRef]
- Shen, S.Y.; Fulthorpe, R. Seasonal variation of bacterial endophytes in urban trees. Front. Microbiol. 2015, 6, 427. [Google Scholar] [CrossRef] [PubMed]
- Ou, T.; Xu, W.F.; Wang, F.; Strobel, G.; Zhou, Z.Y.; Xiang, Z.H.; Liu, J.; Xie, J. A microbiome study reveals seasonal variation in endophytic bacteria among different mulberry cultivars. Comput. Struct. Biotechnol. J. 2019, 17, 1091–1100. [Google Scholar] [CrossRef]
- Chen, A.L.; Zhao, W.Q.; Ruan, Y.Q.; Guo, C.C.; Zhang, W.G.; Shi, J.M.; Yang, G.Y. Pattern of emergence and degradation of Phyllostachys edulis ‘Pachyloen’ shoot and the changes of nutrient composition during degradation. Sci. Silvae Sin. 2019, 55, 32–40. [Google Scholar]
- Chen, Y.H.; Song, D.Q. A study on growth regularity of Phyllostachys nidularia Munro in shooting period. J. Nanjing For. Univ. (Nat. Sci. Ed.) 2005, 29, 109–112. [Google Scholar]
- Gu, D.X.; Chen, S.L.; Zheng, W.M. Summarize on bamboo’s ecologicol adaptability. J. Bamboo Res. 2010, 29, 17–23. [Google Scholar]
- Li, R.; He, M.X.; Dao, D.W.; Xiang, M.H.; Sun, A.L.; Yang, Q. The bamboo shooting and young bamboo growth rhythm of Dendrocalamus hamiltonii. Genom. Appl. Biol. 2010, 29, 735–739. [Google Scholar]
- Zhong, Y.B.; Jiang, X.; Lou, C. Research progress in growth rhythm of bamboos. World Bamboo Rattan 2014, 12, 35–44. [Google Scholar]
- Yang, H.Q.; Sun, M.S.; Mao, W.; Yang, Y.M.; Li, D.Z. Cephalostachyum pingbianense (Poaceae: Bambusoideae), comb. nova. Ann. Bot. Fenn. 2008, 45, 394–395. [Google Scholar] [CrossRef]
- Li, L.S.; Xia, T.Z.; Li, B.; Yang, H.Q. Hormone and carbohydrate metabolism associated genes play important roles in rhizome bud full-year germination of Cephalostachyum pingbianense. Physiol. Plant. 2022, 174, e13674. [Google Scholar] [CrossRef] [PubMed]
- Mao, W.; Bao, C.Y.; Cheng, Q.; Liang, N.; Wang, L.C.; Yang, H.Q. All-Year high IAA and ABA contents in rhizome buds may contribute to natural four-season shooting in woody bamboo Cephalostachyum pingbianense. Plants 2024, 13, 410. [Google Scholar] [CrossRef]
- Chaudhary, P.; Agri, U.; Chaudhary, A.; Kumar, A.; Kumar, G. Endophytes and their potential in biotic stress management and crop production. Front. Microbiol. 2022, 13, 933017. [Google Scholar] [CrossRef]
- Li, L.S.; Xia, T.Z.; Yang, H.Q. Seasonal patterns of rhizosphere microorganisms suggest carbohydrate-degrading and nitrogen-fixing microbes contribute to the attribute of full-year shooting in woody bamboo Cephalostachyum pingbianense. Front. Microbiol. 2022, 13, 1033293. [Google Scholar] [CrossRef]
- Tauro, T.P.; Mtambanengwe, F.; Mpepereki, S.; Mapfumo, P. Soil fungal community structure and seasonal diversity following application of organic amendments of different quality under maize cropping in Zimbabwe. PLoS ONE 2021, 16, e0258227. [Google Scholar] [CrossRef]
- Li, B.; Shi, H.X.; Liu, L.L.; Zhang, X.; Yang, P.; Chen, H.; Feng, Y.; Chen, X.M. Structure and diversity of bacterial community in different niches of garden plant Ligustrum lucidum Ait. Acta Microbiol. Sin. 2022, 62, 686–702. [Google Scholar]
- Wang, R.T.; Zhang, Q.C.; Ju, M.X.; Yan, S.Y.; Zhang, Q.Q.; Gu, P.W. The endophytic fungi diversity, community structure, and ecological function prediction of Sophora alopecuroides in Ningxia, China. Microorganisms 2022, 10, 2099. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Louca, S.; Parfrey, L.W.; Doebeli, M. Decoupling function and taxonomy in the global ocean microbiome. Science 2016, 353, 1272–1277. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Papik, J.; Folkmanova, M.; Polivkova-Majorova, M.; Suman, J.; Uhlik, O. The invisible life inside plants: Deciphering the riddles of endophytic bacterial diversity. Biotechnol. Adv. 2020, 44, 107614. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Xia, Y.; Zhang, H.; Cui, J.L. The microscopic mechanism between endophytic fungi and host plants: From recognition to building stable mutually beneficial relationships. Microbiol. Res. 2022, 261, 127056. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zheng, R.; Wang, Z.H.; Li, H.B.; Shi, Y.J.; Pan, Z.J.; Liu, M. Leaf health status regulates endophytic microbial community structure, network complexity, and assembly processes in the leaves of the rare and endangered plant species Abies fanjingshanensis. Microorganisms 2024, 12, 1254. [Google Scholar] [CrossRef]
- Li, L.; Wang, Y.; Yu, C.; Li, S.; Lin, T.; Han, S.; Zhu, T.; Li, S. Seasonal changes in the abundance Fusarium proliferatium, microbial endophytes and nutrient levels in the roots of hybrid bamboo Bambusa pervariabilis × Dendrocalamopsis grandis. Front. Plant Sci. 2023, 14, 1185449. [Google Scholar] [CrossRef]
- Campisano, A.; Albanese, D.; Yousaf, S.; Pancher, M.; Donati, C.; Pertot, I. Temperature drives the assembly of endophytic communities’ seasonal succession. Environ. Microbiol. 2017, 19, 3353–3364. [Google Scholar] [CrossRef]
- Gomes, T.; Pereira, J.A.; Benhadi, J.; Lino-Neto, T.; Baptista, P. Endophytic and epiphytic phyllosphere fungal communities are shaped by different environmental factors in a Mediterranean ecosystem. Microb. Ecol. 2018, 76, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Mocali, S.; Bertelli, E.; Di Cello, F.; Mengoni, A.; Sfalanga, A.; Viliani, F.; Caciotti, A.; Tegli, S.; Surico, G.; Fani, R. Fluctuation of bacteria isolated from elm tissues during different seasons and from different plant organs. Res. Microbiol. 2003, 154, 105–114. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.T.; Yu, L.; Han, B.; Liu, K.; Shao, X.Q.; Jia, S.G. Spatial variations of root-associated bacterial communities of alpine plants in the Qinghai-Tibet Plateau. Sci. Total Environ. 2022, 839, 156086. [Google Scholar] [CrossRef]
- Suryanarayanan, T.; Kumaresan, V. Endophytic fungi of some halophytes from an estuarine mangrove forest. Mycol. Res. 2000, 104, 1465–1467. [Google Scholar] [CrossRef]
- Aleynova, O.A.; Nityagovsky, N.N.; Suprun, A.R.; Ananev, A.A.; Dubrovina, A.S.; Kiselev, K.V. The diversity of fungal endophytes from wild grape Vitis amurensis Rupr. Plants 2022, 11, 2897. [Google Scholar] [CrossRef]
- Oita, S.; Ibáñez, A.; Lutzoni, F.; Miadlikowska, J.; Geml, J.; Lewis, L.A.; Hom, E.F.Y.; Carbone, I.; U’Ren, J.M.; Arnold, A.E. Climate and seasonality drive the richness and composition of tropical fungal endophytes at a landscape scale. Commun. Biol. 2021, 4, 313. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant-microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Paungfoo-Lonhienne, C.; Lonhienne, T.G.A.; Yeoh, Y.K.; Webb, R.I.; Lakshmanan, P.; Chan, C.X.; Lim, P.-E.; Ragan, M.A.; Schmidt, S.; Hugenholtz, P. A new species of Burkholderia isolated from sugarcane roots promotes plant growth. Microb. Biotechnol. 2014, 7, 142–154. [Google Scholar] [CrossRef]
- Reis, V.M.; Santos, P.E.-d.L.; Tenorio-Salgado, S.; Vogel, J.; Stoffels, M.; Guyon, S.; Mavingui, P.; Baldani, V.L.D.; Schmid, M.; Baldani, J.I.; et al. Burkholderia tropica sp. nov., a novel nitrogen-fixing, plant-associated bacterium. Int. J. Syst. Evol. Microbiol. 2004, 54, 2155–2162. [Google Scholar] [CrossRef]
- Mattos, K.A.; Pádua, V.L.M.; Romeiro, A.; Hallack, L.F.; Neves, B.C.; Ulisses, T.M.U.; Barros, C.F.; Todeschini, A.R.; Previato, J.O.; Mendonça-Previato, L. Endophytic colonization of rice (Oryza sativa L.) by the diazotrophic bacterium Burkholderia kururiensis and its ability to enhance plant growth. An. Acad. Bras. Cienc. 2008, 80, 477–493. [Google Scholar] [CrossRef]
- Banik, A.; Mukhopadhaya, S.K.; Dangar, T.K. Characterization of N2-fixing plant growth promoting endophytic and epiphytic bacterial community of Indian cultivated and wild rice (Oryza spp.) genotypes. Planta 2016, 243, 799–812. [Google Scholar] [CrossRef] [PubMed]
- Yu, P.; He, X.M.; Baer, M.; Beirinckx, S.; Tian, T.; Moya, Y.A.T.; Zhang, X.C.; Deichmann, M.; Frey, F.P.; Bresgen, V.; et al. Plant flavones enrich rhizosphere Oxalobacteraceae to improve maize performance under nitrogen deprivation. Nat. Plants 2021, 7, 481–499. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.M.; Lin, A.B.; Liu, P.; Fan, X.J.; Wu, C.J.; Li, N.; Wei, L.; Wang, W.; Wei, D.Z. Trichoderma reesei ACE4, a novel transcriptional activator involved in the regulation of cellulase genes during growth on cellulose. Appl. Environ. Microbiol. 2021, 87, e0059321. [Google Scholar] [CrossRef] [PubMed]
- Tyśkiewicz, R.; Nowak, A.; Ozimek, E.; Jaroszuk-Ściseł, J. Trichoderma: The current status of its application in agriculture for the biocontrol of fungal phytopathogens and stimulation of plant growth. Int. J. Mol. Sci. 2022, 23, 2329. [Google Scholar] [CrossRef]
- Guzmán-Guzmán, P.; Porras-Troncoso, M.D.; Olmedo-Monfil, V.; Herrera-Estrella, A. Trichoderma species: Versatile plant symbionts. Phytopathology 2019, 109, 6–16. [Google Scholar] [CrossRef]
- Genre, A.; Lanfranco, L.; Perotto, S.; Bonfante, P. Unique and common traits in mycorrhizal symbioses. Nat. Rev. Microbiol. 2020, 18, 649–660. [Google Scholar] [CrossRef]
- Zhang, L.; Zhou, J.C.; George, T.S.; Limpens, E.; Feng, G. Arbuscular mycorrhizal fungi conducting the hyphosphere bacterial orchestra. Trends Plant Sci. 2022, 27, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.L.; Yuan, Y.S.; Liu, Q.; Yin, H.J. Plant nitrogen acquisition from inorganic and organic sources via root and mycelia pathways in ectomycorrhizal alpine forests. Soil Biol. Biochem. 2019, 136, 107517. [Google Scholar] [CrossRef]
- Tang, Z.; Zhao, F.J. The roles of membrane transporters in arsenic uptake, translocation and detoxification in plants. Crit. Rev. Environ. Sci. Technol. 2021, 51, 2449–2484. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Hurek, T. Life in grasses: Diazotrophic endophytes. Trends Microbiol. 1998, 6, 139–144. [Google Scholar] [CrossRef]
- Carvalho, T.L.G.; Balsemão-Pires, E.; Saraiva, R.M.; Ferreira, P.C.G.; Hemerly, A.S. Nitrogen signalling in plant interactions with associative and endophytic diazotrophic bacteria. J. Exp. Bot. 2014, 65, 5631–5642. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.C.; Melo, J.; de Souza, S.B.; Bertolazi, A.A.; Silva, R.A.; Rodrigues, W.P.; Campostrini, E.; Olivares, F.L.; Eutrópio, F.J.; Cruz, C.; et al. Inoculation with the endophytic bacterium Herbaspirillum seropedicae promotes growth, nutrient uptake and photosynthetic efficiency in rice. Planta 2020, 252, 87. [Google Scholar] [CrossRef] [PubMed]
- Irineu, L.E.S.d.S.; Soares, C.d.P.; Soares, T.S.; Almeida, F.A.d.; Almeida-Silva, F.; Gazara, R.K.; Meneses, C.H.S.G.; Canellas, L.P.; Silveira, V.; Venancio, T.M.; et al. Multiomic approaches reveal hormonal modulation and nitrogen uptake and assimilation in the initial growth of maize inoculated with Herbaspirillum seropedicae. Plants 2022, 12, 48. [Google Scholar] [CrossRef] [PubMed]
- Ingleby, K.; Wilson, J.; Munro, R.; Cavers, S. Mycorrhizas in agroforestry: Spread and sharing of arbuscular mycorrhizal fungi between trees and crops: Complementary use of molecular and microscopic approaches. Plant Soil 2007, 294, 125–136. [Google Scholar] [CrossRef]
- Merckx, V.; Gomes, S.I.F.; Wang, D.; Verbeek, C.; Jacquemyn, H.; Zahn, F.E.; Gebauer, G.; Bidartondo, M.I. Mycoheterotrophy in the wood-wide web. Nat. Plants 2024, 10, 710–718. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, L.; Li, B.; Li, Q.; Wang, L.; Yang, H. Root Endophytic Microorganisms Contribute to the Attribute of Full-Year Shooting in Woody Bamboo Cephalostachyum pingbianense. Microorganisms 2024, 12, 1927. https://doi.org/10.3390/microorganisms12091927
Li L, Li B, Li Q, Wang L, Yang H. Root Endophytic Microorganisms Contribute to the Attribute of Full-Year Shooting in Woody Bamboo Cephalostachyum pingbianense. Microorganisms. 2024; 12(9):1927. https://doi.org/10.3390/microorganisms12091927
Chicago/Turabian StyleLi, Lushuang, Bin Li, Qing Li, Lianchun Wang, and Hanqi Yang. 2024. "Root Endophytic Microorganisms Contribute to the Attribute of Full-Year Shooting in Woody Bamboo Cephalostachyum pingbianense" Microorganisms 12, no. 9: 1927. https://doi.org/10.3390/microorganisms12091927
APA StyleLi, L., Li, B., Li, Q., Wang, L., & Yang, H. (2024). Root Endophytic Microorganisms Contribute to the Attribute of Full-Year Shooting in Woody Bamboo Cephalostachyum pingbianense. Microorganisms, 12(9), 1927. https://doi.org/10.3390/microorganisms12091927





