Bacteriophages: A Challenge for Antimicrobial Therapy
Abstract
1. Introduction
2. Phage Classification
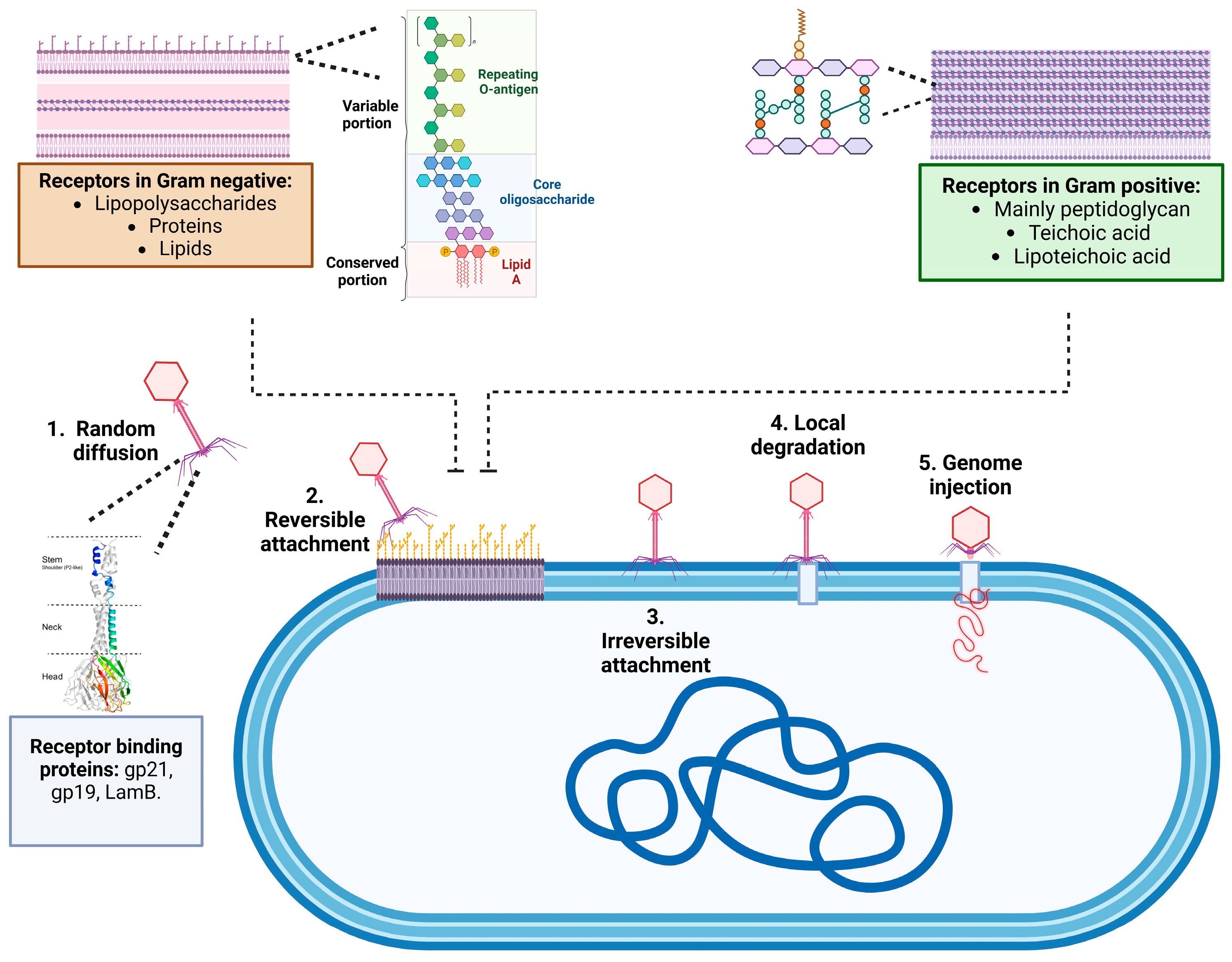
2.1. Mechanisms of Adsorption and Infection
| Phage | Receptor-Binding Protein (RBP) | Host Receptor | Bacterial Species | Gram Type | References |
|---|---|---|---|---|---|
| Phage T4 | Hoc, J, and K proteins | Lipopolysaccharides (LPS) | E. coli | Gram-negative | [37] |
| Phage λ | LamB (Maltose porin) | Maltose/maltodextrin transporters | E. coli | Gram-negative | [38] |
| Phage DMS3 | Pilus-binding protein | Type IV pili | Pseudomonas aeruginosa | Gram-negative | [39] |
| Phage MS2 | Coat protein (F-pilus recognition) | F-pili (Fertility pilus) | E. coli | Gram-negative | [40] |
| Phage TLS | Receptor-binding protein | TolC | E. coli | Gram-negative | [41] |
| Phage PM2 | P10 protein | Sugar moieties on the cell surface | Pseudoalteromonas | Gram-negative | [42] |
| Phage Ω8 | Receptor-binding protein | Polysaccharide, outer membrane proteins | E. coli | Gram-negative | [43] |
| Phage S16 | Tail fibers | Outer membrane protein (OmpC) | Salmonella | Gram-negative | [44] |
| Phage ϕ29 | gp12 | Teichoic acids, cell wall peptidoglycan | Bacillus subtilis | Gram-positive | [45] |
| Phage SPP1 | Tailspike protein | Teichoic acid | Bacillus subtilis | Gram-positive | [46] |
| Phage Bam35 | Coat proteins | N-acetyl-muramic acid (MurNAc) of peptidoglycan in the cell wall | Bacillus thuringiensis | Gram-positive | [47] |
| Phage φLC3 | Receptor-binding protein | Cell wall polysaccharides | Lactococcus lactis | Gram-positive | [48] |
| Phage A511 | Tailspike protein | Peptidoglycan | Listeria monocytogenes | Gram-positive | [49] |
| Phage φ812 | Tail protein | Anionic backbone of wall teichoic acid | Staphylococcus aureus | Gram-positive | [50] |
| Phage φSLT | Phage tail tip | Poly(glycerophosphate) moiety of lipoteichoic acid (LTA) | Staphylococcus aureus | Gram-positive | [51] |
| Phage T3 | Tailspike protein | Teichoic acid | Corynebacterium glutamicum | Gram-positive | [50] |
2.2. Life Cycle of Bacteriophages
3. Structure and Organization of Bacteriophage Genomes
3.1. Key Genes for the Replication and Assembly of Phages
3.2. Viral Versatility: How Bacteriophages Exploit Host Defenses and Drive Evolutionary Change
3.3. Genetic Modification of Bacteriophages
4. Therapeutic Applications
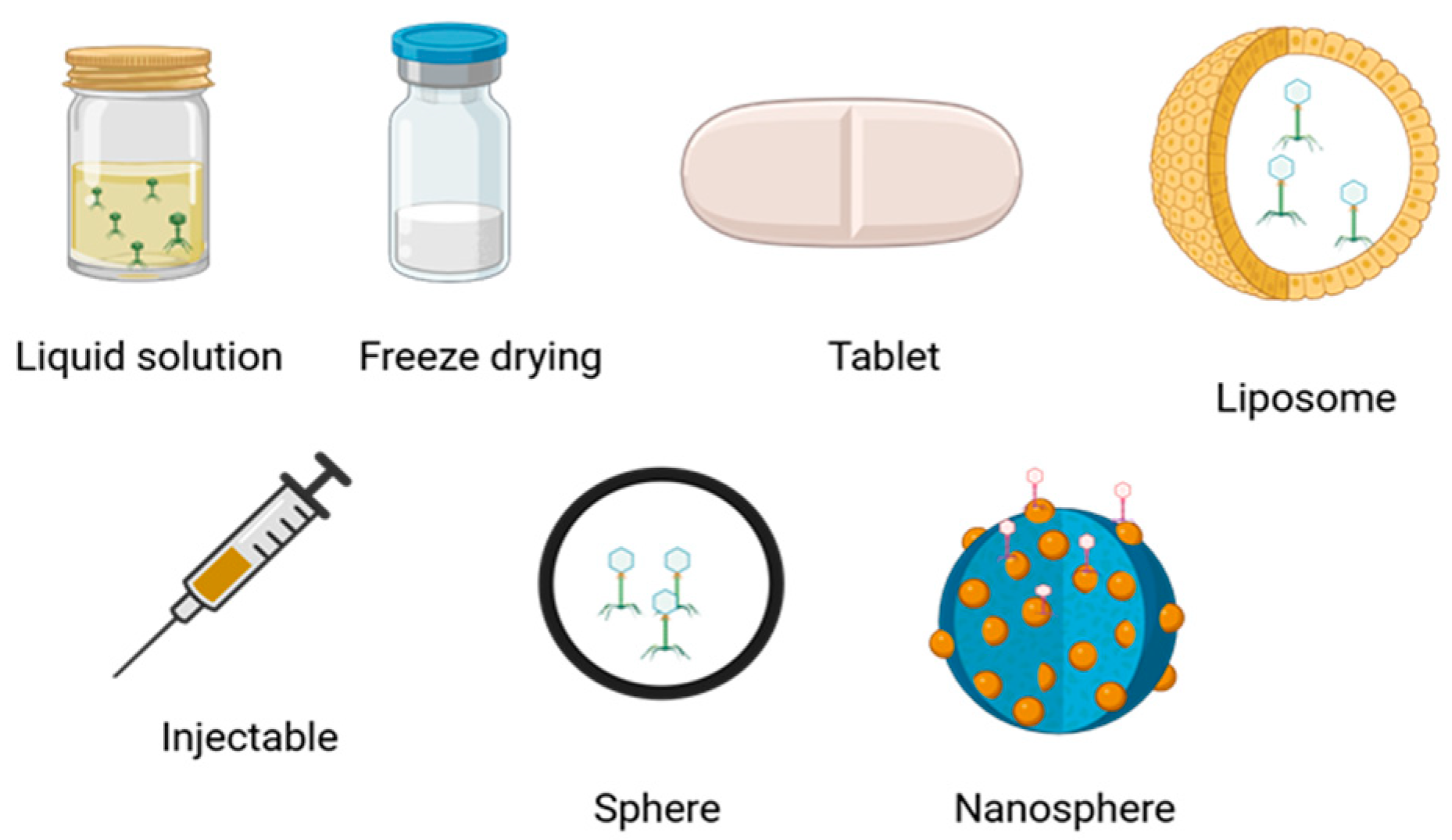
5. Pharmacokinetic Aspects of Bacteriophages
6. Pharmacodynamic Aspects of Bacteriophages
7. Regulation and Challenges of Phage Therapy
8. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Breitbart, M.; Rohwer, F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005, 13, 278–284. [Google Scholar] [CrossRef]
- Gauthier, C.H.; Hatfull, G.F. A Bioinformatic Ecosystem for Bacteriophage Genomics: PhaMMSeqs, Phamerator, pdm_utils, PhagesDB, DEPhT, and PhamClust. Viruses 2024, 16, 1278. [Google Scholar] [CrossRef] [PubMed]
- Loannou, P.; Bailou, S.; Samonis, G. Bacteriophages in Infectious Diseases and Beyond—A Narrative Review. Antibiotics 2023, 12, 1012. [Google Scholar] [CrossRef]
- Andrade-Martínez, J.S.; Valera, L.C.C.; Cárdenas, L.A.C.; Forero-Junco, L.; López-Leal, G.; Moreno-Gallego, J.L.; Rangel-Pineros, G.; Reyes, A. Computational Tools for the Analysis of Uncultivated Phage Genomes. Microbiol. Mol. Biol. Rev. 2022, 86, e00004-21. [Google Scholar] [CrossRef] [PubMed]
- Dutilh, B.E.; Cassman, N.; McNair, K.; Sanchez, S.E.; Silva, G.G.; Boling, L.; Barr, J.J.; Speth, D.R.; Seguritan, V.; Aziz, R.K.; et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 2014, 5, 4498. [Google Scholar] [CrossRef] [PubMed]
- Camarillo-Guerrero, L.F.; Almeida, A.; Rangel-Pineros, G.; Finn, R.D.; Lawley, T.D. Massive expansion of human gut bacteriophage diversity. Cell 2021, 184, 1098–1109.e9. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Review on Antimicrobial Resistance Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations. Lond. Rev. Antimicrob. Resist. 2014, 8–16. [Google Scholar]
- Schooley, R.T.; Biswas, B.; Gill, J.J.; Hernandez-Morales, A.; Lancaster, J.; Lessor, L.; Barr, J.J.; Reed, S.L.; Rohwer, F.; Benler, S.; et al. Development and Use of Personalized Bacteriophage-Based Therapeutic Cocktails to Treat a Patient with a Disseminated Resistant Acinetobacter baumannii Infection. Antimicrob. Agents Chemother. 2017, 61, 14. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.H.; Wade, E. Classification of bacterial viruses: Characteristics of the T1,D20 species of coli-dysentery phages. J. Bacteriol. 1955, 70, 253–259. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, A.A.; Hellerqvist, C.G. Bacteriophage attachment sites, serological specificity, and chemical composition of the lipopolysaccharides of semirough and rough mutants of Salmonella typhimurium. J. Bacteriol. 1971, 105, 57–64. [Google Scholar] [CrossRef]
- Pieroni, P.; Rennie, R.P.; Ziola, B.; Deneer, H.G. The use of bacteriophages to differentiate serologically cross-reactive isolates of Klebsiella pneumoniae. J. Med. Microbiol. 1994, 41, 423–429. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.C.; Fraser, D. Morphology of the seven T-bacteriophages. J. Bacteriol. 1953, 66, 458–464. [Google Scholar] [CrossRef]
- Simmonds, P.; Adriaenssens, E.M.; Zerbini, F.M.; Abrescia, N.G.A.; Aiewsakun, P.; Alfenas-Zerbini, P.; Bao, Y.; Barylski, J.; Drosten, C.; Duffy, S.; et al. Four principles to establish a universal virus taxonomy. PLoS Biol. 2023, 21, e3001922. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, P.; Alewsakun, P. Virus Classification-where do you daw the line? Arch. Virol. 2018, 163, 2037–2046. [Google Scholar] [CrossRef] [PubMed]
- Cieślik, M.; Bagińska, N.; Jończyk-Matysiak, E.; Węgrzyn, A.; Węgrzyn, G.; Górski, A. Temperate Bacteriophages—The Powerful Indirect Modulators of Eukaryotic Cells and Immune Functions. Viruses 2021, 13, 1013. [Google Scholar] [CrossRef] [PubMed]
- Nazir, A.; Ali, A.; Qing, H.; Tong, Y. Emerging Aspects of Jumbo Bacteriophages. Infect. Drug Resist. 2021, 14, 5041–5055. [Google Scholar] [CrossRef] [PubMed]
- Turner, D.; Kropinski, A.M.; Adriaenssens, E.M. A Roadmap for Genome-Based Phage Taxonomy. Viruses 2021, 13, 506. [Google Scholar] [CrossRef] [PubMed]
- Moraru, C.; Varsani, A.; Kropinski, A.M. VIRIDIC-A Novel Tool to Calculate the Intergenomic Similarities of Prokaryote-Infecting Viruses. Viruses 2020, 12, 1268. [Google Scholar] [CrossRef]
- Moraru, C. VirClust-A Tool for Hierarchical Clustering, Core Protein Detection and Annotation of (Prokaryotic) Viruses. Viruses 2023, 15, 1007. [Google Scholar] [CrossRef] [PubMed]
- Bolduc, B.; Jang, H.B.; Doulcier, G.; You, Z.Q.; Roux, S.; Sullivan, M.B. vConTACT: An iVirus tool to classify double-stranded DNA viruses that infect Archaea and Bacteria. PeerJ 2017, 5, e3243. [Google Scholar] [CrossRef]
- Turner, D.; Shkoporov, A.N.; Lood, C.; Millard, A.D.; Dutilh, B.E.; Alfenas-Zerbini, P.; van Zyl, L.J.; Aziz, R.K.; Oksanen, H.M.; Poranen, M.M.; et al. Abolishment of morphology-based taxa and change to binomial species names: 2022 taxonomy update of the ICTV bacterial viruses subcommittee. Arch. Virol. 2023, 168, 74. [Google Scholar] [CrossRef] [PubMed]
- De Paepe, M.; Hutinet, G.; Son, O.; Amarir-Bouhram, J.; Schbath, S.; Petit, M.A. Temperate phages acquire DNA from defective prophages by relaxed homologous recombination: The role of Rad52-like recombinases. PLoS Genet. 2014, 10, e1004181. [Google Scholar] [CrossRef]
- Piña-González, A.M.; Castelán-Sánchez, H.G.; Hurtado-Ramírez, J.M.; López-Leal, G. prophage diversity reveals pervasive recombination between prophages from different species. Microbiol. Spectr. 2024, 12, e02795-23. [Google Scholar] [CrossRef]
- Yahara, K.; Lehours, P.; Vale, F.F. Analysis of genetic recombination and the pan-genome of a highly recombinogenic bacteriophage species. Microb. Genom. 2019, 5, e000282. [Google Scholar] [CrossRef] [PubMed]
- Perez-Losada, M.; Arenas, M.; Galan, J.C.; Palero, F.; Gonzalez-Candelas, F. Recombination in viruses: Mechanisms, methods of study, and evolutionary consequences. Infect. Genet. Evol. 2015, 30, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Storms, Z.J.; Sauvageau, D. Modeling tailed bacteriophage adsorption: Insight into mechanisms. Virology 2015, 485, 355–362. [Google Scholar] [CrossRef]
- Casjens, S.R.; Molineux, I.J. Short noncontractile tail machines: Adsorption and DNA delivery by podoviruses. Adv. Exp. Med. Biol. 2012, 726, 143–179. [Google Scholar] [CrossRef] [PubMed]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. Fems. Microbiol. Lett. 2016, 363, fnw002. [Google Scholar] [CrossRef]
- Pires, D.P.; Oliveira, H.; Melo, L.D.; Sillankorva, S.; Azeredo, J. Bacteriophage-encoded depolymerases: Their diversity and biotechnological applications. Appl. Microbiol. Biotechnol. 2016, 100, 2141–2151. [Google Scholar] [CrossRef]
- Davidson, A.R.; Cardarelli, L.; Pell, L.G.; Radford, D.R.; Maxwell, K.L. Long noncontractile tail machines of bacteriophages. Adv. Exp. Med. Biol. 2012, 726, 115–142. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, H.; Melo, L.D.; Santos, S.B.; Nóbrega, F.L.; Ferreira, E.C.; Cerca, N.; Azeredo, J.; Kluskens, L.D. Molecular aspects and comparative genomics of bacteriophage endolysins. Virol. J. 2013, 87, 4558–4570. [Google Scholar] [CrossRef] [PubMed]
- Scheurwater, E.; Reid, C.W.; Clarke, A.J. Lytic transglycosylases: Bacterial space-making autolysins. Int. J. Biochem. Cell. Biol. 2008, 40, 586–591. [Google Scholar] [CrossRef]
- Rakhuba, D.V.; Kolomiets, E.I.; Dey, E.S.; Novik, G.I. Bacteriophage receptors, mechanisms of phage adsorption and penetration into host cell. Pol. J. Microbiol. 2010, 59, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Shaw, D.R.; Chatterjee, A.N. O-Acetyl groups as a component of the bacteriophage receptor on Staphylococcus aureus cell walls. J. Bacteriol. 1971, 108, 584–585. [Google Scholar] [CrossRef] [PubMed]
- Xia, G.; Corrigan, R.M.; Winstel, V.; Goerke, C.; Grundling, A.; Peschel, A. Wall teichoic Acid-dependent adsorption of staphylococcal siphovirus and myovirus. J. Bacteriol. 2011, 193, 4006–4009. [Google Scholar] [CrossRef]
- McGee, L.W.; Barhoush, Y.; Shima, R.; Hennessy, M. Phage-resistant mutations impact bacteria susceptibility to future phage infections and antibiotic response. Ecol. Evol. 2023, 13, e9712. [Google Scholar] [CrossRef]
- Kutter, E.; De-Vos, D.; Gvasalia, G.; Alavidze, Z.; Gogokhia, L.; Kuhl, S.; Abedon, S.T. Phage therapy in clinical practice: Treatment of human infections. Curr. Pharm. Biotechnol. 2010, 11, 69–86. [Google Scholar] [CrossRef]
- Chatterjee, S.; Rothenberg, E. Interaction of Bacteriophage l with Its E. coli Receptor, LamB. Viruses 2012, 4, 3162–3168. [Google Scholar] [CrossRef] [PubMed]
- Budzik, J.M.; Rosche, W.A.; Rietsch3, A.; O’Toole, G.A. Isolation and Characterization of a Generalized Transducing Phage for Pseudomonas aeruginosa Strains PAO1 and PA14. J. Bacteriol. 2004, 186, 3270–3273. [Google Scholar] [CrossRef]
- Dai, X.; Li, Z.; Lai, M.; Shu, S.; Du, Y.; Hong, Z.; Sun, R. In situ structures of the genome and genome delivery apparatus in a single-stranded RNA virus. Lett. Res. 2017, 112, 112–116. [Google Scholar] [CrossRef]
- German, G.J.; Misra, R. The TolC protein of Escherichia coli serves as a cell-surface receptor for the newly characterized TLS bacteriophage. J. Mol. Biol. 2001, 308, 579–585. [Google Scholar] [CrossRef]
- Kivelä, H.M.; Madonna, S.; Krupovìč, M.; Tutino, M.L.; Bamford, J.K.H. Genetics for Pseudoalteromonas Provides Tools To Manipulate Marine Bacterial Virus PM2. J. Bacteriol. 2008, 190, 1298–1307. [Google Scholar] [CrossRef]
- Reske, K.; Wallenfels, B.; Jann, K. Enzymatic Degradation of O-Antigenic Lipopolysaccharides by Coliphage Ω8. Eur. J. Biochem. 1973, 36, 167–171. [Google Scholar] [CrossRef]
- Marti, R.; Zurfluh, K.; Hagens, S.; Pianezzi, J.; Klumpp, J.; Loessner, M.J. T4-like Salmonella phage S16. Mol. Microbiol. 2013, 87, 818–834. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Leiman, Y.G.; Li, L.; Grimes, S.; Anderson, D.L.; Rossmann1, M.G. Crystallographic Insights into the Autocatalytic Assembly Mechanism of a Bacteriophage Tail Spike. Moll. Cell 2009, 34, 375–386. [Google Scholar] [CrossRef] [PubMed]
- Baptista, C.; Santos, M.A.; São-José, C. Phage SPP1 Reversible Adsorption to Bacillus subtilis Cell Wall Teichoic Acids Accelerates Virus Recognition of Membrane Receptor YueB. J. Bacteriol. 2008, 190, 4989–4996. [Google Scholar] [CrossRef] [PubMed]
- Gaidelyte, A.; Cvirkaite-Krupovic, V.; Daugelavicius, R.; Bamford, J.K.; Bamford, D.H. The entry mechanism of membrane-containing phage Bam35 infecting Bacillus thuringiensis. J. Bacteriol. 2006, 188, 5925–5934. [Google Scholar] [CrossRef] [PubMed]
- Ainsworth, S.; Sadovskaya, I.; Vinogradov, E.; Courtin, P.; Guerardel, Y.; Mahony, J.; Grard, T.; Cambillau, C.; Chapot-Chartier, M.; van Sinderen, D. Differences in Lactococcal Cell Wall Polysaccharide Structure Are Major Determining Factors in Bacteriophage Sensitivity. mBio 2014, 5. [Google Scholar] [CrossRef]
- Wendlinger, G.; Loessner, M.J.; Scherer, S. Bacteriophage receptors on Listeria monocytogenes cells are the N-acetylglucosamine and rhamnose substituents of teichoic acids or the peptidoglycan itself. Microbiology 1996, 142, 985–992. [Google Scholar] [CrossRef] [PubMed]
- Kaneko, J.; Narita-Yamada, S.; Wakabayashi, Y.; Kamio, Y. Identification of ORF636 in Phage φSLT Carrying Panton-Valentine Leukocidin Genes, Acting as an Adhesion Protein for a Poly(Glycerophosphate) Chain of Lipoteichoic Acid on the Cell Surface of Staphylococcus aureus. J. Bacteriol. 2009, 191, 4674–4680. [Google Scholar] [CrossRef]
- Maurice, C.F.; Bouvier, C.; de Wit, R.; Bouvier, T. Linking the lytic and lysogenic bacteriophage cycles to environmental conditions, host physiology and their variability in coastal lagoons. Environ. Microbiol. 2013, 15, 2463–2475. [Google Scholar] [CrossRef] [PubMed]
- Young, R. Phage lysis: Do we have the hole story yet? Curr. Opin. Microbiol. 2013, 16, 790–797. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.R.; Monteiro, R.; Azeredo, J. Genomic analysis of Acinetobacter baumannii prophages reveals remarkable diversity and suggests profound impact on bacterial virulence and fitness. Sci. Rep. 2018, 8, 15346. [Google Scholar] [CrossRef] [PubMed]
- López-Leal, G.; Santamaria, R.I.; Cevallos, M.A.; Gonzalez, V.; Castillo-Ramírez, S. Prophages Encode Antibiotic Resistance Genes in. Microb. Drug Resist. 2020, 26, 1275–1277. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, R.W.; Smith, M.C.M.; Burns, R.N.; Ford, M.E.; Hatfull, G.F. Evolutionary relationships among diverse bacteriophages and prophages: All the world’s a phage. Proc. Natl. Acad. Sci. USA 1999, 96, 2192–2197. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Leal, G.; Camelo-Valera, L.C.; Hurtado-Ramirez, J.M.; Verleyen, J.; Castillo-Ramirez, S.; Reyes-Munoz, A. Mining of Thousands of Prokaryotic Genomes Reveals High Abundance of Prophages with a Strictly Narrow Host Range. mSystems 2022, 7, e0032622. [Google Scholar] [CrossRef]
- Smeal, S.W.; Schmitt, M.A.; Pereira, R.R.; Prasad, A.; Fisk, J.D. Simulation of the M13 life cycle I: Assembly of a genetically-structured deterministic chemical kinetic simulation. Virology 2017, 500, 259–274. [Google Scholar] [CrossRef] [PubMed]
- Poranen, M.M.; Daugelavicius, R.; Bamford, D.H. Common principles in viral entry. Annu. Rev. Microbiol. 2002, 56, 521–538. [Google Scholar] [CrossRef]
- Bardhan, N.M.; Ghosh, D.; Belcher, A.M. M13 virus based detection of bacterial infections in living hosts. J. Biophotonics 2014, 7, 617–623. [Google Scholar] [CrossRef]
- Chameettachal, A.; Mustafa, F.; Rizvi, T.A. Understanding Retroviral Life Cycle and its Genomic RNA Packaging. J. Mol. Biol. 2023, 435, 167924. [Google Scholar] [CrossRef]
- Lima-Mendez, G.; Van Helden, J.; Toussaint, A.; Leplae, R. Reticulate representation of evolutionary and functional relationships between phage genomes. Mol. Biol. Evol. 2008, 25, 762–777. [Google Scholar] [CrossRef] [PubMed]
- Albà, M.M. Replicative DNA polymerases. Genome. Biol. 2001, 2. [Google Scholar] [CrossRef] [PubMed]
- Rothwell, P.J.; Waksman, G. Structure and mechanism of DNA polymerases. Adv. Protein Chem. 2005, 71, 401–440. [Google Scholar] [CrossRef]
- Morcinek-Orlowska, J.; Zdrojewska, K.; Wegrazyn, A. Bacteriophage-Encoded DNA Polymerases-Beyond the Traditional View of Polymerase Activities. Int. J. Mol. Sci. 2022, 23, 635. [Google Scholar] [CrossRef]
- Fuller, C.W.; Richardson, C.C. Replication of Bacteriophage-T7 Deoxyribonucleic-Acid 30. Initiation of DNA-Replication at the Primary Origin of Bacteriophage-T7 by Purified Proteins—Site and Direction of Initial DNA-Synthesis. J. Biol. Chem. 1985, 260, 3185–3196. [Google Scholar] [CrossRef] [PubMed]
- Hinkle, D.C. Evidence for Direct Involvement of T7 Rna-Polymerase in Bacteriophage DNA-Replication. J. Virol. 1980, 34, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Richardson, C.C. Bacteriophage-T7—Minimal Requirements for the Replication of a Duplex DNA Molecule. Cell 1983, 33, 315–317. [Google Scholar] [CrossRef]
- Hermoso, J.M.; Freire, R.; Bravo, A.; Gutierrez, C.; Serrano, M.; Salas, M. DNA-Structure in the Nucleoprotein Complex That Activates Replication of Phage Ø/29. Biophys. Chem. 1994, 50, 183–189. [Google Scholar] [CrossRef]
- Mendez, J.; Blanco, L.; Esteban, J.A.; Bernad, A.; Salas, M. Initiation of phi 29 DNA replication occurs at the second 3’ nucleotide of the linear template: A sliding-back mechanism for protein-primed DNA replication. Proc. Natl. Acad. Sci. USA. 1992, 89, 9579–9583. [Google Scholar] [CrossRef]
- Luder, A.; Mosig, G. Two alternative mechanisms for initiation of DNA replication forks in bacteriophage T4: Priming by RNA polymerase and by recombination. Proc. Natl. Acad. Sci. USA 1982, 79, 1101–1105. [Google Scholar] [CrossRef]
- Bleuit, J.S.; Ma, Y.; Munro, J.; Morrical, S.W. Mutations in a conserved motif inhibit single-stranded DNA binding and recombination mediator activities of bacteriophage T4 UvsY protein. J. Biol. Chem. 2004, 279, 6077–6086. [Google Scholar] [CrossRef] [PubMed]
- Kreuzer, K.N. Recombination-dependent DNA replication in phage T4. Trends. Biochem. Sci. 2000, 25, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Mosig, G. Recombination and recombination-dependent DNA replication in bacteriophage T4. Annu. Rev. Genet. 1998, 32, 379–413. [Google Scholar] [CrossRef]
- Mosig, G.; Gewin, J.; Luder, A.; Colowick, N.; Vo, D. Two recombination-dependent DNA replication pathways of bacteriophage T4, and their roles in mutagenesis and horizontal gene transfer. Proc. Natl. Acad. Sci. USA 2001, 98, 8306–8311. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V. Genome replication/expression strategies of positive-strand RNA viruses: A simple version of a combinatorial classification and prediction of new strategies. Virus Genes 1991, 5, 273–281. [Google Scholar] [CrossRef]
- Koskella, B.; Brockhurst, M.A. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. Fems. Microbiol. Rev. 2014, 38, 916–931. [Google Scholar] [CrossRef] [PubMed]
- Makarova, K.S.; Haft, D.H.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Horvath, P.; Moineau, S.; Mojica, F.J.M.; Wolf, Y.I.; Yakunin, A.F.; et al. Evolution and classification of the CRISPR-Cas systems. Nat. Rev. Microbiol. 2011, 9, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Pozhydaieva, N.; Billau, F.A.; Wolfram-Schauerte, M.; Rojas, A.A.R.; Paczia, N.; Schindler, D.; Höfer, K. Temporal epigenome modulation enables efficient bacteriophage engineering and functional analysis of phage DNA modifications. PLoS Genet. 2024, 20, e1011384. [Google Scholar] [CrossRef]
- Bobay, L.M.; Touchon, M.; Rocha, E.P.C. Pervasive domestication of defective prophages by Bacteriaproc. Natl. Acad. Sci. USA 2014, 111, 12127–12132. [Google Scholar] [CrossRef] [PubMed]
- Ramisetty, B.C.M.; Sudhakari, P.A. Bacterial ‘Grounded’ Prophages: Hotspots for Genetic Renovation and Innovation. Front. Genet. 2019, 10, 65. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.P.; Bondy-Denomy, J. Anti-CRISPRs go viral: The infection biology of CRISPR-Cas inhibitors. Cell Host Microbe. 2021, 29, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Davidson, A.R.; Lu, W.R.; Stanley, Y.S.; Wang, J.; Mejdani, M.; Trost, C.; Hicks, T.B.; Lee, J.; Sontheimer, S.J. Anti-CRISPRs: Protein Inhibitors of CRISPR-Cas Systems. Annu. Rev. Biochem. 2020, 89, 309–332. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, N.; Tandi, D.; Verma, R.K.; Yadav, V.K.; Dhingra, N.; Ghosh, T.; Choudhary, M.; Gaur, R.K.; Abdellatif, M.H.; Gacem, A.; et al. A comprehensive appraisal of mechanism of anti-CRISPR proteins: An advanced genome editor to amend the CRISPR gene editing. Front. Plant Sci. 2023, 14, 1164–1461. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.E.; Li, D.; Gao, A.; Macrae, R.K.; Zhang, F. Phage-triggered reverse transcription assembles a toxic repetitive gene from a noncoding RNA. Science 2024, 386, eadq3977. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Altae-Tran, H.; Bohning, F.; Makarova, K.S.; Segel, M.; Schmid-Burgk, J.L.; Koob, J.; Wolf, Y.I.; Koonin, E.V.; Zhang, F. Diverse enzymatic activities mediate antiviral immunity in prokaryotes. Science 2020, 369, 1077–1084. [Google Scholar] [CrossRef]
- Bobonis, J.; Mitosch, K.; Mateus, A.; Karcher, N.; Kritikos, G.; Selkrig, J.; Zietek, M.; Monzon, V.; Pfalz, B.; Garcia-Santamarina, S.; et al. Bacterial retrons encode phage-defending tripartite toxin-antitoxin systems. Nature 2022, 609, 144–150. [Google Scholar] [CrossRef] [PubMed]
- Alqahtani, A. Bacteriophage treatment as an alternative therapy for multidrug-resistant bacteria. Saudi Med. J. 2023, 44, 1222–1231. [Google Scholar] [CrossRef]
- Aranaga, C.; Pantoja, L.D.; Martínez, E.A.; Falco, A. Phage Therapy in the Era of Multidrug Resistance in Bacteria: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 4577. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.M.; Chen, J.C.; Zhao, X.Y.; Luo, Y.N.; Jin, M.L.; Fan, F.X.; Park, C.; Yang, X.M.; Sun, C.Q.; Yan, J.; et al. Genetic and Chemical Engineering of Phages for Controlling Multidrug-Resistant Bacteria. Antibiotics 2021, 10, 202. [Google Scholar] [CrossRef]
- Brüssow, H. Hurdles for Phage Therapy to Become a Reality-An Editorial Comment. Viruses 2019, 11, 557. [Google Scholar] [CrossRef]
- Gibb, B.; Hyman, P.; Schneider, C. The Many Applications of Engineered Bacteriophages-An Overview. Pharmaceuticals 2021, 14, 634. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Shi, D.L.; Li, Y.X.; Xiao, Y.Y.; Chen, M.M.; Chen, L.; Du, H.; Zhang, W. Recombination of T4-like Phages and Its Activity against Pathogenic in n Planktonic and Biofilm Forms. Virol. Sin. 2020, 35, 651–661. [Google Scholar] [CrossRef] [PubMed]
- da Silva, J.L.; Piuri, M.; Broussard, G.; Marinelli, L.J.; Bastos, G.M.; Hirata, R.D.C.; Hatfull, G.F.; Hirata, M.H. Application of BRED technology to construct recombinant D29 reporter phage expressing EGFP. Fems. Microbiol. Lett. 2013, 344, 166–172. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bazan, J.; Calkosinski, I.; Gamian, A. Phage display-A powerful technique for immunotherapy 2. Vaccine delivery. Hum. Vacc. Immunother. 2012, 8, 1829–1835. [Google Scholar] [CrossRef] [PubMed]
- McCafferty, J.; Griffiths, A.D.; Winter, G.; Chiswell, D.J. Phage antibodies: Filamentous phage displaying antibody variable domains. Nature 1990, 348, 552–554. [Google Scholar] [CrossRef]
- Bessette, P.H.; Rice, J.J.; Daugherty, P.S. Rapid isolation of high-affinity protein binding peptides using bacterial display. Protein Eng. Des. Sel. 2004, 17, 731–739. [Google Scholar] [CrossRef]
- De Palma, G.D.; Colavita, I.; Zambrano, G.; Giglio, M.C.; Maione, F.; Luglio, G.; Sarnelli, G.; Rispo, A.; Schettino, P.; D’Armiento, F.P.; et al. Detection of colonic dysplasia in patients with ulcerative colitis using a targeted fluorescent peptide and confocal laser endomicroscopy: A pilot study. PLoS ONE 2017, 12, e0180509. [Google Scholar] [CrossRef] [PubMed]
- Lan, L.; Hin, C.C. TCP-1 as a novel phage-display peptide targeting colon cancer. Faseb. J. 2013, 27, 1093.19. [Google Scholar] [CrossRef]
- Miller, S.J.; Joshi, B.P.; Feng, Y.; Gaustad, A.; Fearon, E.R.; Wang, T.D. In vivo fluorescence-based endoscopic detection of colon dysplasia in the mouse using a novel peptide probe. PLoS ONE 2011, 6, e17384. [Google Scholar] [CrossRef]
- Lowy, F.D. Antimicrobial resistance: The example of. J. Clin. Investig. 2003, 111, 1265–1273. [Google Scholar] [CrossRef] [PubMed]
- Canning, J.S.; Laucirica, D.R.; Ling, K.M.; Nicol, M.P.; Stick, S.M.; Kicic, A. Phage therapy to treat cystic fibrosis Burkholderia cepacia complex lung infections: Perspectives and challenges. Front. Microbiol. 2024, 15, 1476041. [Google Scholar] [CrossRef] [PubMed]
- Cislo, M.; Dabrowski, M.; Weberdabrowska, B.; Woyton, A. Bacteriophage Treatment of Suppurative Skin Infections. Arch. Immunol. Ther. Exp. 1987, 35, 175–183. [Google Scholar]
- Cano, E.J.; Caflisch, K.M.; Bollyky, P.L.; Van Belleghem, J.D.; Patel, R.; Fackler, J.; Brownstein, M.J.; Horne, B.; Biswas, B.; Henry, M.; et al. Phage Therapy for Limb-threatening Prosthetic Knee Infection: Case Report and in vitro Characterization of Anti-biofilm Activity. Clin. Infect. Dis. 2021, 73, E144–E151. [Google Scholar] [CrossRef]
- Merabishvili, M.; Pirnay, J.P.; Verbeken, G.; Chanishvili, N.; Tediashvili, M.; Lashkhi, N.; Glonti, T.; Krylov, V.; Mast, J.; Van Parys, L.; et al. Quality-Controlled Small-Scale Production of a Well-Defined Bacteriophage Cocktail for Use in Human Clinical Trials. PLoS ONE 2009, 4, e4944. [Google Scholar] [CrossRef] [PubMed]
- Suh, G.A.; Lodise, T.P.; Tamma, P.D.; Knisely, J.M.; Alexander, J.; Aslam, S.; Barton, K.D.; Bizzell, E.; Totten, K.M.C.; Campbell, J.L.; et al. Considerations for the Use of Phage Therapy in Clinical Practice. Antimicrob. Agents Chemother. 2022, 66, e0207121. [Google Scholar] [CrossRef]
- Strathdee, S.A.; Hatfull, G.F.; Mutalik, V.K.; Schooley, R.T. Phage therapy: From biological mechanisms to future directions. Cell 2023, 186, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.; Hawkins, C.H.; Anggard, E.E.; Harper, D.R. A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 2009, 34, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Cruz, J.C.; Huelgas-Méndez, D.; Jimenez-Zuniga, J.S.; Rebollar-Juarez, X.; Hernandez-Garnica, M.; Fernandez-Presas, A.M.; Husain, F.M.; Alenazy, R.; Alqasmi, M.; Albalawi, T.; et al. Myriad applications of bacteriophages beyond phage therapy. PeerJ 2023, 11, e15272. [Google Scholar] [CrossRef] [PubMed]
- Juul, F.E.; Garborg, K.; Bretthauer, M.; Skudal, H.; Oines, M.N.; Wiig, H.; Rose, O.; Seip, B.; Lamont, J.T.; Midtvedt, T.; et al. Fecal Microbiota Transplantation for Primary Infection. N. Engl. J. Med. 2018, 378, 2535. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Liang, J.; Zhang, Y.; Hu, L.; Gong, P.; Cai, R.; Zhang, L.; Zhang, H.; Ge, J.; Ji, Y.; et al. The Bacteriophage EF-P29 Efficiently Protects against Lethal Vancomycin-Resistant Enterococcus faecalis and Alleviates Gut Microbiota Imbalance in a Murine Bacteremia Model. Front. Microbiol. 2017, 8, 837. [Google Scholar] [CrossRef]
- de Melo, A.C.C.; da Mata Gomes, A.; Melo, F.L.; Ardisson-Araujo, D.M.P.; de Vargas, A.P.C.; Ely, V.L.; Kitajima, E.W.; Ribeiro, B.M.; Wolff, J.L.C. Characterization of a bacteriophage with broad host range against strains of Pseudomonas aeruginosa isolated from domestic animals. BMC Microbiol. 2019, 19, 134. [Google Scholar] [CrossRef] [PubMed]
- Khalifa, L.; Brosh, Y.; Gelman, D.; Coppenhagen-Glazer, S.; Beyth, S.; Poradosu-Cohen, R.; Que, Y.A.; Beyth, N.; Hazan, R. Targeting Enterococcus faecalis biofilms with phage therapy. Appl. Environ. Microbiol. 2015, 81, 2696–2705. [Google Scholar] [CrossRef]
- Sarker, S.A.; McCallin, S.; Barretto, C.; Berger, B.; Pittet, A.C.; Sultana, S.; Krause, L.; Huq, S.; Bibiloni, R.; Bruttin, A.; et al. Oral T4-like phage cocktail application to healthy adult volunteers from Bangladesh. Virology 2012, 434, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Cinquerrui, S.; Mancuso, F.; Vladisavljevic, G.T.; Bakker, S.E.; Malik, D.J. Nanoencapsulation of Bacteriophages in Liposomes Prepared Using Microfluidic Hydrodynamic Flow Focusing. Front. Microbiol. 2018, 9, 2172. [Google Scholar] [CrossRef] [PubMed]
- Merabishvili, M.; Vervaet, C.; Pirnay, J.P.; De Vos, D.; Verbeken, G.; Mast, J.; Chanishvili, N.; Vaneechoutte, M. Stability of Staphylococcus aureus phage ISP after freeze-drying (lyophilization). PLoS ONE 2013, 8, e68797. [Google Scholar] [CrossRef]
- Szymczak, M.; Pankowski, J.A.; Kwiatek, A.; Grygorcewicz, B.; Karczewska-Golec, J.; Sadowska, K.; Golec, P. An effective antibiofilm strategy based on bacteriophages armed with silver nanoparticles. Sci. Rep. 2024, 14, 9088. [Google Scholar] [CrossRef] [PubMed]
- Colom, J.; Cano-Sarabia, M.; Otero, J.; Cortes, P.; Maspoch, D.; Llagostera, M. Liposome-Encapsulated Bacteriophages for Enhanced Oral Phage Therapy against Salmonella spp. Appl. Environ. Microbiol. 2015, 81, 4841–4849. [Google Scholar] [CrossRef]
- Barros, J.A.R.; de Melo, L.D.R.; da Silva, R.A.R.; Ferraz, M.P.; Azeredo, J.C.V.D.; Pinheiro, V.M.D.; Colaço, B.J.A.; Fernandes, M.H.R.; Gomes, P.D.; Monteiro, F.J. Encapsulated bacteriophages in alginate-nanohydroxyapatite hydrogel as a novel delivery system to prevent orthopedic implant-associated infections. Nanomed. Nanotechnol. Biol. Med. 2020, 24, 102145. [Google Scholar] [CrossRef] [PubMed]
- Chambers, H.F.; Deleo, F.R. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 2009, 7, 629–641. [Google Scholar] [CrossRef]
- Gondil, V.S.; Chhibber, S. Bacteriophage and Endolysin Encapsulation Systems: A Promising Strategy to Improve Therapeutic Outcomes. Front. Pharmacol. 2021, 12, 675440. [Google Scholar] [CrossRef] [PubMed]
- Sawa, T.; Moriyama, K.; Kinoshita, M. Current status of bacteriophage therapy for severe bacterial infections. J. Intensive Care 2024, 12, 44. [Google Scholar] [CrossRef] [PubMed]
- Pirnay, J.P.; Djebara, S.; Steurs, G.; Griselain, J.; Cochez, C.; De Soir, S.; Glonti, T.; Spiessens, A.; Vanden Berghe, E.; Green, S.; et al. Personalized bacteriophage therapy outcomes for 100 consecutive cases: A multicentre, multinational, retrospective observational study. Nat. Microbiol. 2024, 9, 1434–1453. [Google Scholar] [CrossRef] [PubMed]
- Aslam, S.; Roach, D.; Nikolich, M.P.; Biswas, B.; Schooley, R.T.; Lilly-Bishop, K.A.; Rice, G.K.; Cer, R.Z.; Hamilton, T.; Henry, M.; et al. Pseudomonas aeruginosa ventricular assist device infections: Findings from ineffective phage therapies in five cases. Antimicrob. Agents Chemother. 2024, 68, e0172823. [Google Scholar] [CrossRef]
- Bichet, M.C.; Adderley, J.; Avellaneda-Franco, L.; Magnin-Bougma, I.; Torriero-Smith, N.; Gearing, L.J.; Deffrasnes, C.; David, C.; Pepin, G.; Gantier, M.P.; et al. Mammalian cells internalize bacteriophages and use them as a resource to enhance cellular growth and survival. PLoS Biol. 2023, 21, e3002341. [Google Scholar] [CrossRef] [PubMed]
- Nang, S.C.; Lin, Y.W.; Fabijan, A.P.; Chang, R.Y.K.; Rao, G.G.; Iredell, J.; Chan, H.K.; Li, J. Pharmacokinetics/pharmacodynamics of phage therapy: A major hurdle to clinical translation. Clin. Microbiol. Infect. 2023, 29, 702–709. [Google Scholar] [CrossRef]
- Kang, D.Y.; Bagchi, D.; Chen, I.A. Pharmacokinetics and Biodistribution of Phages and their Current Applications in Antimicrobial Therapy. Adv. Ther. 2024, 7, 2300355. [Google Scholar] [CrossRef] [PubMed]
- Costantine, M.M. Physiologic and pharmacokinetic changes in pregnancy. Front. Pharmacol. 2014, 5, 65. [Google Scholar] [CrossRef]
- Bhoomandla, S.; Pandiri, S.; Premalatha, A.; Kumar, K.S. Pharmacokinetics and Pharmacodynamics: Current Concepts and Applications. Lat. Am. J. Pharm. 2023, 42, 1525–1532. [Google Scholar]
- Dlusskaya, E.A.; Atrazhev, A.M.; Ashbolt, N.J. Colloid chemistry pitfall for flow cytometric enumeration of viruses in water. Water Res. X 2019, 2, 100025. [Google Scholar] [CrossRef] [PubMed]
- Goldsmith, C.S.; Miller, S.E. Modern uses of electron microscopy for detection of viruses. Clin. Microbiol. Rev. 2009, 22, 552–563. [Google Scholar] [CrossRef] [PubMed]
- Anderson, B.; Rashid, M.H.; Carter, C.; Pasternack, G.; Rajanna, C.; Revazishvili, T.; Dean, T.; Senecal, A.; Sulakvelidze, A. Enumeration of bacteriophage particles: Comparative analysis of the traditional plaque assay and real-time QPCR- and nanosight-based assays. Bacteriophage 2011, 1, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Abedon, S.T. How Simple Maths Can Inform Our Basic Understanding of Phage Therapy. Clin. Infect. Dis. 2023, 77 (Suppl. S5), S401–S406. [Google Scholar] [CrossRef]
- Wommack, K.E.; Williamson, K.E.; Helton, R.R.; Bench, S.R.; Winget, D.M. Methods for the isolation of viruses from environmental samples. Methods Mol. Biol. 2009, 501, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Sada, T.S.; Tessema, T.S. Isolation and characterization of lytic bacteriophages from various sources in Addis Ababa against antimicrobial-resistant diarrheagenic Escherichia coli strains and evaluation of their therapeutic potential. BMC Infect. Dis. 2024, 24, 310. [Google Scholar] [CrossRef] [PubMed]
- Stacey, H.J.; De Soir, S.; Jones, J.D. The Safety and Efficacy of Phage Therapy: A Systematic Review of Clinical and Safety Trials. Antibiotics 2022, 11, 1340. [Google Scholar] [CrossRef]
- Zaczek, M.; Weber-Dabrowska, B.; Miedzybrodzki, R.; Lusiak-Szelachowska, M.; Górski, A. Phage Therapy in Poland—A Centennial Journey to the First Ethically Approved Treatment Facility in Europe. Front. Microbiol. 2020, 11, 1056. [Google Scholar] [CrossRef]
- Kulangara, A.C.; Sellers, M.I. Passage of bacteriophages from mother to foetus in the rat. Proc. Soc. Exp. Biol. Med. 1959, 101, 207–211. [Google Scholar] [CrossRef] [PubMed]
- Soutourina, O.A.; Monot, M.; Boudry, P.; Saujet, L.; Pichon, C.; Sismeiro, O.; Semenova, E.; Severinov, K.; Le Bouguenec, C.; Coppee, J.Y.; et al. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium Difficile. PLoS Genet. 2013, 9, e1003493. [Google Scholar] [CrossRef] [PubMed]
- Willy, C.; Bugert, J.J.; Classen, A.Y.; Deng, L.; Düchting, A.; Gross, J.; Hammerl, J.A.; Korf, I.H.E.; Kühn, C.; Lieberknecht-Jouy, S.; et al. Phage Therapy in Germany-Update 2023. Viruses 2023, 15, 588. [Google Scholar] [CrossRef]
- Ling, H.; Lou, X.; Luo, Q.; He, Z.; Sun, M.; Sun, J. Recent advances in bacteriophage-based therapeutics: Insight into the post-antibiotic era. Acta Pharm. Sin. B 2022, 12, 4348–4364. [Google Scholar] [CrossRef]
- Steele, A.; Stacey, H.J.; de Soir, S.; Jones, J.D. The Safety and Efficacy of Phage Therapy for Superficial Bacterial Infections: A Systematic Review. Antibiotics 2020, 9, 754. [Google Scholar] [CrossRef]
- Delbridge, A.R.; Strasser, A. The BCL-2 protein family, BH3-mimetics and cancer therapy. Cell Death Differ. 2015, 22, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Payaslian, F.; Gradaschi, V.; Salazar, L.R.; Dieterle, M.E.; Urdániz, E.; Di Paola, M.; Zon, F.; Allievi, M.; Rivas, C.S.; Raya, R.R.; et al. Tightening Bonds in Latin America Through Phagel Discovery. Phage-Ther. Appl. Res. 2021, 2, 7–10. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Segundo-Arizmendi, N.; Arellano-Maciel, D.; Rivera-Ramírez, A.; Piña-González, A.M.; López-Leal, G.; Hernández-Baltazar, E. Bacteriophages: A Challenge for Antimicrobial Therapy. Microorganisms 2025, 13, 100. https://doi.org/10.3390/microorganisms13010100
Segundo-Arizmendi N, Arellano-Maciel D, Rivera-Ramírez A, Piña-González AM, López-Leal G, Hernández-Baltazar E. Bacteriophages: A Challenge for Antimicrobial Therapy. Microorganisms. 2025; 13(1):100. https://doi.org/10.3390/microorganisms13010100
Chicago/Turabian StyleSegundo-Arizmendi, Nallelyt, Dafne Arellano-Maciel, Abraham Rivera-Ramírez, Adán Manuel Piña-González, Gamaliel López-Leal, and Efren Hernández-Baltazar. 2025. "Bacteriophages: A Challenge for Antimicrobial Therapy" Microorganisms 13, no. 1: 100. https://doi.org/10.3390/microorganisms13010100
APA StyleSegundo-Arizmendi, N., Arellano-Maciel, D., Rivera-Ramírez, A., Piña-González, A. M., López-Leal, G., & Hernández-Baltazar, E. (2025). Bacteriophages: A Challenge for Antimicrobial Therapy. Microorganisms, 13(1), 100. https://doi.org/10.3390/microorganisms13010100








