Cultivable and Non-Cultivable Approach to Bacteria from Undisturbed Soil with Plant Growth-Promoting Capacity
Abstract
:1. Introduction
2. Materials and Methods
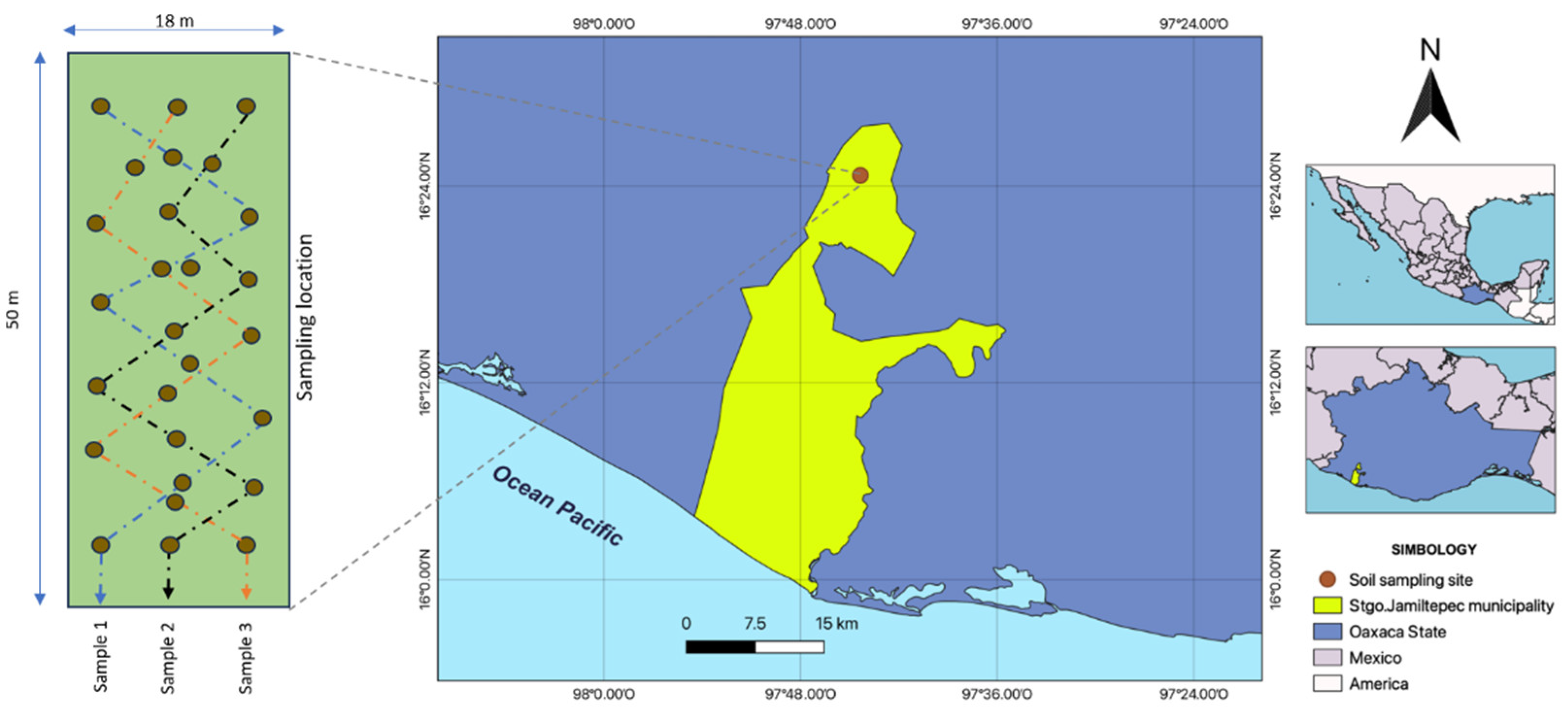
2.1. Description of the Study Area and Soil Sample
2.2. Isolation of Culturable Bacteria
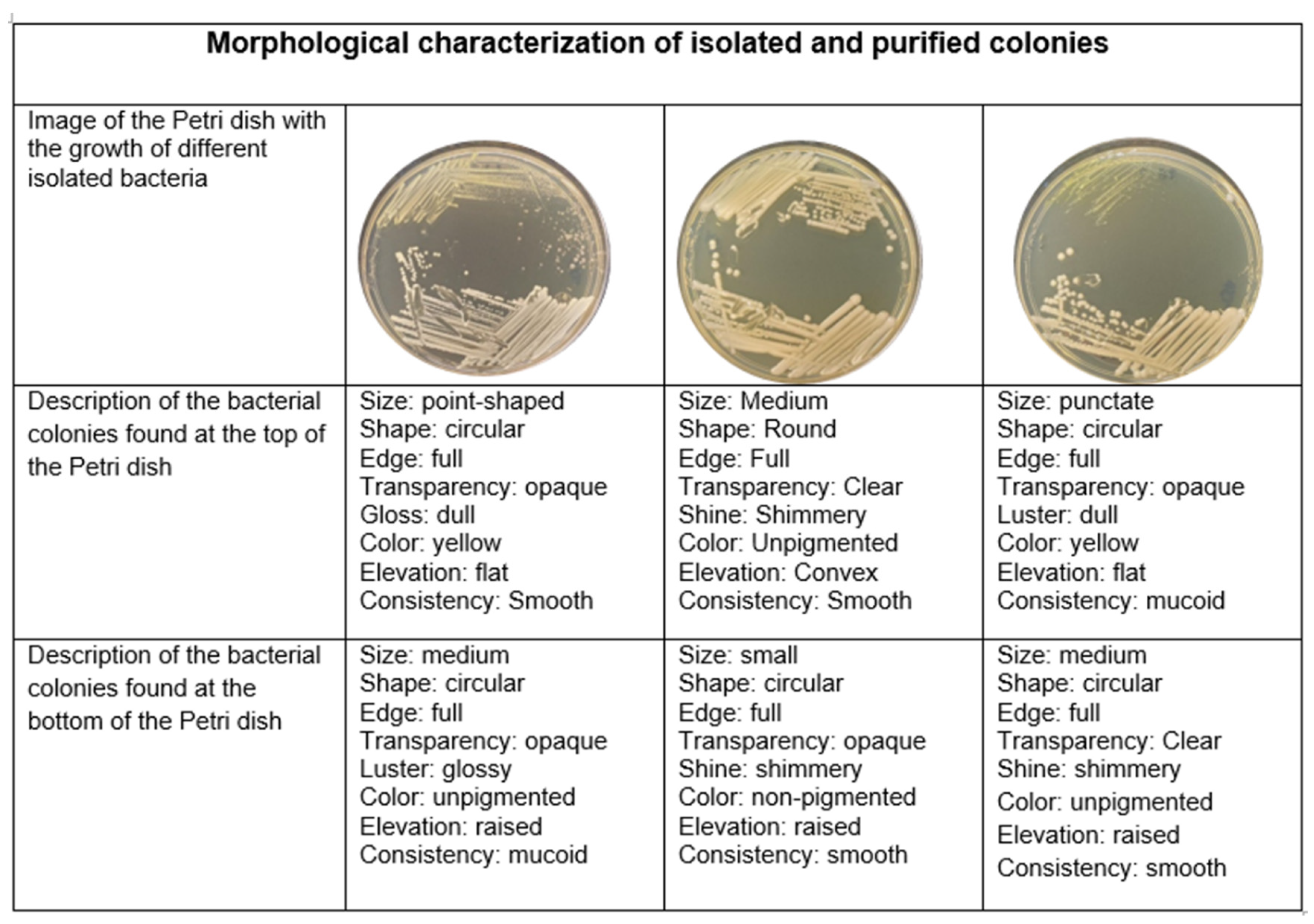
2.3. Morphological and Biochemical Characterization of Rhizobacteria
2.4. Phosphate Solubilization
2.5. Potassium Solubilization
2.6. Siderophore Production
2.7. Jensen’s N-Free Medium
2.8. Growth Promotion in Corn Seedlings by Promoter Bacteria
2.9. Description of No Culture Soil Bacteria
2.9.1. DNA Extraction and Preparation of Libraries and Sequencing Procedure Metabarcoding Approach
2.9.2. Bioinformatics Analysis
2.10. Evaluating Co-Occurrence Networks Between Genera
3. Results and Discussion
3.1. Growth Promotion In Vitro Evaluation
3.2. Identification of PGPB from Biochemical Tests
3.3. Evaluation of Growth Promotion in Corn Seedlings
3.4. Analysis of the Non-Cultivable Microbiota Associated with Undisturbed Soil
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Labouyrie, M.; Ballabio, C.; Romero, F.; Panagos, P.; Jones, A.; Schmid, M.W.; Mikryukov, V.; Dulya, O.; Tedersoo, L.; Bahram, M.; et al. Patterns in Soil Microbial Diversity across Europe. Nat. Commun. 2023, 14, 3311. [Google Scholar] [CrossRef] [PubMed]
- Van Der Heijden, M.G.A.; Bardgett, R.D.; Van Straalen, N.M. The Unseen Majority: Soil Microbes as Drivers of Plant Diversity and Productivity in Terrestrial Ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Baquerizo, M.; Reich, P.B.; Trivedi, C.; Eldridge, D.J.; Abades, S.; Alfaro, F.D.; Bastida, F.; Berhe, A.A.; Cutler, N.A.; Gallardo, A.; et al. Multiple Elements of Soil Biodiversity Drive Ecosystem Functions across Biomes. Nat. Ecol. Evol. 2020, 4, 210–220. [Google Scholar] [CrossRef]
- Schoenborn, L.; Yates, P.S.; Grinton, B.E.; Hugenholtz, P.; Janssen, P.H. Liquid Serial Dilution Is Inferior to Solid Media for Isolation of Cultures Representative of the Phylum-Level Diversity of Soil Bacteria. Appl. Environ. Microbiol. 2004, 70, 4363–4366. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Plant Growth-Promoting Bacteria: Mechanisms and Applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Sheflin, A.M.; Manter, D.K.; Vivanco, J.M. Manipulating the Soil Microbiome to Increase Soil Health and Plant Fertility. Biol. Fertil. Soils 2012, 48, 489–499. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the Unknown: Disentangling the Complexities of the Soil Microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Gómez-Godínez, L.J.; Martínez-Romero, E.; Banuelos, J.; Arteaga-Garibay, R.I. Tools and Challenges to Exploit Microbial Communities in Agriculture. Curr. Res. Microb. Sci. 2021, 2, 100062. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Six, J. Soil Structure and Microbiome Functions in Agroecosystems. Nat. Rev. Earth Environ. 2022, 4, 4–18. [Google Scholar] [CrossRef]
- Gamalero, E.; Glick, B.R. Mechanisms Used by Plant Growth-Promoting Bacteria. In Bacteria in Agrobiology: Plant Nutrient Management; Maheshwari, D.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 17–46. ISBN 978-3-642-21060-0. [Google Scholar]
- Gómez-Godínez, L.J.; Aguirre-Noyola, J.L.; Martínez-Romero, E.; Arteaga-Garibay, R.I.; Ireta-Moreno, J.; Ruvalcaba-Gómez, J.M. A Look at Plant-Growth-Promoting Bacteria. Plants 2023, 12, 1668. [Google Scholar] [CrossRef]
- Karnwal, A. Pseudomonas Spp., a Zinc-Solubilizing Vermicompost Bacteria with Plant Growth-Promoting Activity Moderates Zinc Biofortification in Tomato. Int. J. Veg. Sci. 2021, 27, 398–412. [Google Scholar] [CrossRef]
- Cook, J.; Degon, Z.; Ruiz, D.; Pope, J.; Rahmatallah, Y.; Mukherjee, A. The Plant Growth-Promoting Bacteria, Azospirillum Brasilense, Induce a Diverse Array of Genes in Rice Shoots and Promote Their Growth. Plant Growth Regul. 2022, 97, 143–155. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, S.; Mukherjee, A.; Rastogi, R.P.; Verma, J.P. Salt-Tolerant Plant Growth-Promoting Bacillus Pumilus Strain JPVS11 to Enhance Plant Growth Attributes of Rice and Improve Soil Health under Salinity Stress. Microbiol. Res. 2021, 242, 126616. [Google Scholar] [CrossRef]
- Tsotetsi, T.; Nephali, L.; Malebe, M.; Tugizimana, F. Bacillus for Plant Growth Promotion and Stress Resilience: What Have We Learned? Plants 2022, 11, 2482. [Google Scholar] [CrossRef]
- Salazar-Garcia, G.; Balaguera-Lopez, H.E.; Hernandez, J.P. Effect of Plant Growth-Promoting Bacteria Azospirillum Brasilense on the Physiology of Radish (Raphanus Sativus L.) under Waterlogging Stress. Agronomy 2022, 12, 726. [Google Scholar] [CrossRef]
- Singh, D.P. Microbiome Bioprospecting for Sustainable Agrobiome and Circular Bioeconomy. Anthr. Sci. 2024, 3, 113–121. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren Van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing Structure and Assembly Cues for Arabidopsis Root-Inhabiting Bacterial Microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and Heritability of the Maize Rhizosphere Microbiome under Field Conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef] [PubMed]
- Cordovez, V.; Dini-Andreote, F.; Carrión, V.J.; Raaijmakers, J.M. Ecology and Evolution of Plant Microbiomes. Annu. Rev. Microbiol. 2019, 73, 69–88. [Google Scholar] [CrossRef]
- Arunrat, N.; Sansupa, C.; Sereenonchai, S.; Hatano, R. Stability of Soil Bacteria in Undisturbed Soil and Continuous Maize Cultivation in Northern Thailand. Front. Microbiol. 2023, 14, 1285445. [Google Scholar] [CrossRef]
- Wang, J.; Rhodes, G.; Huang, Q.; Shen, Q. Plant Growth Stages and Fertilization Regimes Drive Soil Fungal Community Compositions in a Wheat-Rice Rotation System. Biol. Fertil. Soils 2018, 54, 731–742. [Google Scholar] [CrossRef]
- Hua, H.; Sui, X.; Liu, Y.; Liu, X.; Chang, Q.; Xu, R.; Li, M.; Mu, L. Effects of Land Use Type Transformation on the Structure and Diversity of Soil Bacterial Communities. Life 2024, 14, 252. [Google Scholar] [CrossRef] [PubMed]
- Williams, D.R.; Alvarado, F.; Green, R.E.; Manica, A.; Phalan, B.; Balmford, A. Land-use Strategies to Balance Livestock Production, Biodiversity Conservation and Carbon Storage in Yucatán, Mexico. Glob. Change Biol. 2017, 23, 5260–5272. [Google Scholar] [CrossRef]
- Meng, M.; Lin, J.; Guo, X.; Liu, X.; Wu, J.; Zhao, Y.; Zhang, J. Impacts of Forest Conversion on Soil Bacterial Community Composition and Diversity in Subtropical Forests. CATENA 2019, 175, 167–173. [Google Scholar] [CrossRef]
- Gouda, S.; Kerry, R.G.; Das, G.; Paramithiotis, S.; Shin, H.-S.; Patra, J.K. Revitalization of Plant Growth Promoting Rhizobacteria for Sustainable Development in Agriculture. Microbiol. Res. 2018, 206, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, T.; Rashid, M.I.; Maire, V.; Barot, S.; Perveen, N.; Alvarez, G.; Mougin, C.; Fontaine, S. Root Penetration in Deep Soil Layers Stimulates Mineralization of Millennia-Old Organic Carbon. Soil Biol. Biochem. 2018, 124, 150–160. [Google Scholar] [CrossRef]
- Eickhorst, T.; Tippkötter, R. Detection of Microorganisms in Undisturbed Soil by Combining Fluorescence in Situ Hybridization (FISH) and Micropedological Methods. Soil Biol. Biochem. 2008, 40, 1284–1293. [Google Scholar] [CrossRef]
- Wydro, U. Soil Microbiome Study Based on DNA Extraction: A Review. Water 2022, 14, 3999. [Google Scholar] [CrossRef]
- DeFord, L.; Yoon, J.-Y. Soil Microbiome Characterization and Its Future Directions with Biosensing. J. Biol. Eng. 2024, 18, 50. [Google Scholar] [CrossRef]
- Nkongolo, K.K.; Narendrula-Kotha, R. Advances in Monitoring Soil Microbial Community Dynamic and Function. J. Appl. Genet. 2020, 61, 249–263. [Google Scholar] [CrossRef]
- Boers, S.A.; Jansen, R.; Hays, J.P. Understanding and Overcoming the Pitfalls and Biases of Next-Generation Sequencing (NGS) Methods for Use in the Routine Clinical Microbiological Diagnostic Laboratory. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1059–1070. [Google Scholar] [CrossRef] [PubMed]
- INEGI. Anuario estadístico y geográfico de Oaxaca 2016, 1st ed.; Instituto Nacional de Estadística y Geográfica: Aguascalientes, México, 2016; ISBN 978-607-739-966-7. [Google Scholar]
- Norma Oficial Mexicana 110 (NOM-110); Bienes y Servicios. Preparación y Dilución de Muestras de Alimentos para su Análisis Microbiológico. Secretaria de Salud (SSA), Diario Oficial de la Federacion: Mexico City, Mexico, 1994.
- Clarke, P.H.; Cowan, S.T. Biochemical Methods for Bacteriology. J. Gen. Microbiol. 1952, 6, 187–197. [Google Scholar] [CrossRef]
- Nautiyal, C.S. An Efficient Microbiological Growth Medium for Screening Phosphate Solubilizing Microorganisms. FEMS Microbiol. Lett. 1999, 170, 265–270. [Google Scholar] [CrossRef] [PubMed]
- Velázquez-Gurrola, A.; Ramos-Alegría, M. Beneficios de Microorganismos Solubilizadores de P y K En La Recuperación y Mantenimiento de Suelos Agrícolas. In Proceedings of the VIII Congreso Mundial de la Palta 2015, Lima, Perú, 3–18 September 2015; pp. 495–499. [Google Scholar]
- Schwyn, B.; Neilands, J.B. Universal Chemical Assay for the Detection and Determination of Siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Ali, A.A.; El-Kholy, A.S. Isolation and Characterization of Endophytic Kosakonia Radicincitans to Stimulate Wheat Growth in Saline Soil. J. Adv. Microbiol. 2022, 22, 115–126. [Google Scholar] [CrossRef]
- Gómez-Godínez, L.J.; Fernandez-Valverde, S.L.; Martinez Romero, J.C.; Martínez-Romero, E. Metatranscriptomics and Nitrogen Fixation from the Rhizoplane of Maize Plantlets Inoculated with a Group of PGPRs. Syst. Appl. Microbiol. 2019, 42, 517–525. [Google Scholar] [CrossRef] [PubMed]
- Calvillo-Aguilar, F.F.; Cruz-Cárdenas, C.I.; Chávez-Díaz, I.F.; Sandoval-Cancino, G.; Ruiz-Ramírez, S.; Bautista-Ramírez, E.; Ramos-Garza, J.; Hernández-Rodríguez, C.H.; Zelaya-Molina, L.X. Germination Test for the Evaluation of Plant-Growth Promoting Microorganisms. J. Microbiol. Methods 2023, 207, 106708. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 4 January 2025).
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet.journal 2011, 17, 10. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Liu, C.; Li, C.; Jiang, Y.; Zeng, R.J.; Yao, M.; Li, X. A Guide for Comparing Microbial Co-occurrence Networks. iMeta 2023, 2, e71. [Google Scholar] [CrossRef] [PubMed]
- Csardi, G.; Nepusz, T. The Igraph Software. Complex Syst 2006, 1695, 1–9. [Google Scholar]
- Marasco, R.; Mosqueira, M.J.; Fusi, M.; Ramond, J.-B.; Merlino, G.; Booth, J.M.; Maggs-Kölling, G.; Cowan, D.A.; Daffonchio, D. Rhizosheath Microbial Community Assembly of Sympatric Desert Speargrasses Is Independent of the Plant Host. Microbiome 2018, 6, 215. [Google Scholar] [CrossRef]
- Wood, W.A.; Krieg, N.R. Methods for General and Molecular Bacteriology; American Society for Microbiology: Washington, DC, USA, 1989; pp. 1–9. [Google Scholar]
- Sullivan, T.S.; Gadd, G.M. Metal Bioavailability and the Soil Microbiome. In Advances in Agronomy; Elsevier: Amsterdam, The Netherlands, 2019; Volume 155, pp. 79–120. ISBN 978-0-12-817408-1. [Google Scholar]
- Kloepper, J.W.; Leong, J.; Teintze, M.; Schroth, M.N. Pseudomonas Siderophores: A Mechanism Explaining Disease-Suppressive Soils. Curr. Microbiol. 1980, 4, 317–320. [Google Scholar] [CrossRef]
- Sharma, A.; Johri, B.N. Growth Promoting Influence of Siderophore-Producing Pseudomonas Strains GRP3A and PRS9 in Maize (Zea Mays L.) under Iron Limiting Conditions. Microbiol. Res. 2003, 158, 243–248. [Google Scholar] [CrossRef]
- Gu, S.; Wei, Z.; Shao, Z.; Friman, V.-P.; Cao, K.; Yang, T.; Kramer, J.; Wang, X.; Li, M.; Mei, X.; et al. Competition for Iron Drives Phytopathogen Control by Natural Rhizosphere Microbiomes. Nat. Microbiol. 2020, 5, 1002–1010. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, G.; Huang, Y.; Guo, M.; Song, J.; Zhang, T.; Long, Y.; Wang, B.; Liu, H. A Potential Biofertilizer—Siderophilic Bacteria Isolated from the Rhizosphere of Paris Polyphylla Var. Yunnanensis. Front. Microbiol. 2022, 13, 870413. [Google Scholar] [CrossRef]
- Dong, X.; Lv, L.; Wang, W.; Liu, Y.; Yin, C.; Xu, Q.; Yan, H.; Fu, J.; Liu, X. Differences in Distribution of Potassium-Solubilizing Bacteria in Forest and Plantation Soils in Myanmar. Int. J. Environ. Res. Public. Health 2019, 16, 700. [Google Scholar] [CrossRef]
- Sarikhani, M.R.; Oustan, S.; Ebrahimi, M.; Aliasgharzad, N. Isolation and Identification of Potassium-releasing Bacteria in Soil and Assessment of Their Ability to Release Potassium for Plants. Eur. J. Soil Sci. 2018, 69, 1078–1086. [Google Scholar] [CrossRef]
- Rosenblueth, M.; Ormeño-Orrillo, E.; López-López, A.; Rogel, M.A.; Reyes-Hernández, B.J.; Martínez-Romero, J.C.; Reddy, P.M.; Martínez-Romero, E. Nitrogen Fixation in Cereals. Front. Microbiol. 2018, 9, 1794. [Google Scholar] [CrossRef]
- Nag, P.; Shriti, S.; Das, S. Microbiological Strategies for Enhancing Biological Nitrogen Fixation in Nonlegumes. J. Appl. Microbiol. 2020, 129, 186–198. [Google Scholar] [CrossRef]
- Mukherjee, R.; Sen, S. Role of Biological Nitrogen Fixation (BNF) in Sustainable Agriculture: A Review. Int. J. Adv. Life Sci. Res. 2021, 4, 1–7. [Google Scholar] [CrossRef]
- Singh, P.; Singh, R.K.; Li, H.-B.; Guo, D.-J.; Sharma, A.; Lakshmanan, P.; Malviya, M.K.; Song, X.-P.; Solanki, M.K.; Verma, K.K.; et al. Diazotrophic Bacteria Pantoea Dispersa and Enterobacter Asburiae Promote Sugarcane Growth by Inducing Nitrogen Uptake and Defense-Related Gene Expression. Front. Microbiol. 2021, 11, 600417. [Google Scholar] [CrossRef]
- Ghosh, A.; Pramanik, K.; Bhattacharya, S.; Mondal, S.; Ghosh, S.K.; Maiti, T.K. A Potent Cadmium Bioaccumulating Enterobacter Cloacae Strain Displays Phytobeneficial Property in Cd-Exposed Rice Seedlings. Curr. Res. Microb. Sci. 2022, 3, 100101. [Google Scholar] [CrossRef]
- Kang, S.-M.; Khan, M.-A.; Hamayun, M.; Kim, L.-R.; Kwon, E.-H.; Kang, Y.-S.; Kim, K.-Y.; Park, J.-J.; Lee, I.-J. Phosphate-Solubilizing Enterobacter Ludwigii AFFR02 and Bacillus Megaterium Mj1212 Rescues Alfalfa’s Growth under Post-Drought Stress. Agriculture 2021, 11, 485. [Google Scholar] [CrossRef]
- Ortega-Ortega, Y.; Sarmiento-López, L.G.; Baylón-Palomino, A.; Vázquez-Lee, J.; Maldonado-Bonilla, L.D.; Flores-Olivas, A.; Valenzuela-Soto, J.H. Enterobacter sp. DBA51 Produces ACC Deaminase and Promotes the Growth of Tomato (Solanum Lycopersicum L.) and Tobacco (Nicotiana Tabacum L.) Plants under Greenhouse Condition. Curr. Res. Microb. Sci. 2024, 6, 100207. [Google Scholar] [CrossRef]
- Anzuay, M.S.; Prenollio, A.; Ludueña, L.M.; Morla, F.D.; Cerliani, C.; Lucero, C.; Angelini, J.G.; Taurian, T. Enterobacter sp. J49: A Native Plant Growth-Promoting Bacteria as Alternative to the Application of Chemical Fertilizers on Peanut and Maize Crops. Curr. Microbiol. 2023, 80, 85. [Google Scholar] [CrossRef]
- Awolope, O.K.; O’Driscoll, N.H.; Di Salvo, A.; Lamb, A.J. The Complete Genome Sequence of Hafnia Alvei A23BA; a Potential Antibiotic-Producing Rhizobacterium. BMC Res. Notes 2021, 14, 8. [Google Scholar] [CrossRef]
- Rana, A.; Saharan, B.; Kabi, S.R.; Prasanna, R.; Nain, L. Providencia, a PGPR with Biocontrol Potential Elicits Defense Enzymes in Wheat. Ann. Plant Prot. Sci. 2011, 19, 138–141. [Google Scholar]
- Kersters, K.; De Vos, P.; Gillis, M.; Swings, J.; Vandamme, P.; Stackebrandt, E. Introduction to the Proteobacteria. In The Prokaryotes; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 3–37. ISBN 978-0-387-25495-1. [Google Scholar]
- Mukhopadhya, I.; Hansen, R.; El-Omar, E.M.; Hold, G.L. IBD—What Role Do Proteobacteria Play? Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 219–230. [Google Scholar] [CrossRef]
- Janssen, P.H. Identifying the Dominant Soil Bacterial Taxa in Libraries of 16S rRNA and 16S rRNA Genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef]
- Kielak, A.M.; Barreto, C.C.; Kowalchuk, G.A.; Van Veen, J.A.; Kuramae, E.E. The Ecology of Acidobacteria: Moving beyond Genes and Genomes. Front. Microbiol. 2016, 7, 744. [Google Scholar] [CrossRef]
- Yoon, J.; Yasumoto-Hirose, M.; Katsuta, A.; Sekiguchi, H.; Matsuda, S.; Kasai, H.; Yokota, A. Coraliomargarita akajimensis Gen. Nov., sp. Nov., a Novel Member of the Phylum ‘Verrucomicrobia’ Isolated from Seawater in Japan. Int. J. Syst. Evol. Microbiol. 2007, 57, 959–963. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.; Matsuo, Y.; Adachi, K.; Nozawa, M.; Matsuda, S.; Kasai, H.; Yokota, A. Description of Persicirhabdus sediminis Gen. Nov., sp. Nov., Roseibacillus ishigakijimensis Gen. Nov., sp. Nov., Roseibacillus ponti sp. Nov., Roseibacillus persicicus sp. Nov., Luteolibacter pohnpeiensis Gen. Nov., sp. Nov. and Luteolibacter algae sp. Nov., Six Marine Members of the Phylum “Verrucomicrobia”, and Emended Descriptions of the Class Verrucomicrobiae, the Order Verrucomicrobiales and the Family Verrucomicrobiaceae. Int. J. Syst. Evol. Microbiol. 2008, 58, 998–1007. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Horn, M. The Planctomycetes, Verrucomicrobia, Chlamydiae and Sister Phyla Comprise a Superphylum with Biotechnological and Medical Relevance. Curr. Opin. Biotechnol. 2006, 17, 241–249. [Google Scholar] [CrossRef]
- Blümel, S.; Mark, B.; Busse, H.J.; Kämpfer, P.; Stolz, A. Pigmentiphaga kullae Gen. Nov., sp. Nov., a Novel Member of the Family Alcaligenaceae with the Ability to Decolorize Azo Dyes Aerobically. Int. J. Syst. Evol. Microbiol. 2001, 51, 1867–1871. [Google Scholar] [CrossRef] [PubMed]
- Kämpfer, P.; Busse, H.-J.; Criscuolo, A.; Bizet, C.; Clermont, D.; McInroy, J.A.; Glaeser, S.P. Pigmentiphaga humi sp. Nov., Isolated from Soil Amended with Humic Acid. Int. J. Syst. Evol. Microbiol. 2019, 69, 1573–1578. [Google Scholar] [CrossRef]
- Wang, G.; Yue, W.; Liu, Y.; Li, F.; Xiong, M.; Zhang, H. Biodegradation of the Neonicotinoid Insecticide Acetamiprid by Bacterium Pigmentiphaga sp. Strain AAP-1 Isolated from Soil. Bioresour. Technol. 2013, 138, 359–368. [Google Scholar] [CrossRef]
- Xiong, J.-X.; Du, L.-S.; Li, N.-N.; Wu, X.-T.; Xiang, Y.; Li, S.; Zou, L.; Liu, D.; Huang, D.; Xie, Z.F.; et al. Pigmentiphaga Kullae CHJ604 Improved the Growth of Tobacco by Degrading Allelochemicals and Xenobiotics in Continuous Cropping Obstacles. J. Hazard. Mater. 2024, 465, 133466. [Google Scholar] [CrossRef]
- Hiraishi, A.; Ueda, Y. Rhodoplanes Gen. Nov., a New Genus of Phototrophic Bacteria Including Rhodopseudomonas Rosea as Rhodoplanes Roseus Comb. Nov. and Rhodoplanes elegans sp. Nov. Int. J. Syst. Bacteriol. 1994, 44, 665–673. [Google Scholar] [CrossRef]
- Zhang, H.; Shi, Y.; Dong, Y.; Lapen, D.R.; Liu, J.; Chen, W. Subsoiling and Conversion to Conservation Tillage Enriched Nitrogen Cycling Bacterial Communities in Sandy Soils under Long-Term Maize Monoculture. Soil Tillage Res. 2022, 215, 105197. [Google Scholar] [CrossRef]
- Ward, N.L.; Challacombe, J.F.; Janssen, P.H.; Henrissat, B.; Coutinho, P.M.; Wu, M.; Xie, G.; Haft, D.H.; Sait, M.; Badger, J.; et al. Three Genomes from the Phylum Acidobacteria Provide Insight into the Lifestyles of These Microorganisms in Soils. Appl. Environ. Microbiol. 2009, 75, 2046–2056. [Google Scholar] [CrossRef] [PubMed]
- Challacombe, J.F.; Eichorst, S.A.; Hauser, L.; Land, M.; Xie, G.; Kuske, C.R. Biological Consequences of Ancient Gene Acquisition and Duplication in the Large Genome of Candidatus Solibacter Usitatus Ellin6076. PLoS ONE 2011, 6, e24882. [Google Scholar] [CrossRef]
- Dwivedi, M. Gluconobacter. In Beneficial Microbes in Agro-Ecology; Elsevier: Amsterdam, The Netherlands, 2020; pp. 521–544. ISBN 978-0-12-823414-3. [Google Scholar]
- Gupta, A.; Singh, V.K.; Qazi, G.; Kumar, A. Gluconobacter Oxydans: Its Biotechnological Applications. J. Mol. Microbiol. Biotechnol. 2001, 3, 445–456. [Google Scholar]
- Yamada, Y.; Yukphan, P. Genera and Species in Acetic Acid Bacteria. Int. J. Food Microbiol. 2008, 125, 15–24. [Google Scholar] [CrossRef]
- Cavalcante, V.A.; Dobereiner, J. A New Acid-Tolerant Nitrogen-Fixing Bacterium Associated with Sugarcane. Plant Soil 1988, 108, 23–31. [Google Scholar] [CrossRef]
- Fuentes-Ramírez, L.E.; Bustillos-Cristales, R.; Tapia-Hernández, A.; Jiménez-Salgado, T.; Wang, E.T.; Martínez-Romero, E.; Caballero-Mellado, J. Novel Nitrogen-Fixing Acetic Acid Bacteria, Gluconacetobacter johannae sp. Nov. and Gluconacetobacter azotocaptans sp. Nov., Associated with Coffee Plants. Int. J. Syst. Evol. Microbiol. 2001, 51, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Maruoka, M.; Nanatani, K.; Hidaka, M.; Abe, N.; Kaneko, J.; Sakai, Y.; Abe, K.; Yokota, A.; Yabe, S. High Cellulolytic Potential of the Ktedonobacteria Lineage Revealed by Genome-Wide Analysis of CAZymes. J. Biosci. Bioeng. 2021, 131, 622–630. [Google Scholar] [CrossRef]
- Felske. Akkermans Prominent Occurrence of Ribosomes from an Uncultured Bacterium of the Verrucomicrobiales Cluster in Grassland Soils. Lett. Appl. Microbiol. 1998, 26, 219–223. [Google Scholar] [CrossRef]
- Brewer, T.E.; Handley, K.M.; Carini, P.; Gilbert, J.A.; Fierer, N. Genome Reduction in an Abundant and Ubiquitous Soil Bacterium ‘Candidatus Udaeobacter Copiosus’ . Nat. Microbiol. 2016, 2, 16198. [Google Scholar] [CrossRef]
- Weon, H.-Y.; Anandham, R.; Kim, B.-Y.; Hong, S.-B.; Jeon, Y.-A.; Kwon, S.-W. Dyella soli sp. Nov. and Dyella terrae sp. Nov., Isolated from Soil. Int. J. Syst. Evol. Microbiol. 2009, 59, 1685–1690. [Google Scholar] [CrossRef] [PubMed]
- Dethier, L.; Jespersen, J.R.P.; Lloyd, J.; Pupi, E.; Zhou, W.; Liu, F.; Bai, Y.; Halkier, B.A.; Xu, D. Isolation of a Novel Plant Growth-Promoting Dyella sp. from a Danish Natural Soil. bioRxiv 2025. [Google Scholar] [CrossRef]
- Zhang, M.; Liang, G.; Ren, S.; Li, L.; Li, C.; Li, Y.; Yu, X.; Yin, Y.; Liu, T.; Liu, X. Responses of Soil Microbial Community Structure, Potential Ecological Functions, and Soil Physicochemical Properties to Different Cultivation Patterns in Cucumber. Geoderma 2023, 429, 116237. [Google Scholar] [CrossRef]
- Hall, C.M.; Busch, J.D.; Shippy, K.; Allender, C.J.; Kaestli, M.; Mayo, M.; Sahl, J.W.; Schupp, J.M.; Colman, R.E.; Keim, P.; et al. Diverse Burkholderia Species Isolated from Soils in the Southern United States with No Evidence of B. Pseudomallei. PLoS ONE 2015, 10, e0143254. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wong, P. Effect of Burkholderia (Pseudomonas) Cepacia and Soil Type on the Control of Crown Rot in Wheat. Plant Soil 1998, 203, 103–108. [Google Scholar] [CrossRef]
- Saxena, A.K.; Kumar, M.; Chakdar, H.; Anuroopa, N.; Bagyaraj, D.J. Bacillus Species in Soil as a Natural Resource for Plant Health and Nutrition. J. Appl. Microbiol. 2020, 128, 1583–1594. [Google Scholar] [CrossRef]
- Mandic-Mulec, I.; Stefanic, P.; Van Elsas, J.D. Ecology of Bacillaceae. In The Bacterial Spore; Driks, A., Eichenberger, P., Eds.; ASM Press: Washington, DC, USA, 2016; pp. 59–85. ISBN 978-1-68367-078-0. [Google Scholar] [CrossRef]
- Liu, J.; Cui, X.; Liu, Z.; Guo, Z.; Yu, Z.; Yao, Q.; Sui, Y.; Jin, J.; Liu, X.; Wang, G. The Diversity and Geographic Distribution of Cultivable Bacillus-Like Bacteria Across Black Soils of Northeast China. Front. Microbiol. 2019, 10, 1424. [Google Scholar] [CrossRef]




| Bacteria | Phosphorus Solubilization | Production of Siderophores | Jensen’s N-Free Medium | Potassium Solubilization |
|---|---|---|---|---|
| US-B1 | − | − | + | + |
| US-B2 | − | − | − | + |
| US-B3 | − | − | − | + |
| US-B4 | − | − | + | + |
| US-B5 | − | − | + | + |
| US-B6 | − | − | + | + |
| US-B7 | − | − | + | + |
| US-B8 | − | + | + | + |
| US-B9 | − | − | + | − |
| US-B10 | + | + | + | − |
| US-B11 | − | − | + | − |
| US-B12 | − | − | + | + |
| US-B13 | − | − | + | + |
| US-B14 | − | − | + | − |
| US-B15 | + | + | + | + |
| US-B16 | − | + | + | + |
| US-B17 | − | − | + | − |
| US-B18 | − | − | + | + |
| US-B19 | − | − | − | + |
| US-B20 | + | + | − | − |
| US-B21 | − | + | + | + |
| US-B22 | + | + | + | − |
| US-B23 | − | + | + | + |
| US-B24 | − | + | + | + |
| US-B25 | − | + | − | − |
| US-B26 | − | − | − | − |
| US-B27 | − | − | − | − |
| US-B28 | − | − | − | − |
| US-B29 | − | − | − | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-Godínez, L.J.; Cisneros-Saguilán, P.; Toscano-Santiago, D.D.; Santiago-López, Y.E.; Fonseca-Pérez, S.N.; Ruiz-Rivas, M.; Aguirre-Noyola, J.L.; García, G. Cultivable and Non-Cultivable Approach to Bacteria from Undisturbed Soil with Plant Growth-Promoting Capacity. Microorganisms 2025, 13, 909. https://doi.org/10.3390/microorganisms13040909
Gómez-Godínez LJ, Cisneros-Saguilán P, Toscano-Santiago DD, Santiago-López YE, Fonseca-Pérez SN, Ruiz-Rivas M, Aguirre-Noyola JL, García G. Cultivable and Non-Cultivable Approach to Bacteria from Undisturbed Soil with Plant Growth-Promoting Capacity. Microorganisms. 2025; 13(4):909. https://doi.org/10.3390/microorganisms13040909
Chicago/Turabian StyleGómez-Godínez, Lorena Jacqueline, Pedro Cisneros-Saguilán, Dulce Darina Toscano-Santiago, Yair Eduardo Santiago-López, Saúl Neftalí Fonseca-Pérez, Magali Ruiz-Rivas, José Luis Aguirre-Noyola, and Gabriel García. 2025. "Cultivable and Non-Cultivable Approach to Bacteria from Undisturbed Soil with Plant Growth-Promoting Capacity" Microorganisms 13, no. 4: 909. https://doi.org/10.3390/microorganisms13040909
APA StyleGómez-Godínez, L. J., Cisneros-Saguilán, P., Toscano-Santiago, D. D., Santiago-López, Y. E., Fonseca-Pérez, S. N., Ruiz-Rivas, M., Aguirre-Noyola, J. L., & García, G. (2025). Cultivable and Non-Cultivable Approach to Bacteria from Undisturbed Soil with Plant Growth-Promoting Capacity. Microorganisms, 13(4), 909. https://doi.org/10.3390/microorganisms13040909







