The Clinical Validity and Utility of PCR Compared to Conventional Culture and Sensitivity Testing for the Management of Complicated Urinary Tract Infections in Adults: A Secondary (Ad Hoc) Analysis of Pathogen Detection, Resistance Profiles, and Impact on Clinical Outcomes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Laboratory Testing Procedures
2.3. Interpretation of Resistance Markers and Treatment Selection
2.4. Treatment Decisions and Polymicrobial Considerations
2.5. Study and Analysis Population
2.6. Statistical Analysis
3. Results
3.1. Test Performance Across Clinical and Demographic Subpopulations
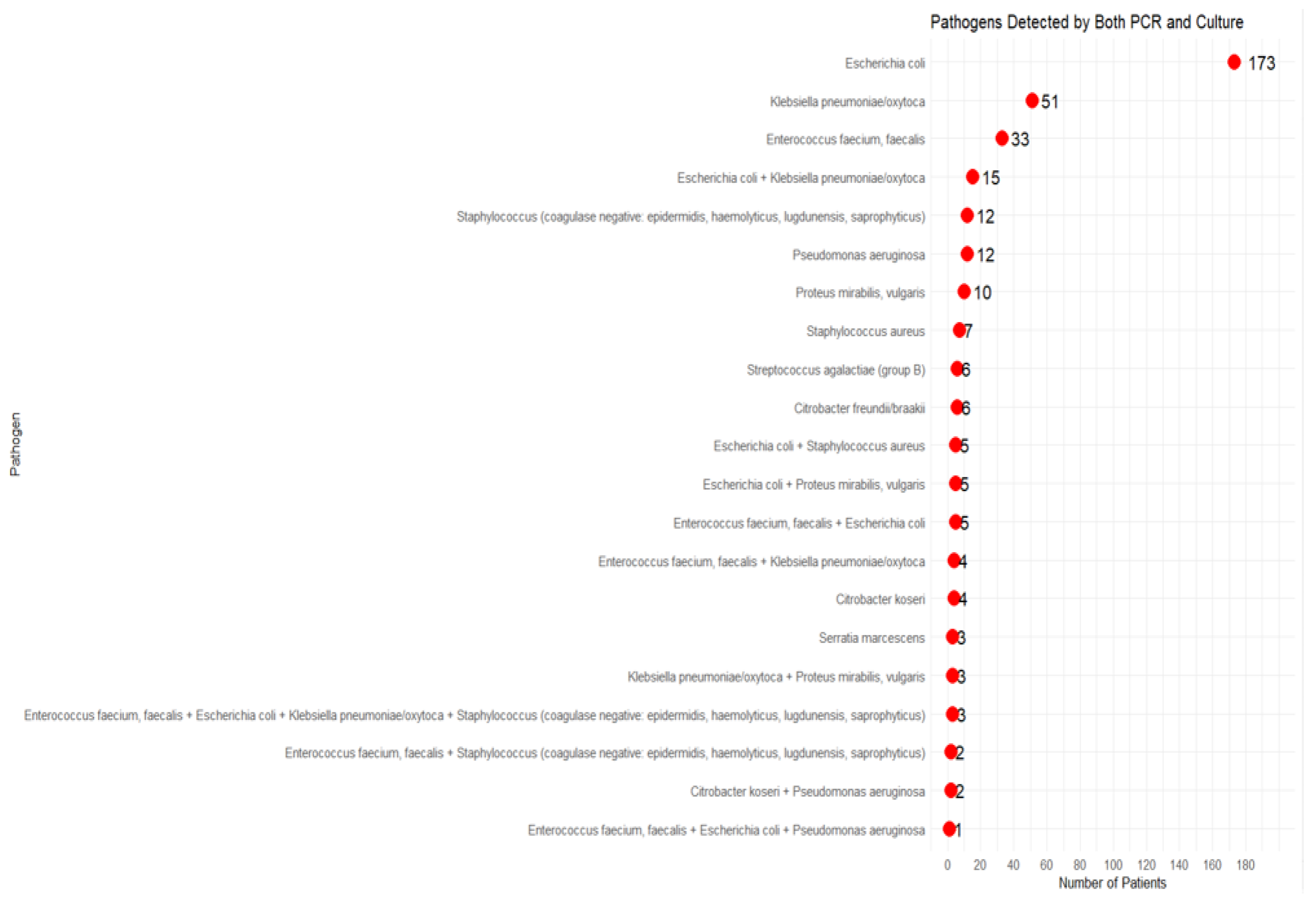
3.2. Polymicrobial Detection and Impact on Clinical Outcomes
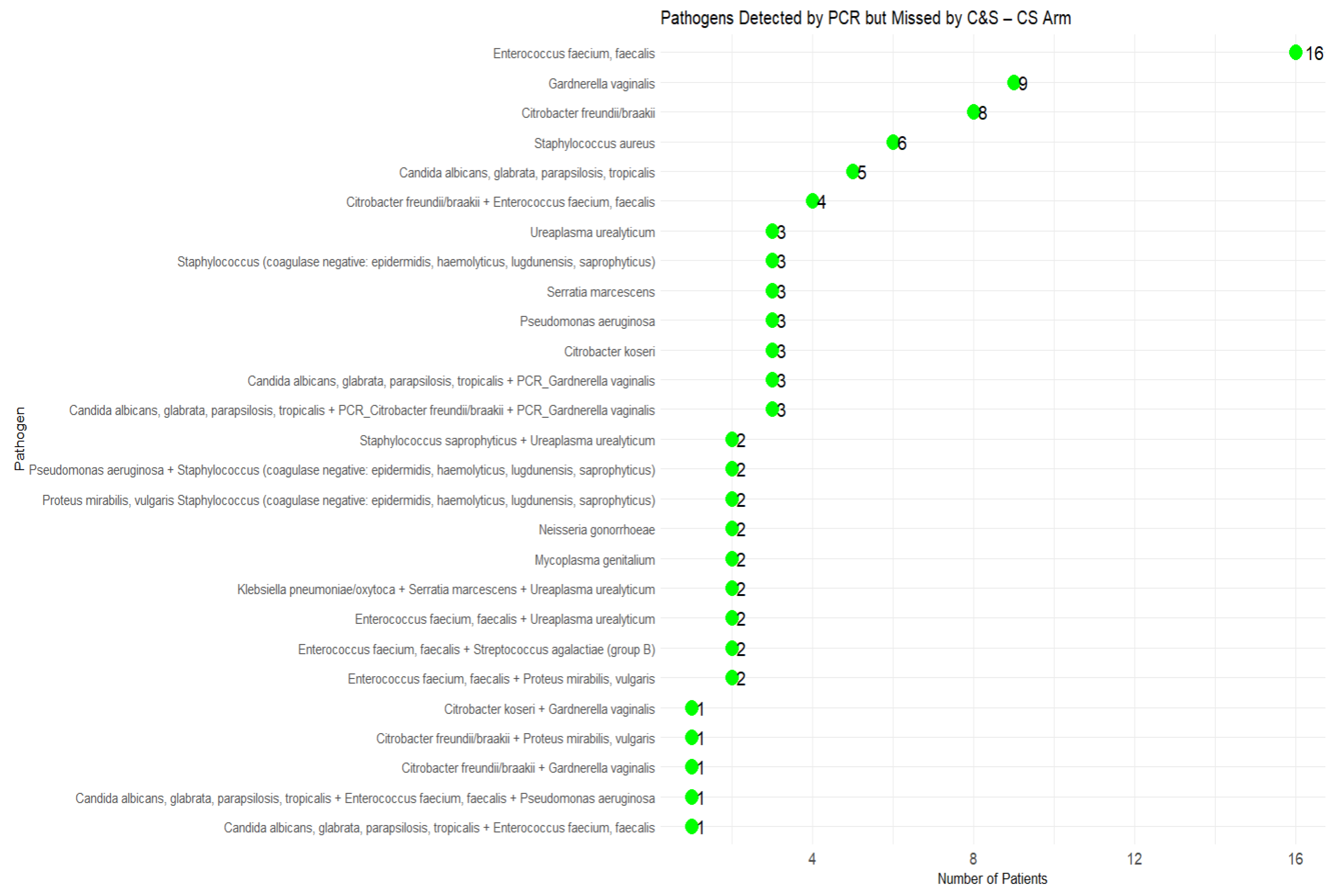
3.3. Missed Pathogens by C&S and Impact on Clinical Outcomes
3.4. Undetected Phenotypic Resistance by C&S and Its Impact on Antibiotic Selection and Clinical Outcomes
4. Discussion
4.1. Test Performance Across Clinical and Demographic Subpopulations
4.2. Polymicrobial Detection and Impact on Clinical Outcomes
4.3. Pathogens Missed by C&S and Impact on Clinical Outcomes
4.4. Undetected Phenotypic Resistance by C&S and Its Impact on Antibiotic Selection and Clinical Outcomes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Laudisio, A.; Marinosci, F.; Fontana, D.; Gemma, A.; Zizzo, A.; Coppola, A.; Rodano, L.; Incalzi, R.A. The burden of comorbidity is associated with symptomatic polymicrobial urinary tract infection among institutionalized elderly. Aging Clin. Exp. Res. 2015, 27, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Deebel, N.; Casals, R.; Dutta, R.; Mirzazadeh, M. A New Gold Rush: A review of current and developing diagnostic tools for urinary tract infections. Diagnostics 2021, 11, 479. [Google Scholar] [CrossRef] [PubMed]
- Grey, B.; Upton, M.; Joshi, L.T. Urinary tract infections: A review of the current diagnostics landscape. J. Med Microbiol. 2023, 72, 001780. [Google Scholar] [CrossRef]
- Spangler, F.L.; Williams, C.; Aberger, M.E.; Wilson, B.A.; Ajib, K.; Gholami, S.S.; Goodwin, H.N.; Park, L.Y.; Kardjadj, M.; Derrick, D.; et al. Clinical utility of PCR compared to conventional culture and sensitivity testing for the management of complicated urinary tract infections in adults: Part I. Assessment of clinical outcomes, investigator satisfaction scores, and turnaround times. Diagn. Microbiol. Infect. Dis. 2025, 111, 116601. [Google Scholar] [CrossRef] [PubMed]
- Spangler, F.L.; Williams, C.; Aberger, M.E.; Wilson, B.A.; Ajib, K.; Gholami, S.S.; Goodwin, H.N.; Park, L.Y.; Kardjadj, M.; Derrick, D.; et al. Clinical utility of PCR compared to conventional culture and sensitivity testing for management of complicated urinary tract infections in adults: Part II.Evaluation of diagnostic concordance, discordant results, and antimicrobial selection efficacy. Diagn. Microbiol. Infect. Dis. 2025, 111, 116646. [Google Scholar] [CrossRef]
- Festa, R.A.; Cockerill, F.R.; Pesano, R.L.; Haley, E.; Luke, N.; Mathur, M.; Chen, X.; Havrilla, J.; Percaccio, M.; Magallon, J.; et al. Pooled antibiotic susceptibility testing for polymicrobial UTI performs within CLSI validation standards. Antibiotics 2025, 14, 143. [Google Scholar] [CrossRef]
- Yoo, J.-J.; Shin, H.B.; Song, J.S.; Kim, M.; Yun, J.; Kim, Z.; Lee, Y.M.; Lee, S.W.; Lee, K.W.; Bin Kim, W.; et al. Urinary microbiome characteristics in female patients with acute uncomplicated cystitis and recurrent cystitis. J. Clin. Med. 2021, 10, 1097. [Google Scholar] [CrossRef]
- Akhlaghpour, M.; Haley, E.; Parnell, L.; Luke, N.; Mathur, M.; Festa, R.A.; Percaccio, M.; Magallon, J.; Remedios-Chan, M.; Rosas, A.; et al. Urine biomarkers individually and as a consensus model show high sensitivity and specificity for detecting UTIs. BMC Infect. Dis. 2024, 24, 1–11. [Google Scholar] [CrossRef]
- Haley, E.; Luke, N.; Mathur, M.; Festa, R.A.; Wang, J.; Jiang, Y.; Anderson, L.; Baunoch, D. Comparison shows that multiplex polymerase chain reaction identifies infection-associated urinary biomarker–positive urinary tract infections that are missed by standard urine culture. Eur. Urol. Open Sci. 2023, 58, 73–81. [Google Scholar] [CrossRef]
- Robledo, X.G.; Arcila, K.V.O.; Riascos, S.H.M.; García-Perdomo, H.A. Accuracy of molecular diagnostic techniques in patients with a confirmed urine culture: A systematic review and meta-analysis. Can Urol Assoc J. 2022, 16, E484–E489. [Google Scholar] [CrossRef]
- Szlachta-McGinn, A.; Douglass, K.M.; Chung, U.Y.R.; Jackson, N.J.; Nickel, J.C.; Ackerman, A.L. Molecular diagnostic methods versus conventional urine culture for diagnosis and treatment of urinary tract infection: A systematic review and meta-analysis. Eur Urol Open Sci. 2022, 44, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Price, T.K.; Dune, T.; Hilt, E.E.; Thomas-White, K.J.; Kliethermes, S.; Brincat, C.; Brubaker, L.; Wolfe, A.J.; Mueller, E.R.; Schreckenberger, P.C. The clinical urine culture: Enhanced techniques improve detection of clinically relevant microorganisms. J. Clin. Microbiol. 2016, 54, 1216–1222. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Cognetti, M.; Patel, C.; Jean-Charles, N.; Tumati, A.; Burch-Smith, R.; Holton, M.; Kapoor, D.A. The essential role of PCR and PCR panel size in comparison with urine culture in identification of polymicrobial and fastidious organisms in patients with complicated urinary tract infections. Int. J. Mol. Sci. 2023, 24, 14269. [Google Scholar] [CrossRef]
- Sun, Z.; Liu, W.; Zhang, J.; Wang, S.; Yang, F.; Fang, Y.; Jiang, W.; Ding, L.; Zhao, H.; Zhang, Y. The direct semi-quantitative detection of 18 pathogens and simultaneous screening for nine resistance genes in clinical urine samples by a high-throughput multiplex genetic detection system. Front. Cell. Infect. Microbiol. 2021, 11. [Google Scholar] [CrossRef] [PubMed]
- Band, V.I.; Weiss, D.S. Heteroresistance: A cause of unexplained antibiotic treatment failure? PLOS Pathog. 2019, 15, e1007726. [Google Scholar] [CrossRef]
- Xu, L.; Mo, X.; Zhang, H.; Wan, F.; Luo, Q.; Xiao, Y. Epidemiology, mechanisms, and clinical impact of bacterial heteroresistance. npj Antimicrob. Resist. 2025, 3, 1–9. [Google Scholar] [CrossRef]
- Zhang, L.; Huang, W.; Zhang, S.; Li, Q.; Wang, Y.; Chen, T.; Jiang, H.; Kong, D.; Lv, Q.; Zheng, Y.; et al. Rapid detection of bacterial pathogens and antimicrobial resistance genes in clinical urine samples with urinary tract infection by metagenomic nanopore sequencing. Front. Microbiol. 2022, 13, 858777. [Google Scholar] [CrossRef]
- Zhao, M.; Qi, S.; Sun, Y.; Zheng, X. Comparison of polymerase chain reaction and next-generation sequencing with conventional urine culture for the diagnosis of urinary tract infections: A meta-analysis. Open Med. 2024, 19, 20240921. [Google Scholar] [CrossRef]
- Ho, H.J.; Tan, M.X.; Chen, M.I.; Tan, T.Y.; Koo, S.H.; Koong, A.Y.L.; Ng, L.P.; Hu, P.L.; Tan, K.T.; Moey, P.K.S.; et al. Interaction between antibiotic resistance, resistance genes, and treatment response for urinary tract infections in primary care. J. Clin. Microbiol. 2019, 57, e00143-19. [Google Scholar] [CrossRef]
- Somorin, Y.M.; Weir, N.-J.M.; Pattison, S.H.; Crockard, M.A.; Hughes, C.M.; Tunney, M.M.; Gilpin, D.F. Antimicrobial resistance in urinary pathogens and culture-independent detection of trimethoprim resistance in urine from patients with urinary tract infection. BMC Microbiol. 2022, 22, 1–8. [Google Scholar] [CrossRef]
- Wojno, K.J.; Baunoch, D.; Luke, N.; Opel, M.; Korman, H.; Kelly, C.; Jafri, S.M.A.; Keating, P.; Hazelton, D.; Hindu, S.; et al. Multiplex PCR-based urinary tract infection (UTI) analysis compared to traditional urine culture in identifying significant pathogens in symptomatic patients. Urology. 2020, 136, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Elia, J.; Hafron, J.; Holton, M.; Ervin, C.; Hollander, M.B.; Kapoor, D.A. The impact of polymerase chain reaction urine testing on clinical decision-making in the management of complex urinary tract infections. Int. J. Mol. Sci. 2024, 25, 6616. [Google Scholar] [CrossRef] [PubMed]
- Hughes, D.; I Andersson, D. Environmental and genetic modulation of the phenotypic expression of antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 374–391. [Google Scholar] [CrossRef] [PubMed]
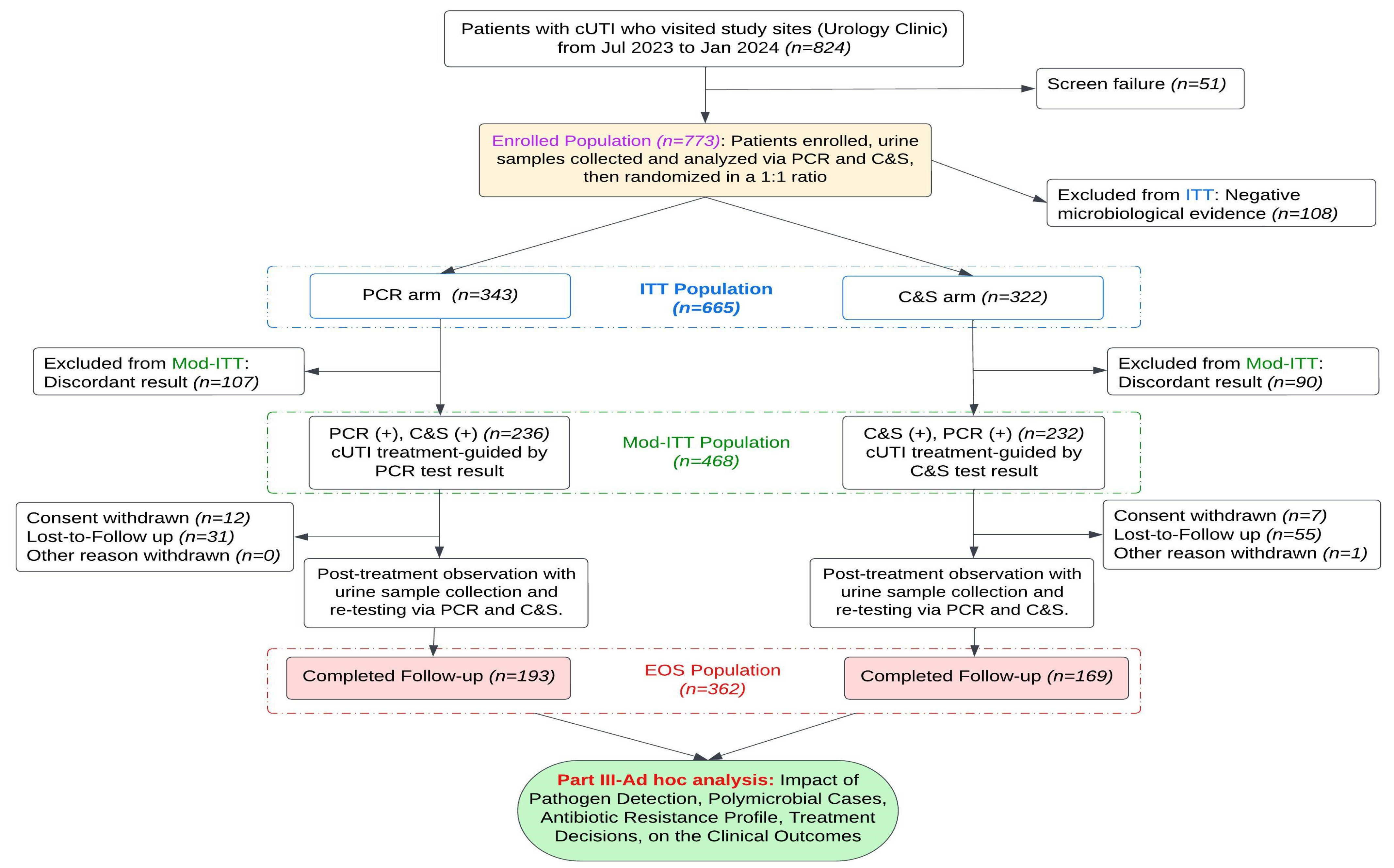

| Enrolled Population (n = 773) | EOS Population (n = 362) | ||||
|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | ||
| Sites | AUG | 94.37% | 40.23% | 94.16% | 63.25% |
| ALB | 96.12% | 36.24% | 85.62% | 62.88% | |
| NRM | 92.66% | 38.15% | 87.51% | 62.95% | |
| PSJ | 97.25% | 39.45% | 88.22% | 64.55% | |
| SCV | 95.24% | 38.12% | 91.35% | 61.22% | |
| SET | 94.66% | 37.69% | 94.23% | 61.35% | |
| Age | Elderly (65 yrs) | 94.47% | 38.35% | 85.12% | 62.45% |
| Non-Elderly (<65 yrs) | 96.32% | 39.12% | 88.74% | 63.88% | |
| Sex at birth | Male | 95.71% | 37.23% | 86.72% | 61.58% |
| Female | 95.42% | 39.47% | 84.71% | 64.25% | |
| cUTI Symptoms’ association | 02 Symptoms | 94.50% | 37.55% | 87.18% | 62.22% |
| 03 Symptoms | 95.43% | 38.73% | 85.23% | 62.57% | |
| 04 Symptoms | 95.83% | 38.66% | 98.33% | 63.45% | |
| cUTI event | Mono-Infection | 95.12% | 38.56% | 88.23% | 62.55% |
| Poly-Infection | 96.66% | 37.89% | 87.57% | 63.89% | |
| Overall | 95.32% | 38.30% | 88.06% | 62.91% | |
| C&S Arm (n = 169) | PCR Arm (n = 193) | ||||
|---|---|---|---|---|---|
| Pathogen detection | CF | FCl | Pathogen detection | CF | FCl |
| Mono C&S (1 Pathogen) (n = 115) | 25 | 90 | Mono PCR (1 Pathogen) (n = 109) | 16 | 93 |
| PCR 1 pathogen (n = 73) | 11 | 62 | C&S 1 pathogens (n = 109) | 16 | 93 |
| PCR 2 pathogens (n = 42) | 14 | 28 | Poly PCR (2 Pathogens) (n = 34) | 0 | 34 |
| Poly C&S (2 Pathogens) (n = 34) | 8 | 26 | C&S 1 pathogens (n = 34) | 0 | 34 |
| PCR 3 pathogens (n = 17) | 3 | 14 | Poly PCR (3 Pathogens) (n = 22) | 3 | 19 |
| PCR 4 pathogens (n = 15) | 4 | 11 | C&S 1 pathogens (n = 9) | 1 | 8 |
| PCR 5 pathogens (n = 2) | 1 | 1 | C&S 2 pathogens (n = 13) | 2 | 11 |
| Poly C&S (3 Pathogens) (n = 20) | 4 | 16 | Poly PCR (4 Pathogens) (n = 21) | 3 | 18 |
| PCR 3 pathogens (n = 4) | 0 | 4 | C&S 2 pathogens (n = 11) | 3 | 8 |
| PCR 4 pathogens (n = 16) | 4 | 12 | C&S 3 pathogens (n = 10) | 0 | 10 |
| Total (n = 169) | 37 | 132 | Poly PCR (5 Pathogens) (n = 7) | 1 | 6 |
| C&S 3 pathogens (n = 7) | 1 | 6 | |||
| Total (n = 193) | 23 | 170 | |||
| Monomicrobial Cases | Polymicrobial Cases | |||
|---|---|---|---|---|
| Detection | FCl | Detection | FCl | |
| C&S Arm | 115 (68.05%) | 90 (78.26%) | 54 (31.95%) * | 42 (77.78%) # |
| PCR Arm | 109 (56.48%) | 93 (85.32%) | 84 (43.52%) * | 77 (91.67%) # |
| CF in Missed Cases | CF in Concordant Cases | |
|---|---|---|
| C&S Arm | 26/92 (28.26%) * | 11/77 (14.29%) * |
| PCR Arm | 7/84 (8.34%) | 16/109 (14.68%) |
| Case | Pathogen(s) Detected by PCR | Treatment Based on PCR Results | Outcome of PCR-Guided Treatment | Pathogen(s) Detected by Culture | Pathogen(s) Detected by PCR but Missed by Culture | Treatment Chosen if It Was Based on Culture Results Only | Risk of Treatment Failure if Guided by CS? |
|---|---|---|---|---|---|---|---|
| 1 | Escherichia coli + Enterococcus faecalis | Nitrofurantoin | FCl | Escherichia coli | Enterococcus faecalis | Cephalexin | High |
| 2 | Klebsiella pneumoniae + Pseudomonas aeruginosa | Cefepime | FCl | Klebsiella pneumoniae | Pseudomonas aeruginosa | Amoxicillin/Clavulanate | High |
| 3 | Escherichia coli + Proteus mirabilis | Ciprofloxacin | FCl | Escherichia coli | Proteus mirabilis | Nitrofurantoin | High |
| 4 | Enterococcus faecalis + Klebsiella pneumoniae | Nitrofurantoin + Amoxicillin/Clavulanate | FCl | Klebsiella pneumoniae | Enterococcus faecalis | Nitrofurantoin | High |
| 5 | Escherichia coli + Serratia marcescens | Ciprofloxacin | FCl | Escherichia coli | Serratia marcescens | Nitrofurantoin | High |
| 6 | Enterococcus faecalis + Staphylococcus aureus | Amoxicillin/Clavulanate | FCl | Enterococcus faecalis | Staphylococcus aureus | Nitrofurantoin | High |
| 7 | Proteus mirabilis + Klebsiella pneumoniae | Ceftriaxone | FCl | Klebsiella pneumoniae | Proteus mirabilis | Amoxicillin/Clavulanate | High |
| 8 | Escherichia coli + Staphylococcus aureus | Trimethoprim/Sulfamethoxazole | FCl | Escherichia coli | Staphylococcus aureus | Nitrofurantoin | High |
| 9 | Klebsiella pneumoniae + Escherichia coli + Enterococcus faecalis | Fosfomycin | FCl | Klebsiella pneumoniae + Escherichia coli | Enterococcus faecalis | Amoxicillin/Clavulanate | High |
| 10 | Pseudomonas aeruginosa + Proteus mirabilis | Ciprofloxacin | FCl | Proteus mirabilis | Pseudomonas aeruginosa | Trimethoprim/Sulfamethoxazole | High |
| 11 | Serratia marcescens + Escherichia coli | Ciprofloxacin | FCl | Escherichia coli | Serratia marcescens | Nitrofurantoin | High |
| 12 | Enterococcus faecalis + Klebsiella pneumoniae | Amoxicillin/Clavulanate | FCl | Enterococcus faecalis | Klebsiella pneumoniae | Nitrofurantoin | High |
| 13 | Escherichia coli + Staphylococcus aureus | Cephalexin | FCl | Escherichia coli | Staphylococcus aureus | Nitrofurantoin | High |
| 14 | Enterococcus faecalis + Escherichia coli | Nitrofurantoin | FCl | Escherichia coli | Enterococcus faecalis | Cephalexin | High |
| 15 | Proteus mirabilis + Serratia marcescens | Ciprofloxacin | FCl | Proteus mirabilis | Serratia marcescens | Trimethoprim/Sulfamethoxazole | High |
| 16 | Klebsiella pneumoniae + Pseudomonas aeruginosa + Escherichia coli | Cefepime | FCl | Klebsiella pneumoniae + Escherichia coli | Pseudomonas aeruginosa | Amoxicillin/Clavulanate | High |
| 17 | Enterococcus faecalis + Staphylococcus aureus + Escherichia coli | Nitrofurantoin + Cephalexin | FCl | Escherichia coli | Enterococcus faecalis + Staphylococcus aureus | Trimethoprim/Sulfamethoxazole | High |
| 18 | Proteus mirabilis + Escherichia coli | Trimethoprim/Sulfamethoxazole | FCl | Escherichia coli | Proteus mirabilis | Nitrofurantoin | High |
| 19 | Klebsiella pneumoniae + Candida spp. | Meropenem + Fluconazole | FCl | Klebsiella pneumoniae | Candida | Meropenem (no antifungal) | High |
| 20 | Enterococcus faecium + Candida spp. | Linezolid + Fluconazole | FCl | Enterococcus faecium | Candida | Linezolid (no antifungal) | High |
| 21 | Escherichia coli + Candida spp. | Ciprofloxacin + Fluconazole | FCl | Escherichia coli | Candida | Ciprofloxacin (no antifungal) | High |
| 22 | Pseudomonas aeruginosa + Candida spp. | Meropenem + Fluconazole | FCl | Pseudomonas aeruginosa | Candida | Meropenem (no antifungal) | High |
| Case | Pathogen(s) Found in Culture (Guided Treatment) | Antibiotic Prescribed Based on Culture | Outcome of C&S Guided Treatment | Pathogen Detected by PCR | Pathogen(s) Detected by PCR but Missed by Culture | Alternative Antibiotic if Treatment Was Guided by PCR |
|---|---|---|---|---|---|---|
| 1 | Escherichia coli | Ciprofloxacin | CF | Enterococcus faecium/faecalis + Escherichia coli | Enterococcus faecium/faecalis | Fosfomycin or Nitrofurantoin |
| 2 | Escherichia coli | Nitrofurantoin | CF | Pseudomonas aeruginosa + Escherichia coli | Pseudomonas aeruginosa | Ciprofloxacin or Levofloxacin |
| 3 | Enterococcus faecium/faecalis | Nitrofurantoin | CF | Escherichia coli + Klebsiella pneumoniae + Enterococcus faecium/faecalis | Escherichia coli + Klebsiella pneumoniae | Ciprofloxacin or Trimethoprim/Sulfamethoxazole |
| 4 | Escherichia coli | Nitrofurantoin | CF | Proteus mirabilis + Escherichia coli | Proteus mirabilis | Ciprofloxacin or Cephalexin |
| 5 | Escherichia coli | Fosfomycin | CF | Serratia marcescens + Escherichia coli | Serratia marcescens | Ciprofloxacin |
| 6 | Escherichia coli | Cefepime | CF | Escherichia coli + Candida spp. | Candida spp. | + Fluconazole |
| 7 | Klebsiella pneumoniae | Ciprofloxacin | CF | Enterococcus faecium/faecalis + Klebsiella pneumoniae | Enterococcus faecium/faecalis | Nitrofurantoin or Fosfomycin |
| 8 | Klebsiella pneumoniae | Nitrofurantoin | CF | Pseudomonas aeruginosa + Klebsiella pneumoniae | Pseudomonas aeruginosa | Ciprofloxacin or Levofloxacin |
| 9 | Escherichia coli | Trimethoprim/Sulfamethoxazole | CF | Staphylococcus aureus + Escherichia coli | Staphylococcus aureus | Cephalexin or Doxycycline |
| 10 | Proteus mirabilis | Ciprofloxacin | CF | Enterococcus faecium/faecalis + Proteus mirabilis | Enterococcus faecium/faecalis | Nitrofurantoin or Fosfomycin |
| 11 | Klebsiella pneumoniae | Nitrofurantoin | CF | Proteus mirabilis + Klebsiella pneumoniae | Proteus mirabilis | Ciprofloxacin or Cephalexin |
| 12 | Escherichia coli | Trimethoprim/Sulfamethoxazole | CF | Candida glabrata + Escherichia coli | Candida spp. | + Fluconazole |
| 13 | Escherichia coli | Ciprofloxacin | CF | Candida spp. + Enterococcus faecium + Escherichia coli | Candida spp. + Enterococcus faecium | + Fluconazole |
| 14 | Klebsiella pneumoniae | Nitrofurantoin | CF | Escherichia coli + Pseudomonas aeruginosa + Klebsiella pneumoniae | Escherichia coli + Pseudomonas aeruginosa | Ciprofloxacin or Levofloxacin |
| 15 | Proteus mirabilis | Ciprofloxacin | CF | Proteus mirabilis + Serratia marcescens | Serratia marcescens | Cefepime |
| 16 | Escherichia coli | Cefepime | CF | Escherichia coli + Candida spp. | Candida spp. | + Fluconazole |
| 17 | Klebsiella pneumoniae | Nitrofurantoin | CF | Candida krusei + Enterococcus faecium + Klebsiella pneumoniae | Candida spp + Enterococcus faecium | Caspofungin |
| 18 | Escherichia coli | Nitrofurantoin | CF | Pseudomonas aeruginosa + Serratia marcescens + Escherichia coli | Pseudomonas aeruginosa + Serratia marcescens | Ciprofloxacin or Levofloxacin |
| 19 | Escherichia coli | Nitrofurantoin | CF | Enterococcus faecium + Candida albicans + Escherichia coli | Enterococcus faecium + Candida spp. | + Fluconazole |
| 20 | Klebsiella pneumoniae + Escherichia coli | Ciprofloxacin | CF | Enterococcus faecium/faecalis + Klebsiella pneumoniae + Escherichia coli | Enterococcus faecium/faecalis | Fosfomycin or Nitrofurantoin |
| 21 | Klebsiella pneumoniae | Ciprofloxacin | CF | Candida parapsilosis + Pseudomonas aeruginosa + Klebsiella pneumoniae | Candida spp. + Pseudomonas aeruginosa | + Fluconazole |
| 22 | Klebsiella pneumoniae | Nitrofurantoin | CF | Escherichia coli + Proteus mirabilis + Klebsiella pneumoniae | Escherichia coli + Proteus mirabilis | Ciprofloxacin or Cephalexin |
| 23 | Staphylococcus aureus | Vancomycin | CF | Staphylococcus aureus + Enterococcus faecium | Enterococcus faecium | Linezolid or Daptomycin |
| 24 | Escherichia coli | Ciprofloxacin | CF | Candida tropicalis + Enterococcus faecium + Escherichia coli | Candida spp. + Enterococcus faecium | + Fluconazole |
| 25 | Proteus mirabilis | Ciprofloxacin | CF | Proteus mirabilis + Klebsiella pneumoniae | Klebsiella pneumoniae | Cephalexin |
| 26 | Enterococcus faecium/faecalis | Nitrofurantoin | CF | Escherichia coli + Klebsiella pneumoniae + Enterococcus faecium/faecalis | Escherichia coli + Klebsiella pneumoniae | Ciprofloxacin or Trimethoprim/Sulfamethoxazole |
| Resistance | Number of Cases | Affected Antibiotic Class |
|---|---|---|
| ACT, MIR, FOX, ACC Groups (Beta Lactams) | 32 | Cephalosporins (e.g., cefotaxime, ceftazidime), penicillins, monobactams |
| Class A ß-lactamase; blaKPC | 3 | Carbapenems (e.g., meropenem, imipenem, ertapenem), cephalosporins, penicillins |
| Class A ß-lactamase; CTX-M-Group1 | 29 | Cephalosporins (e.g., ceftriaxone, ceftazidime, cefotaxime), penicillins |
| Class B metallo-ß-lactamase; blaNDM | 3 | Carbapenems, cephalosporins, penicillins |
| Class D oxacillinase OXA−48 | 11 | Carbapenems (e.g., imipenem, meropenem), penicillins |
| Class D oxacillinase OXA-−51 | 3 | |
| dfr (A1, A5), sul (1,2) probes (Sulfamethoxazole and trimethoprim) | 142 | Diamino-pyrimidine (Trimethoprim), Sulfonamide, trimethoprim-sulfamethoxazole |
| ermB, C; mefA | 172 | Macrolides (e.g., erythromycin, azithromycin, clarithromycin), lincosamides (clindamycin) |
| IMP, NDM, VIM Groups (Carbapenem) | 20 | Carbapenems, cephalosporins, penicillins |
| MRSA* Mec-A gene | 86 | Beta-lactams, including methicillin, oxacillin, penicillins, and cephalosporins |
| PER−1/VEB−1/GES−1 Groups (ESBL) | 6 | Cephalosporins, penicillins, monobactams |
| qnrA1, A2 | 1 | Fluoroquinolones (e.g., ciprofloxacin, levofloxacin) |
| qnrB | 36 | |
| qnrS | 10 | |
| tetB, tetM | 220 | Tetracyclines (e.g., doxycycline, minocycline, tetracycline) |
| VanA, VanB (Vancomycin) | 6 | Glycopeptide (Vancomycin, Teicoplanin) |
| Total Genetic Resistance Detected | 780 |
| Antibiotic Groups | Phenotypical Resistance Cases |
|---|---|
| Ampicillin/Ampicillin–Sulbactam | 168 |
| Aztreonam/Cefazolin/Cefepime/Cefoxitin/Ceftazidime/Ceftriaxone | 41 |
| Ciprofloxacin/Levofloxacin | 39 |
| Ertapenem/Imipenem/Meropenem | 28 |
| Gentamicin/Tobramycin/Amikacin | 52 |
| Nitrofurantoin | 13 |
| Tigecycline | 127 |
| Trimethoprim/Sulfamethoxazole (TMP-SMX) | 146 |
| Total Phenotypical Resistance Detected | 614 |
| CF for Cases with Undetected Phenotypic Resistance (with the Presence of Their Molecular Equivalent) | CF with Concordant Resistance Detection Cases (C&S and PCR) | |
|---|---|---|
| C&S Arm | 16/42 (50%) * | 21/121 (13.22%) * |
| PCR Arm | 6/48 (12.5%) | 17/155 (10.98%) |
| Case | Pathogen(s) Detected by PCR (with Resistance Marker) | Pathogen(s) Found in Culture | Phenotypic Resistance Detected in Culture | Antibiotic Prescribed Based on Culture | Missed Pathogen(s) by Culture | Undetected Resistance in Missed Pathogen | Outcome of C&S Guided Treatment | Alternative/Additional Treatment Required? |
|---|---|---|---|---|---|---|---|---|
| 1 | Escherichia coli (qnrB, blaCTX-M−15) | Escherichia coli | None detected | Ciprofloxacin | Enterococcus faecium | VanB (VRE) | CF | Linezolid or Daptomycin |
| 2 | Klebsiella pneumoniae (KPC−2) | Klebsiella pneumoniae | Fluoroquinolones, Aminoglycosides | Cefepime | Pseudomonas aeruginosa | OXA−51 (Carbapenemase) | CF | Ceftazidime-Avibactam or Cefiderocol |
| 3 | Enterococcus faecium (VanA) | Enterococcus faecium | Tetracyclines | Ampicillin | Klebsiella pneumoniae | CTX-M−15 (ESBL) | CF | Ceftazidime-Avibactam or Meropenem |
| 4 | Staphylococcus aureus (mecA) | Staphylococcus aureus | Macrolides, Aminoglycosides | Clindamycin | Escherichia coli | qnrB1 (FQ resistance) | CF | Ciprofloxacin or Levofloxacin |
| 5 | Pseudomonas aeruginosa (IMP, OXA−51) | Pseudomonas aeruginosa | Aminoglycosides, Beta-lactams | Cefepime | Serratia marcescens | blaSHV (Beta-lactamase) | CF | Meropenem or Piperacillin-Tazobactam |
| 6 | Escherichia coli (CTX-M−14, qnrB1) | Escherichia coli | Nitrofurantoin, TMP/SMX | Cefepime | Enterococcus faecium | VanB (VRE) | CF | Linezolid or Daptomycin |
| 7 | Morganella morganii (qnrD, blaCMY−2) | Morganella morganii | Fluoroquinolones, TMP/SMX | Cefepime | Proteus mirabilis | qnrS (FQ resistance) | CF | Meropenem or Ciprofloxacin |
| 8 | Providencia stuartii (blaNDM−1, qnrB4) | Providencia stuartii | Beta-lactams, Aminoglycosides | Cefepime | Escherichia coli | CTX-M−27 (ESBL) | CF | Colistin or Cefiderocol |
| 9 | Escherichia coli (CTX-M−15, qnrB1) | Escherichia coli | None detected | Trimethoprim-Sulfamethoxazole | None | CTX-M−15 (ESBL), qnrB1 (FQ resistance) | CF | Meropenem or Fosfomycin |
| 10 | Enterococcus faecium (VanA) | Enterococcus faecium | Tetracyclines | Amoxicillin | None | VanA (Vancomycin Resistance) | CF | Daptomycin or Linezolid |
| 11 | Klebsiella pneumoniae (OXA−48, CTX-M−15) | Klebsiella pneumoniae | Fluoroquinolones | Cephalexin | None | OXA−48 (Carbapenemase), CTX-M−15 (ESBL) | CF | Ceftazidime-Avibactam or Meropenem |
| 12 | Pseudomonas aeruginosa (VIM, OXA−51) | Pseudomonas aeruginosa | Beta-lactams (except Carbapenems) | Levofloxacin | None | VIM (Carbapenemase), OXA−51 (Beta-lactamase) | CF | Cefiderocol or Colistin |
| 13 | Candida glabrata (Azole Resistance) + Escherichia coli (CTX-M−27) | Escherichia coli | Trimethoprim-Sulfamethoxazole | Nitrofurantoin | None | Candida glabrata (Azole Resistance) | CF | Fluconazole or Echinocandins |
| 14 | Klebsiella pneumoniae (KPC−3, blaSHV−12) | Klebsiella pneumoniae | Fluoroquinolones | Amoxicillin-Clavulanate | None | KPC−3 (Carbapenemase), blaSHV−12 (Beta-lactamase) | CF | Ceftazidime-Avibactam or Meropenem |
| 15 | Escherichia coli (OXA−48, qnrS1) | Escherichia coli | Beta-lactams, Trimethoprim-Sulfamethoxazole | Doxycycline | None | OXA−48 (Carbapenemase), qnrS1 (FQ resistance) | CF | Meropenem or Fosfomycin |
| 16 | Enterobacter cloacae (qnrB1, blaCTX-M−15) | Enterobacter cloacae | Fluoroquinolones | Metronidazole | None | qnrB1 (FQ resistance), CTX-M−15 (ESBL) | CF | Meropenem or Fosfomycin |
| Case | Pathogen(s) Detected by PCR (with Resistance Marker) | Pathogen(s) Found in Culture | Phenotypic Resistance Detected in Culture | Antibiotic Prescribed Based on PCR (Guided Treatment) | Outcome of PCR-Guided Treatment | Antibiotic Would Have Been Prescribed Based on CS | Pathogen(s) Detected by PCR but Missed by Culture | Resistance Marker(s) Missed by Culture | Risk of Treatment Failure if Guided by CS? |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Escherichia coli (qnrS, CTX-M−15) | Escherichia coli | Fluoroquinolones, TMP/SMX | Fosfomycin | FCl | Nitrofurantoin | None | qnrS (FQ resistance), CTX-M−15 (ESBL) | High |
| 2 | Klebsiella pneumoniae (CTX-M-Group1) | Klebsiella pneumoniae | Fluoroquinolones, Aminoglycosides | Amoxicillin and Clavulanate Potassium | FCl | Ciprofloxacin | None | CTX-M (Cephalosporin Resistance) | High |
| 3 | Pseudomonas aeruginosa (OXA−50, efflux-mediated FQ resistance) | Pseudomonas aeruginosa | Reduced FQ sensitivity | Levofloxacin | FCl | Ciprofloxacin | None | OXA−50 (Carbapenem Resistance), Efflux-mediated FQ resistance | High |
| 4 | Enterococcus faecium (VanA, ermB) | Enterococcus faecium | Aminoglycosides, Tetracyclines | Doxycycline | FCl | Nitrofurantoin | None | VanA (VRE), ermB (Macrolide resistance) | High |
| 5 | Klebsiella pneumoniae (OXA−48, CTX-M−15) | Klebsiella pneumoniae | Beta-lactams (except Carbapenems), Macrolides | Trimethoprim and Sulfamethoxazole | FCl | Amoxicillin/Clavulanate | None | OXA−48 (Carbapenem Resistance), CTX-M−15 (ESBL) | High |
| 6 | Staphylococcus aureus (mecA, ermC) | Staphylococcus aureus | Macrolides, Tetracyclines | Doxycycline | FCl | Cephalexin | None | mecA (MRSA), ermC (Macrolide resistance) | High |
| 7 | Serratia marcescens (IMP, blaSHV) | Serratia marcescens | Beta-lactams, Aminoglycosides | Levofloxacin | FCl | Ciprofloxacin | None | IMP (Carbapenemase), blaSHV (Beta-lactamase) | High |
| 8 | Escherichia coli + Enterococcus faecalis (VanA) | Escherichia coli | TMP/SMX, Aminoglycosides | Fosfomycin | FCl | Cephalexin | Enterococcus faecalis | VanA (VRE) | High |
| 9 | Klebsiella pneumoniae + Pseudomonas aeruginosa (OXA−50) | Klebsiella pneumoniae | Beta-lactams (except Carbapenems) | Ciprofloxacin | FCl | Amoxicillin/Clavulanate | Pseudomonas aeruginosa | OXA−50 (Carbapenem Resistance) | High |
| 10 | Escherichia coli + Proteus mirabilis (CTX-M−15) | Escherichia coli | TMP/SMX, Aminoglycosides | Nitrofurantoin | FCl | Ciprofloxacin | Proteus mirabilis (missed due to E. coli overgrowth) | CTX-M−15 (ESBL) | High |
| 11 | Enterococcus faecalis + Klebsiella pneumoniae (qnrB) | Klebsiella pneumoniae | Fluoroquinolones, Beta-lactams | Amoxicillin | FCl | Nitrofurantoin | Enterococcus faecalis | qnrB (FQ resistance) | High |
| 12 | Pseudomonas aeruginosa + Escherichia coli (CTX-M−27) | Escherichia coli | TMP/SMX, Cephalosporins | Ciprofloxacin | FCl | Cephalexin | Pseudomonas aeruginosa | CTX-M−27 (ESBL) | High |
| 13 | Candida spp. + Escherichia coli | Escherichia coli | TMP/SMX, Beta-lactams | Metronidazole + Fosfomycin | FCl | Cephalexin | Candida spp. | Antifungal Resistance | High |
| 14 | Proteus mirabilis + Klebsiella pneumoniae (OXA−48) | Klebsiella pneumoniae | Beta-lactams (except Carbapenems), Aminoglycosides | Amoxicillin and Clavulanate Potassium | FCl | Nitrofurantoin | Proteus mirabilis | OXA−48 (Carbapenem Resistance) | High |
| 15 | Escherichia coli + Staphylococcus aureus (ermC) | Escherichia coli | TMP/SMX, Beta-lactams | Trimethoprim and Sulfamethoxazole | FCl | Nitrofurantoin | Staphylococcus aureus | ermC (Macrolide Resistance) | High |
| 16 | Klebsiella pneumoniae + Enterococcus faecalis (VanB) | Klebsiella pneumoniae | Fluoroquinolones, TMP/SMX | Fosfomycin | FCl | Amoxicillin/Clavulanate | Enterococcus faecalis | VanB (VRE) | High |
| 17 | Candida spp. + Enterococcus faecium (VanA) | None (Culture Negative) | None | Metronidazole + Nitrofurantoin | FCl | Cephalexin | Candida spp., Enterococcus faecium | VanA (VRE), Antifungal Resistance | High |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kardjadj, M.; Chang, T.W.; Chavez, R.; Derrick, D.; Spangler, F.L.; Priestly, I.P.; Park, L.Y.; Huard, T.K. The Clinical Validity and Utility of PCR Compared to Conventional Culture and Sensitivity Testing for the Management of Complicated Urinary Tract Infections in Adults: A Secondary (Ad Hoc) Analysis of Pathogen Detection, Resistance Profiles, and Impact on Clinical Outcomes. Microorganisms 2025, 13, 949. https://doi.org/10.3390/microorganisms13040949
Kardjadj M, Chang TW, Chavez R, Derrick D, Spangler FL, Priestly IP, Park LY, Huard TK. The Clinical Validity and Utility of PCR Compared to Conventional Culture and Sensitivity Testing for the Management of Complicated Urinary Tract Infections in Adults: A Secondary (Ad Hoc) Analysis of Pathogen Detection, Resistance Profiles, and Impact on Clinical Outcomes. Microorganisms. 2025; 13(4):949. https://doi.org/10.3390/microorganisms13040949
Chicago/Turabian StyleKardjadj, Moustafa, Tara W. Chang, Roel Chavez, DeAndre Derrick, Frank L. Spangler, Itoe P. Priestly, Lauren Y. Park, and Thomas K. Huard. 2025. "The Clinical Validity and Utility of PCR Compared to Conventional Culture and Sensitivity Testing for the Management of Complicated Urinary Tract Infections in Adults: A Secondary (Ad Hoc) Analysis of Pathogen Detection, Resistance Profiles, and Impact on Clinical Outcomes" Microorganisms 13, no. 4: 949. https://doi.org/10.3390/microorganisms13040949
APA StyleKardjadj, M., Chang, T. W., Chavez, R., Derrick, D., Spangler, F. L., Priestly, I. P., Park, L. Y., & Huard, T. K. (2025). The Clinical Validity and Utility of PCR Compared to Conventional Culture and Sensitivity Testing for the Management of Complicated Urinary Tract Infections in Adults: A Secondary (Ad Hoc) Analysis of Pathogen Detection, Resistance Profiles, and Impact on Clinical Outcomes. Microorganisms, 13(4), 949. https://doi.org/10.3390/microorganisms13040949







