Comparative Transcriptome Analyses during the Vegetative Cell Cycle in the Mono-Cellular Organism Pseudokeronopsis erythrina (Alveolata, Ciliophora)
Abstract
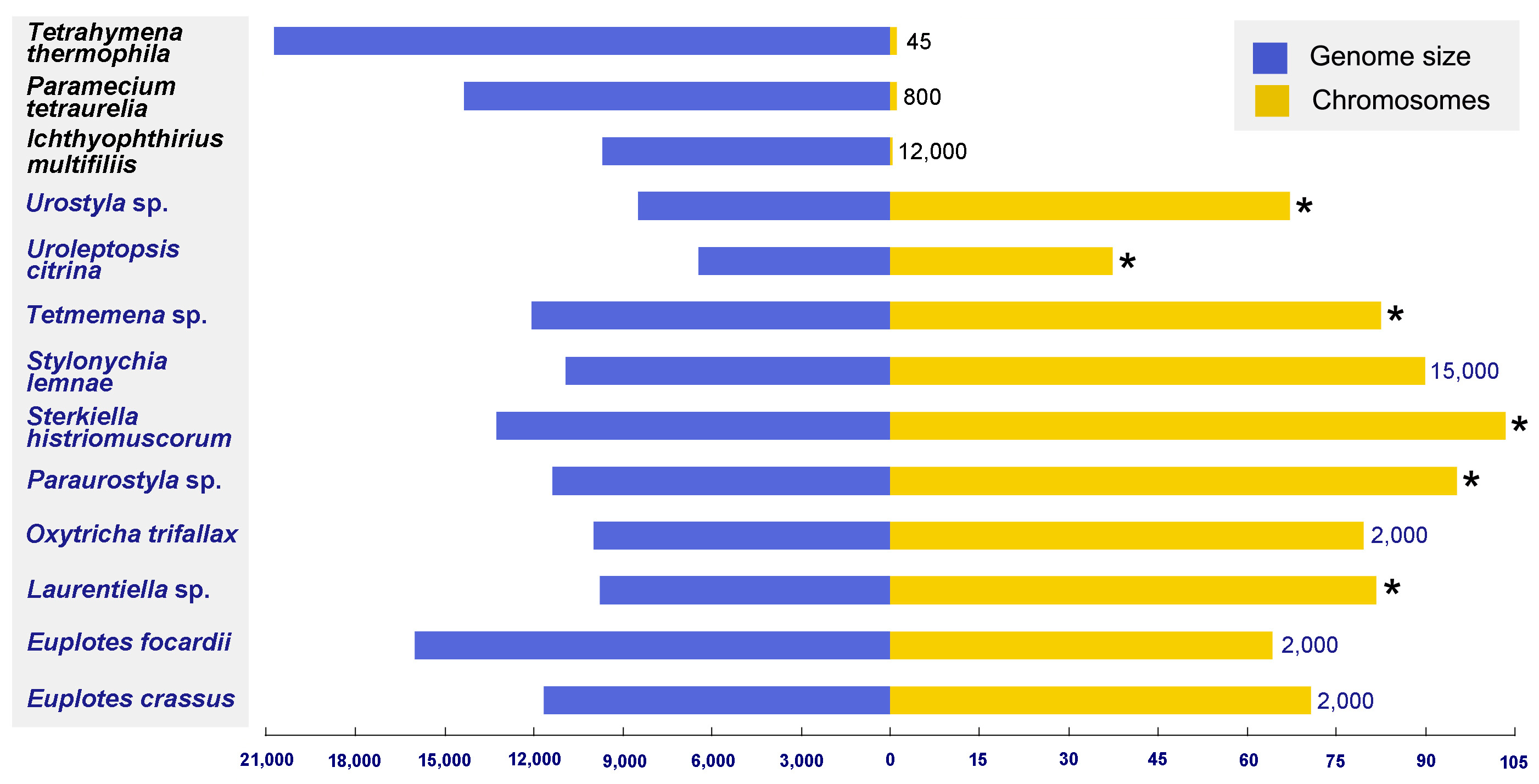
1. Introduction
2. Materials and Methods
2.1. Cell Growth and Sorting
2.2. Single Cell cDNA Amplification and Library Construction
2.3. Sequence Assembly and Analysis
2.4. Differential Gene Expression
2.5. Reverse Transcription Quantitative Polymerase Chain Reaction (RT-qPCR)
3. Results
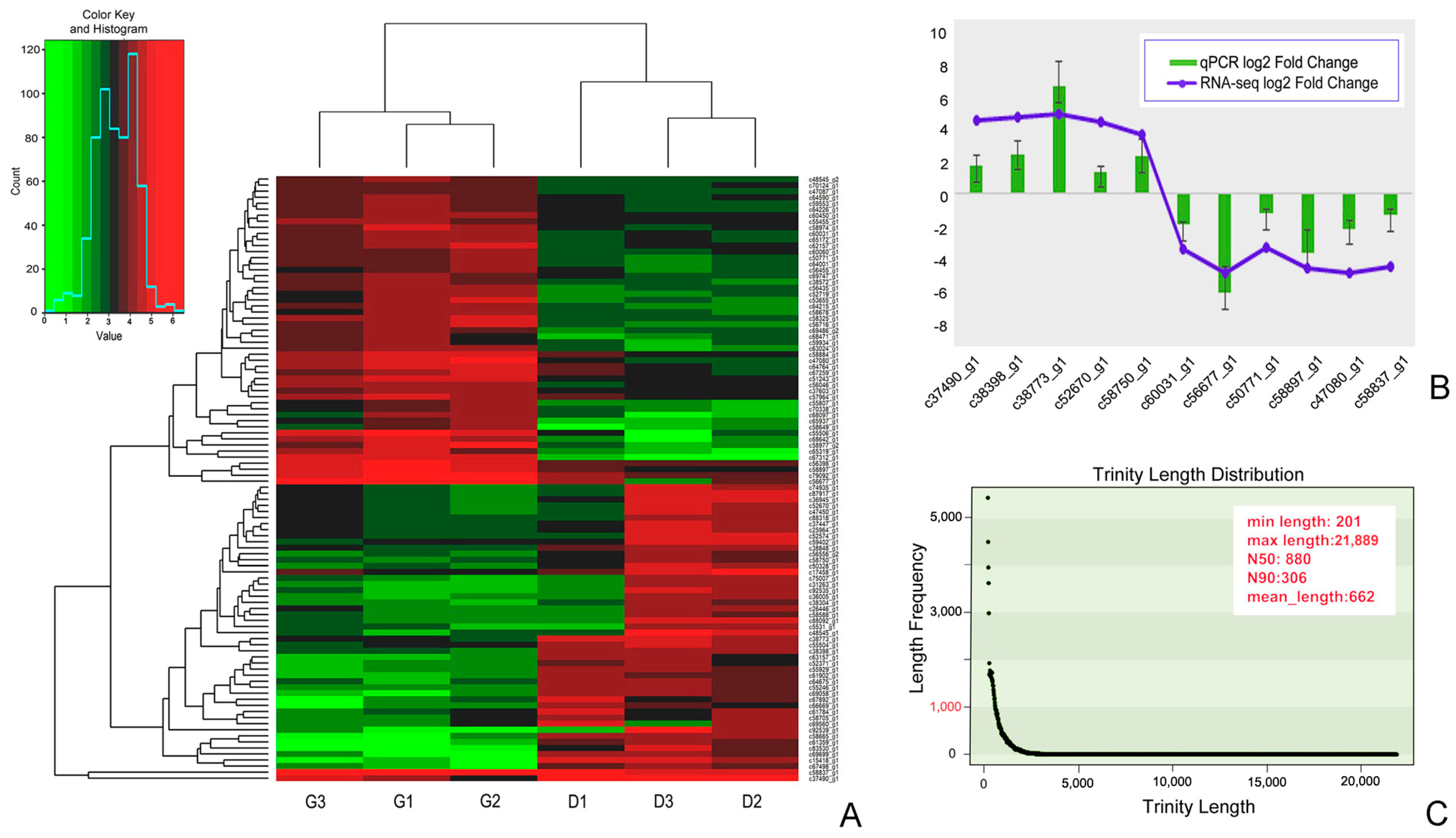
3.1. Transcriptome Sequencing, Assembly Evaluation and Annotation of Unigenes
3.2. Differentially Expressed Genes between the G and D Stages
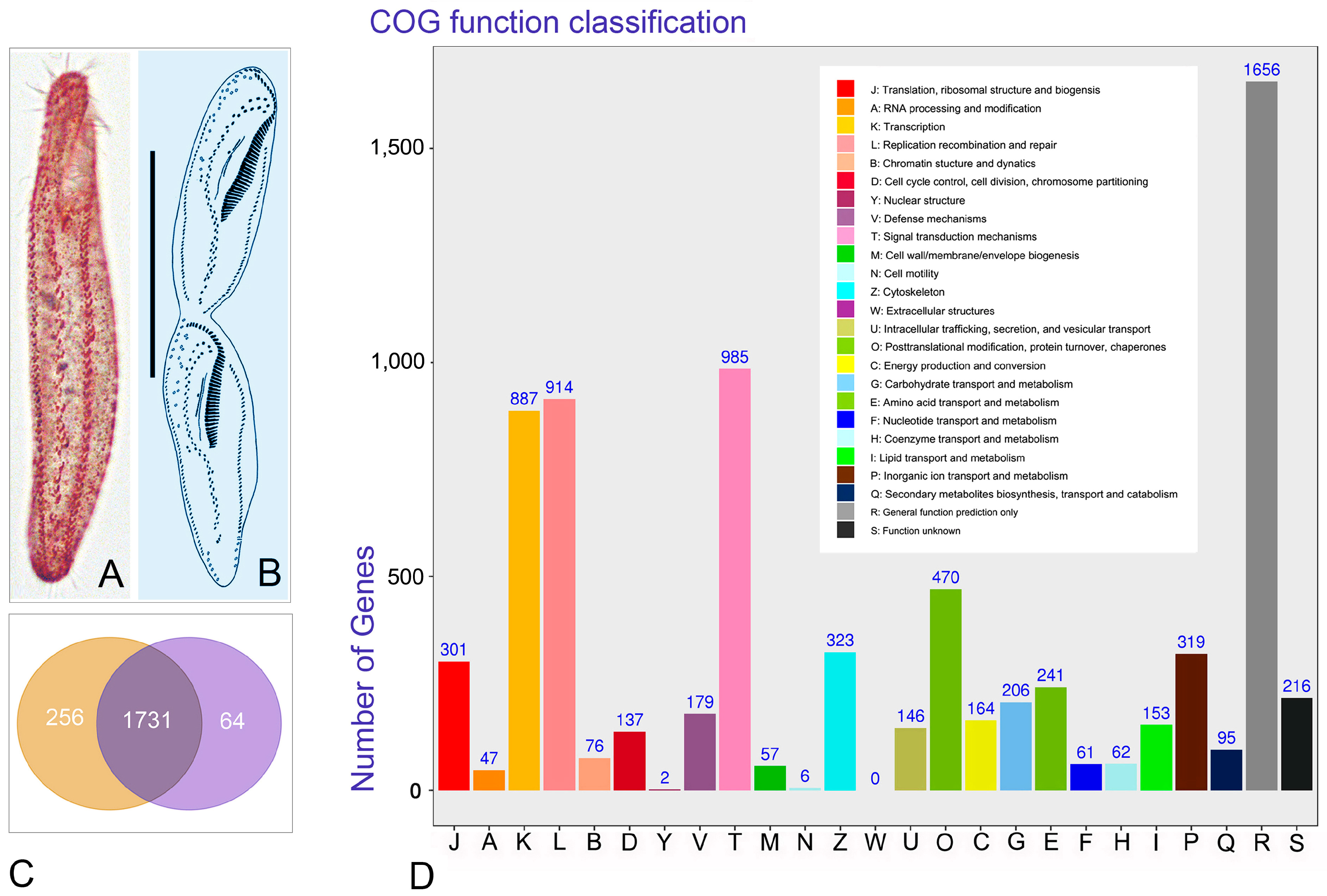
3.3. Annotation of Unigenes Using COG Functional Categories
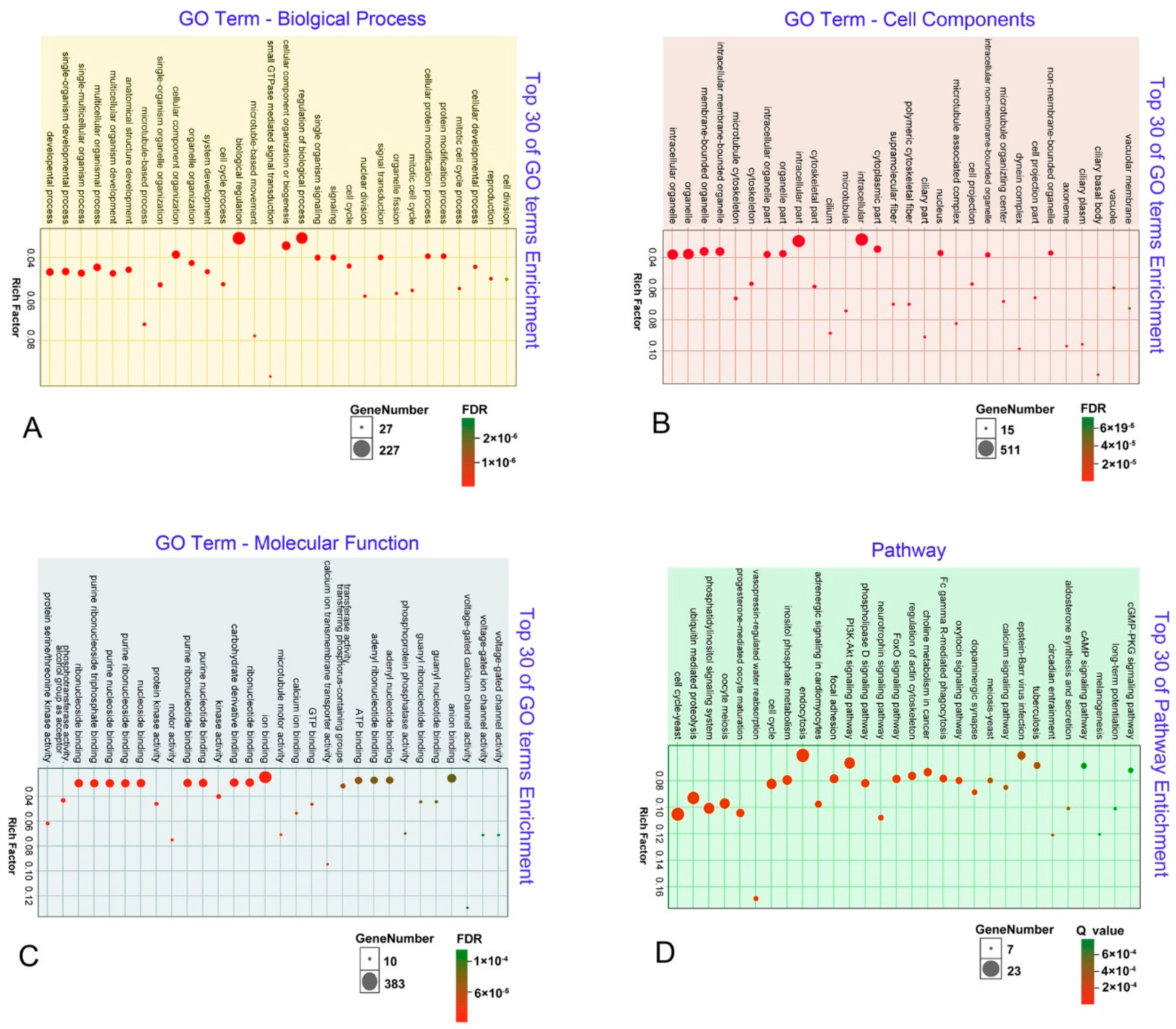
3.4. Annotation of Unigenes Using GO Enrichment
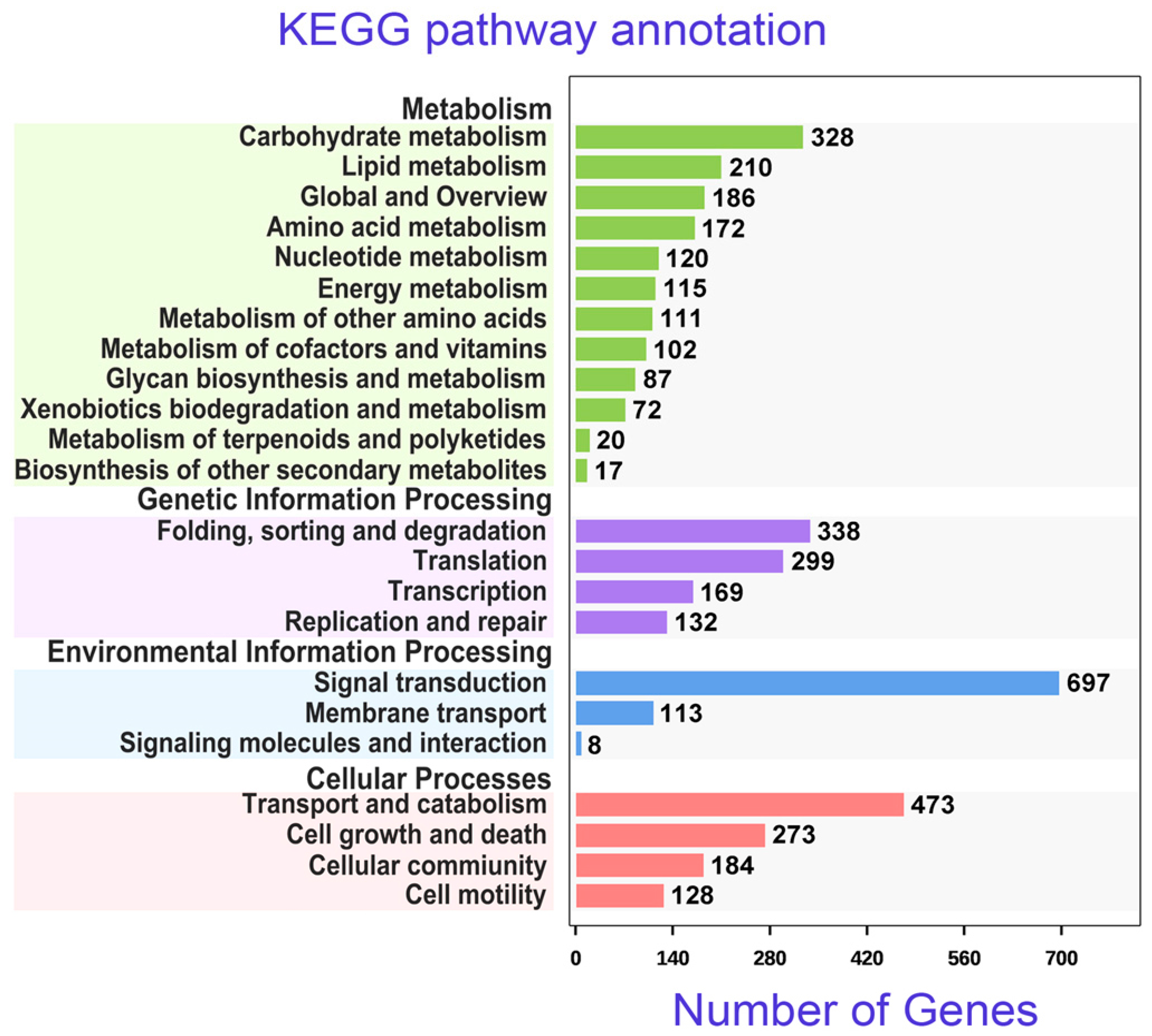
3.5. Annotations of Unigenes Using KEGG
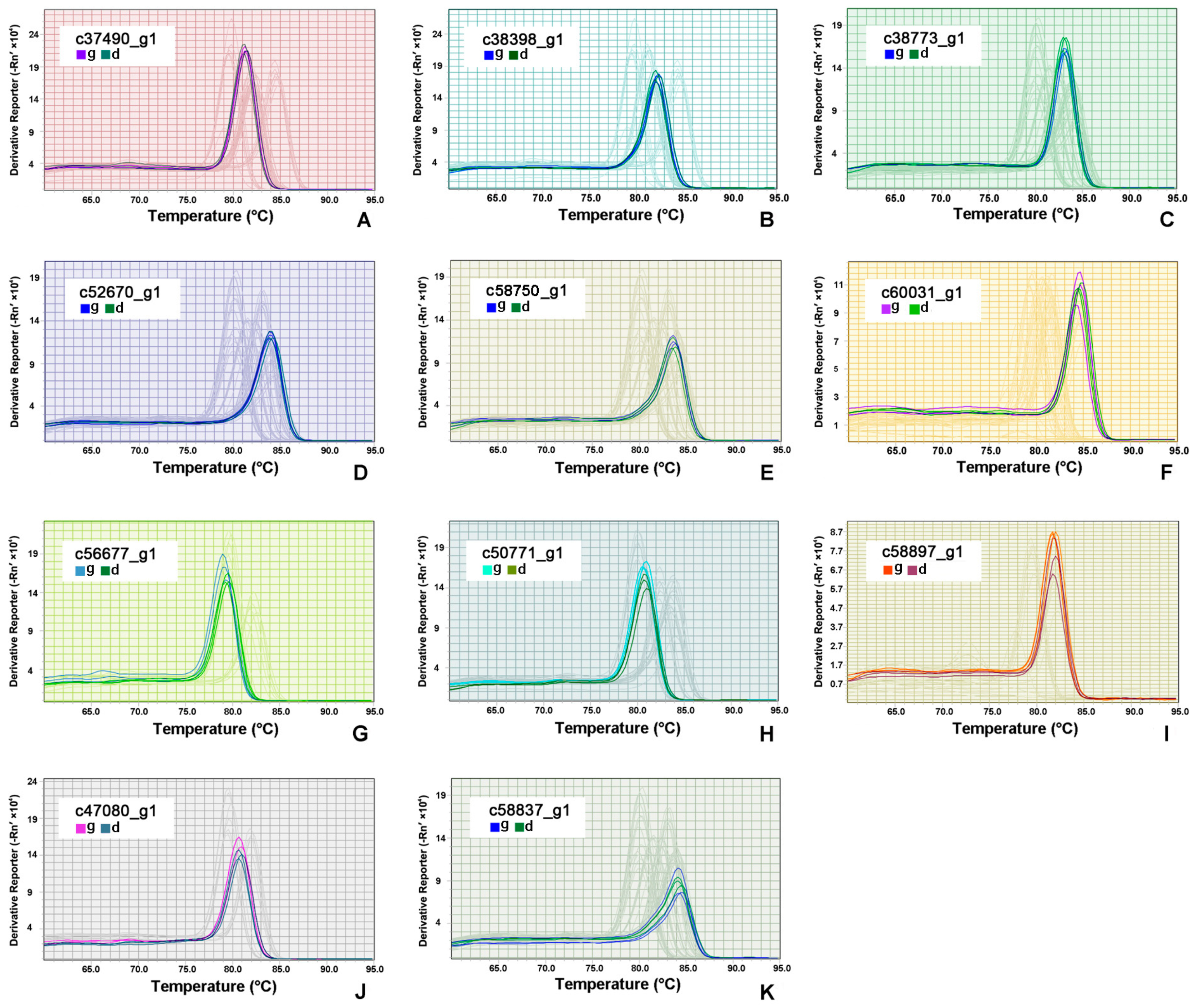
3.6. RT-qPCR Validation of DEGs
4. Discussion
4.1. Ciliates Share Essential Mechanisms of Cell Division with Other Eukaryotic Organisms
4.2. Comparisons with Previous Studies Focusing on the Cell Cycle Regulation of Ciliates
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhai, Z.; Wang, X.; Ding, M. Cell Biology; Science Press: Beijing, China, 2011. [Google Scholar]
- Amarnath, D.; Li, X.; Kato, Y.; Tsunoda, Y. Gene expression in individual bovine somatic cell cloned embryos at the 8-cell and blastocyst stages of preimplantation development. J. Reprod. Dev. 2007, 53, 1247–1263. [Google Scholar] [CrossRef]
- Orford, K.W.; Scadden, D.T. Deconstructing stem cell self-renewal: Genetic insights into cell-cycle regulation. Nat. Rev. Genet. 2008, 9, 115. [Google Scholar] [CrossRef] [PubMed]
- Hi, X.; Ran, L.; Liu, Y.; Zhong, S.H.; Zhou, P.P.; Liao, M.X.; Fang, W. Knockdown of hnRNP A2/B1 inhibits cell proliferation, invasion and cell cycle triggering apoptosis in cervical cancer via PI3K/AKT signaling pathway. Oncol. Rep. 2018, 39, 939–950. [Google Scholar]
- Jiang, Y.H.; Zhang, T.T.; Vallesi, A.; Yang, X.Y.; Gao, F. Time-course analysis of nuclear events during conjugation in the marine ciliate Euplotes vannus and comparison with other ciliates (Protozoa, Ciliophora). Cell Cycle 2019, 18, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Spellman, P.T.; Sherlock, G.; Zhang, M.Q.; Iyer, V.R.; Anders, K.; Eisen, M.B.; Brown, P.O.; Botstein, D.; Futcher, B. Comprehensive identification of cell cycle–regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 1998, 9, 3273–3297. [Google Scholar] [CrossRef] [PubMed]
- Cho, R.J.; Huang, M.; Campbell, M.J.; Dong, H.; Steinmetz, L.; Sapinoso, L.; Hampton, G.; Elledge, S.J.; Davis, R.W.; Lockhart, D.J. Transcriptional regulation and function during the human cell cycle. Nat. Genet. 2001, 27, 48. [Google Scholar] [CrossRef]
- Breyne, P.; Dreesen, R.; Vandepoele, K.; De Veylder, L.; Van Breusegem, F.; Callewaert, L.; Rombauts, S.; Raes, J.; Cannoot, B.; Engler, G.; et al. Transcriptome analysis during cell division in plants. Prod. Natl. Acad. Sci. USA 2002, 99, 14825–14830. [Google Scholar] [CrossRef]
- Xiong, J.; Lu, X.; Zhou, Z.; Chang, Y.; Yuan, D.; Tian, M.; Zhou, Z.; Wang, L.; Fu, C.; Orias, E.; et al. Transcriptome analysis of the model protozoan, Tetrahymena thermophila, using deep RNA sequencing. PLoS ONE 2012, 7, e30630. [Google Scholar] [CrossRef]
- Nolan, T.; Hands, R.E.; Bustin, S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006, 1, 1559. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57. [Google Scholar] [CrossRef]
- Kolisko, M.; Boscaro, V.; Burki, F.; Lynn, D.H.; Keeling, P.J. Single-cell transcriptomics for microbial eukaryotes. Curr. Biol. 2014, 24, R1081–R1082. [Google Scholar] [CrossRef] [PubMed]
- Dzamba, D.; Valihrach, L.; Kubista, M.; Anderova, M. The correlation between expression profiles measured in single cells and in traditional bulk samples. Sci. Rep. 2016, 6, 37022. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.; Wang, Y.Y.; Huang, J.; Chen, X.; Zhao, X.L.; Gao, S.; Song, W.B. Our recent progress in epigenetic research using the model ciliate, Tetrahymena thermophila. Mar. Life Sci. Technol. 2019, 1, 4–14. [Google Scholar] [CrossRef]
- Chen, X.M.; Clamp, J.C.; Song, W.B. Phylogeny and systematic revision of the family Pseudokeronopsidae (Protista, Ciliophora, Hypotricha), with description of a new estuarine species of Pseudokeronopsis. Zool. Scr. 2011, 40, 659–671. [Google Scholar] [CrossRef]
- Chen, X.; Jiang, Y.; Gao, F.; Zheng, W.; Krock, T.J.; Stover, N.A.; Lu, C.; Katz, L.A.; Song, W. Genome analyses of the new model protist Euplotes vannus focusing on genome rearrangement and resistance to environmental stressors. Mol. Ecol. Resour. 2019. [Google Scholar] [CrossRef]
- Gao, S.; Xiong, J.; Zhang, C.; Berquist, B.R.; Yang, R.; Zhao, M.; Molascon, A.J.; Kwiatkowski, S.Y.; Yuan, D.; Qin, Z.; et al. Impaired replication elongation in Tetrahymena mutants deficient in histone H3 Lys 27 monomethylation. Genes Dev. 2013, 27, 1662–1679. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Chen, X.; Sheng, Y.L.; Liu, Y.; Gao, S. N6-adenine DNA methylation is associated with H2A.Z-containing well-positioned nucleosomes in Pol II-transcribed genes in Tetrahymena. Nucleic Acid Res. 2017, 45, 11594–11606. [Google Scholar]
- Wang, Y.R.; Wang, C.; Jiang, Y.; Katz, L.A.; Gao, F.; Yan, Y. Further analyses of variation of ribosome DNA copy number and polymorphism in ciliates provide insights relevant to studies of both molecular ecology and phylogeny. Sci. China Life Sci. 2019, 62, 203–214. [Google Scholar] [CrossRef]
- Zhang, T.T.; Wang, C.D.; Katz, L.A.; Gao, F. A paradox: Rapid evolution rates of germline-limited sequences are associated with conserved patterns of rearrangements in cryptic species of Chilodonella uncinata (Protista, Ciliophora). Sci. China Life Sci. 2018, 61, 1071–1078. [Google Scholar] [CrossRef]
- Zhao, Y.; Yi, Z.Z.; Warren, A.; Song, W.B. Species delimitation for the molecular taxonomy and ecology of the widely distributed microbial eukaryote genus Euplotes (Alveolata, Ciliophora). Proc. R. Soc. B Biol. Sci. 2018, 285. [Google Scholar] [CrossRef]
- Zhao, X.; Xiong, J.; Mao, F.; Sheng, Y.; Chen, X.; Feng, L.; Dui, W.; Yang, W.; Kapusta, A.; Feschotte, C.; et al. RNAi-dependent Polycomb repression controls transposable elements in Tetrahymena. Genes Dev. 2019, 33, 348–364. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.B.; Wang, C.D.; Yan, Y.; Gao, F.; Doak, T.G.; Song, W.B. Insights into an extensively fragmented eukaryotic genome: De novo genome sequencing of the multi-nuclear ciliate Uroleptopsis citrina. Genome Biol. Evol. 2018, 10, 883–894. [Google Scholar] [CrossRef] [PubMed]
- Li, X.H.; Huang, J.; Filker, S.; Stoeck, T.; Bi, Y.H.; Song, W.B. Spatio-temporal patterns of zooplankton in a main-stem dam affected tributary: A case study in the Xiangxi River of the Three Gorges Reservoir. Sci. China Life Sci. 2019. [Google Scholar] [CrossRef] [PubMed]
- Miao, W.; Xiong, J.; Bowen, J.; Wang, W.; Liu, Y.F.; Braguinets, O. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS ONE 2009, 4, e4429. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Wang, R.; Ghanam, A.; Yan, G.; Miao, W.; Song, X. The key role of CYC2 during meiosis in Tetrahymena thermophila. Protein Cell 2016, 7, 236–249. [Google Scholar] [CrossRef][Green Version]
- Xu, J.; Li, X.; Song, W.; Wang, W.; Gao, S. Cyclin Cyc2p is required for micronuclear bouquet formation in Tetrahymena thermophila. Sci. China Life Sci. 2019, 62, 668–680. [Google Scholar] [CrossRef]
- Lipps, H.J.; Nock, A.; Riewe, M.; Steinbruck, G. Chromatin structure in the macronucleus of the ciliate Stylonychia mytilus. Nucleic Acids Res. 1978, 5, 4699–4709. [Google Scholar] [CrossRef][Green Version]
- Prescott, D.M. The DNA of ciliated protozoa. Microbiol. Rev. 1994, 58, 233–267. [Google Scholar] [CrossRef]
- Bender, J.; Klein, A. The telomere binding protein of Euplotes crassus prevents non-specific transcription initiation but has no role in positioning transcription initiation complexes. Nucleic Acids Res. 1997, 25, 2877–2882. [Google Scholar] [CrossRef][Green Version]
- Duerr, H.P.; Eichner, M.; Ammermann, D. Modeling senescence in hypotrichous ciliates. Protist 2004, 155, 45–52. [Google Scholar] [CrossRef]
- Aury, J.M.; Jaillon, O.; Duret, L.; Noel, B.; Jubin, C.; Porcel, B.M.; Ségurens, B.; Daubin, V.; Anthouard, V.; Aiach, N.; et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 2006, 444, 171–178. [Google Scholar] [CrossRef]
- Eisen, J.A.; Coyne, R.S.; Wu, M.; Wu, D.; Thiagarajan, M.; Wortman, J.R.; Badger, J.H.; Ren, Q.; Amedeo, P.; Jones, K.M.; et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006, 4, 1620–1642. [Google Scholar] [CrossRef] [PubMed]
- Coyne, R.S.; Thiagarajan, M.; Jones, K.M.; Wortman, J.R.; Tallon, L.J.; Haas, B.J.; Cassidy-Hanley, D.M.; Wiley, E.A.; Smith, J.J.; Collins, K. Refined annotation and assembly of the Tetrahymena thermophila genome sequence through EST analysis, comparative genomic hybridization, and targeted gap closure. BMC Genom. 2008, 9, 562. [Google Scholar] [CrossRef] [PubMed]
- Estienne, C.; Swart, J.R.; Bracht, V.M.; Patrick, M.; Chen, X. The Oxytricha trifallax macronuclear genome: A complex eukaryotic genome with 16,000 tiny chromosomes. PLoS Biol. 2013, 11, e1001473. [Google Scholar] [CrossRef]
- Chen, X.; Jung, S.; Beh, L.Y.; Eddy, S.R.; Landweber, L.F. Combinatorial DNA rearrangement facilitates the origin of new genes in ciliates. Genome Biol. Evol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Lobanov, A.V.; Heaphy, S.M.; Turanov, A.A.; Gerashchenko, M.V.; Picciarelli, S.; Devaraj, R.R. Position-dependent termination and widespread obligatory frame shifting in Euplotes translation. Nat. Struct. Mol. Biol. 2016. [Google Scholar] [CrossRef]
- Berger, H. Monograph of the Urostyloidea (Ciliophora, Hypotricha). Monogr. Biol. 2006, 85, 1–1304. [Google Scholar]
- Song, W.B.; Shao, C. Ontogenetic Patterns of Hypotrich Ciliates; Science Press: Beijing, China, 2017. (In Chinese) [Google Scholar]
- Numata, O.; Fujiu, K.; Gonda, K. Macronuclear division and cytokinesis in Tetrahymena. Cell Biol. Int. 1999, 23, 849–857. [Google Scholar] [CrossRef]
- Liang, H.X.; Jing, X.U.; Wang, W. Overexpression of RAN1 inhibits macronuclear amitosis in Tetrahymena thermophila. Chin. J. Biochem. Mol. Biol. 2013, 29, 42–48. [Google Scholar]
- Foissner, W. Ontogenesis in ciliated protozoa, with emphasis on stomatogenesis. In Ciliates, Cells as Organisms; Hausmann, K., Bradbury, P.C., Eds.; Gustav Fischer Verlag: Stuttgart, Germany, 1996; pp. 95–177. [Google Scholar]
- Hu, X.Z.; Song, W.B. Morphological redescriotion and morphogenesis of the marine cilaite, Pseudokeronopsis rubra (Ciliophora: Hypotrichida). Acta Protozool. 2001, 40, 107–115. [Google Scholar]
- Yi, Z.; Huang, L.; Yang, R.; Lin, X.; Song, W. Actin evolution in ciliates (Protist, Alveolata) is characterized by high diversity and three duplication events. Mol. Phylogenet. Evol. 2016, 96, 45–54. [Google Scholar] [CrossRef] [PubMed]
- Picelli, S.; Faridani, O.R.; Björklund, Å.K.; Winberg, G.; Sagasser, S.; Sandberg, R. Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 2014, 9, 171. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Y.R.; Sheng, Y.L.; Warren, A.; Gao, S. GPSit: An automated method for evolutionary analysis of nonculturable ciliated microeukaryotes. Mol. Ecol. Resour. 2018, 18, 700–713. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nuclcic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Jaramillo, M.L.; Ammar, D.; Quispe, R.L.; Guzman, F.; Margis, R.; Nazari, E.M.; Müller, Y.M. Identification and evaluation of reference genes for expression studies by RT-qPCR during embryonic development of the emerging model organism, Macrobrachium olfersii. Gene 2017, 598, 97–106. [Google Scholar] [CrossRef]
- Morse, D.; Daoust, P.; Benribague, S. A transcriptome-based perspective of cell cycle regulation in dinoflagellates. Protist 2016, 167, 610–621. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, X.; Tang, L.; Zhang, Q.J.; Frankel, J.; Berger, J.D. A cyclin-dependent protein kinase homologue associated with the basal body domains in the ciliate Tetrahymena thermophile. Biochim. Biophys. Acta Mol. Cell Res. 2002, 1591, 119–128. [Google Scholar] [CrossRef]
- Zhuang, Y.; Zhang, H.; Lin, S. Cyclin B gene and its cell cycle-dependent differential expression in the toxic dinoflagellate Alexandrium fundyense Atama Group, I./Clade, I. Harmful Algae 2013, 26, 71–79. [Google Scholar] [CrossRef]
- Morgan, D.O. The cell cycle: Principles of control. In Niederrheinischen Gesellschaft für Natur-und Heilkunde zu Bonn; New Science Press: London, UK, 2007; pp. 259–263. [Google Scholar]
- Cross, F.R.; Umen, J.G. The Chlamydomonas cell cycle. Plant J. 2015, 82, 370–392. [Google Scholar] [CrossRef] [PubMed]
- Fehér, A.; Magyar, Z. Coordination of cell division and differentiation in plants in comparison to animals. Acta Biol. Szeged. 2015, 59, 275–289. [Google Scholar]
- Ewald, J.C. How yeast coordinates metabolism, growth and division. Curr. Opin. Microbiol. 2018, 45, 1. [Google Scholar] [CrossRef] [PubMed]
- Hammarton, T.C.; Engstler, M.; Mottram, J.C. The Trypanosoma brucei cyclin, CYC2, is required for cell cycle progression through G1 phase and for maintenance of procyclic form cell morphology. J. Biol. Chem. 2004, 279, 24757–24760. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, S.; Gorbsky, G.J. Spatiotemporal regulation of the anaphase-promoting complex in mitosis. Nat. Rev. Mol. Cell Biol. 2015. [Google Scholar] [CrossRef]
- Taira, N.; Mimoto, R.; Kurata, M.; Yamaguchi, T.; Kitagawa, M.; Miki, Y.; Yoshida, K. DYRK2 priming phosphorylation of c-Jun and c-Myc modulates cell cycle progression in human cancer cells. J. Clin. Investig. 2012, 122, 859–872. [Google Scholar] [CrossRef]
- Woods, Y.L.; Cohen, P.; Becker, W.; Jakes, R.; Goedert, M.; Xuemin, W.A.; Proud, C.G. The kinase DYRK phosphorylates protein-synthesis initiation factor eIF2Bɛ at Ser539 and the microtubule-associated protein tau at Thr212: Potential role for DYRK as a glycogen synthase kinase 3-priming kinase. Biochem. J. 2001, 355, 609–615. [Google Scholar] [CrossRef]
- Shimada, H.; Koizumi, M.; Kuroki, K.; Mochizuki, M.; Fujimoto, H.; Ohta, H.; Masuda, T.; Takamiya, K.I. ARC3, a chloroplast division factor, is a chimera of prokaryotic FtsZ and part of eukaryotic phosphatidylinositol-4-phosphate 5-kinase. Plant Cell Physiol. 2004, 45, 960–967. [Google Scholar] [CrossRef]
- Ma, H.; Lou, Y.; Lin, W.H.; Xue, H.W. MORN motifs in plant PIPKs are involved in the regulation of subcellular localization and phospholipid binding. Cell Res. 2006, 16, 466. [Google Scholar] [CrossRef][Green Version]
- Kusano, H.; Testerink, C.; Vermeer, J.E.; Tsuge, T.; Shimada, H.; Oka, A.; Munnik, T.; Aoyama, T. The Arabidopsis phosphatidylinositol phosphate 5-kinase PIP5K3 is a key regulator of root hair tip growth. Plant Cell 2008, 20, 367–380. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Han, C.T.; Hur, Y. Overexpression of BrMORN, a novel ‘membrane occupation and recognition nexus’ motif protein gene from Chinese cabbage, promotes vegetative growth and seed production in Arabidopsis. Mol. Cells 2010, 29, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.J.; Cobb, M.H. Mitogen-activated protein kinase pathways. Curr. Opin. Cell Biol. 1997, 9, 180–186. [Google Scholar] [CrossRef]
- Feng, J.M.; Jiang, C.Q.; Sun, Z.Y.; Hua, C.J.; Wen, J.F.; Miao, W.; Xiong, J. Single-cell transcriptome sequencing of rumen ciliates provides insight into their molecular adaptations to the anaerobic and carbohydrate-rich rumen microenvironment. Mol. Phylogenet. Evol. 2020, 143, 116087. [Google Scholar] [CrossRef]
- Hwang, B.; Lee, J.H.; Bang, D. Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp. Mol. Med. 2018, 50, 96. [Google Scholar] [CrossRef]
- Foissner, W. Life cycle, morphology, ontogenesis, and phylogeny of Bromeliothrix metopoides nov. gen., nov. spec., a peculiar ciliate (Protista, Colpodea) from tank bromeliads (Bromeliaceae). Acta Protozool. 2010, 49, 159–193. [Google Scholar]
- Liu, W.W.; Shao, C.; Gong, J.; Li, J.Q.; Lin, X.F.; Song, W.B. Morphology, morphogenesis, and molecular phylogeny of a new marine urostylid ciliate (Ciliophora, Stichotrichia) from the South China Sea, and a brief overview of the convergent evolution of the midventral pattern within the Spirotrichea. Zool. J. Linn. Soc. 2010, 158, 697–710. [Google Scholar] [CrossRef][Green Version]
- Lu, X.; Huang, J.; Shao, C.; Berger, H. Morphology, cell-division, and phylogeny of Schmidingerothrix elongata spec. nov. (Ciliophora, Hypotricha), and brief guide to hypotrichs with Gonostomum-like oral apparatus. Eur. J. Protistol. 2018, 62, 24–42. [Google Scholar] [CrossRef]
- Keryer, G.; Davis, F.M.; Rao, P.N.; Beisson, J. Protein phosphorylation and dynamics of cytoskeletal structures associated with basal bodies in Paramecium. Cell Motil. Cytoskelet. 1987, 8, 44–54. [Google Scholar] [CrossRef]
- Nussbaum, M. Über spontane und künstliche Zellteilung. Sitzungsberichte der Niederrheinischen Gesellschaft für Natur-und Heilkunde zu Bonn 1884, 41, 259–263. [Google Scholar]
- Tang, S.K.; Marshall, W.F. Self-repairing cells: How single cells heal membrane ruptures and restore lost structures. Science 2017, 356, 1022–1025. [Google Scholar] [CrossRef] [PubMed]
- Onsbring, H.; Jamy, M.; Ettema, T.J. RNA sequencing of Stentor cell fragments reveals transcriptional changes during cellular regeneration. Curr. Biol. 2018, 28, 1281–1288. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.Z.; Warren, A.; Song, W.B. Observations on the morphology and morphogenesis of a new marine hypotrich ciliate (Ciliophora, Hypotrichida) from China. J. Nat. Hist. 2004, 38, 1059–1069. [Google Scholar] [CrossRef]
| Sample | D1 | D2 | D3 | G1 | G2 | G3 |
|---|---|---|---|---|---|---|
| raw read number | 87,103,778 | 84,670,776 | 57,275,044 | 52,963,602 | 60,176,334 | 50,426,634 |
| clean read number | 39,375,244 | 40,285,356 | 41,369,504 | 41,684,596 | 43,051,906 | 35,788,890 |
| raw base number (Mb) | 12,460.30 | 12,112.25 | 8193.26 | 7576.50 | 8608.29 | 7213.59 |
| clean base number (Mb) | 5576.19 | 5721.74 | 5913.74 | 5958.98 | 6154.61 | 5116.24 |
| Q30 (%) | 81.4 | 82.25 | 96.37 | 96.39 | 96.13 | 96.27 |
| Gene Family | Up/Down | Gene_id | Nr Description |
|---|---|---|---|
| CYCs | up | c62398_g1 | Mitotic cyclin-CYC2 [Oxytricha trifallax] |
| up | c67538_g2 | Cyclin [Oxytricha trifallax] | |
| up | c17611_g1 | Cyclin-T [Oxytricha trifallax] | |
| up | c9656_g1 | Cyclin, N-terminal domain-containing protein [Oxytricha trifallax] | |
| up | c36176_g1 | Mitotic cyclin-CYC2, putative [Oxytricha trifallax] | |
| up | c48130_g1 | Cyclin, N-terminal domain-containing protein [Oxytricha trifallax] | |
| up | c54046_g1 | Amine-terminal domain cyclin [Tetrahymena thermophila SB210] | |
| down | c60024_g1 | G1/S-specific cyclin-E1, putative [Oxytricha trifallax] | |
| APCs | up | c57016_g1 | TPR repeat-containing protein [Oxytricha trifallax] |
| up | c75917_g1 | TPR repeat-containing protein [Oxytricha trifallax] | |
| up | c54345_g1 | Anaphase-promoting complex subunit 8-like [Stylonychia lemnae] | |
| up | c58189_g1 | Anaphase-promoting complex subunit 10 [Stylonychia lemnae] | |
| up | c16402_g1 | Anaphase-promoting complex subunit 11 [Stylonychia lemnae] | |
| CDKs | up | c61357_g2 | Cyclin-dependent kinase b2-2-like [Stylonychia lemnae] |
| up | c40281_g1 | Cyclin-dependent kinase regulatory subunit family protein [Stylonychia lemnae] | |
| DYRK | up | c61235_g1 | Putative dual specificity protein phosphatase cdc14 [Oxytricha trifallax] |
| up | c58364_g1 | Dual specificity protein phosphatase [Stylonychia lemnae] | |
| down | c47824_g1 | Dual specificity phosphatase, catalytic domain-containing protein [Oxytricha trifallax] | |
| down | c12202_g1 | Dual specificity catalytic domain-containing protein [Stylonychia lemnae] | |
| MORN | up | c56401_g1 | Putative MORN repeat protein [Oxytricha trifallax] |
| up | c60320_g1 | Putative MORN repeat protein [Oxytricha trifallax] | |
| up | c60776_g1 | Putative MORN repeat protein [Oxytricha trifallax] | |
| up | c48198_g1 | Putative MORN repeat protein [Oxytricha trifallax] | |
| up | c66123_g1 | Putative MORN repeat protein [Oxytricha trifallax] | |
| up | c62710_g1 | MORN repeat protein [Stylonychia lemnae] | |
| up | c60934_g1 | MORN repeat protein [Stylonychia lemnae] | |
| up | c50818_g1 | MORN repeat-containing protein 5 [Stylonychia lemnae] | |
| up | c59470_g1 | MORN repeat protein [Stylonychia lemnae] | |
| up | c26899_g1 | MORN motif protein [Tetrahymena thermophila SB210] | |
| MAPK | up | c57299_g1 | Putative MAPK [Oxytricha trifallax] |
| up | c59004_g1 | Mitogen-activated protein kinase kinase [Oxytricha trifallax] | |
| Tubulin | up | c37490_g1 | Alpha tubulin, putative [Oxytricha trifallax] |
| up | c7781_g1 | Microtubule-associated RP/EB member 2 [Stylonychia lemnae] |
| Gene id | Nr Description | Up/ Down | Fold Change | log2 Fold Change |
|---|---|---|---|---|
| c37490_g1 | Alpha tubulin, putative [Oxytricha trifallax] | up | 3.34 | 1.74 |
| c38398_g1 | 14-3-3 domain-containing protein [Oxytricha trifallax] | up | 5.5 | 2.46 |
| c38773_g1 | Histone variant H3.8 [Stylonychia lemnae] | up | 100.01 | 6.64 |
| c52670_g1 | Electron transferring flavor protein dehydrogenase [Oxytricha trifallax] | up | 2.56 | 1.36 |
| c58750_g1 | Proteasome subunit beta type [Stylonychia lemnae] | up | 4.87 | 2.28 |
| c60031_g1 | Eukaryotic aspartyl protease family protein [Stylonychia lemnae] | down | 0.27 | −1.89 |
| c56677_g1 | EF hand family protein [Stylonychia lemnae] | down | 0.01 | −6.16 |
| c50771_g1 | cAMP-dependent protein kinase catalytic subunit, putative [Oxytricha trifallax] | down | 0.43 | −1.22 |
| c58897_g1 | Citrate synthase [Stylonychia lemnae] | down | 0.09 | −3.68 |
| c47080_g1 | Adenosine kinase [Stylonychia lemnae] | down | 0.22 | −2.17 |
| c58837_g1 | S-adenosyl-L-homocysteine hydrolase [Stylonychia lemnae] | down | 0.39 | −1.35 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Shen, Z.; Gentekaki, E.; Xu, J.; Yi, Z. Comparative Transcriptome Analyses during the Vegetative Cell Cycle in the Mono-Cellular Organism Pseudokeronopsis erythrina (Alveolata, Ciliophora). Microorganisms 2020, 8, 108. https://doi.org/10.3390/microorganisms8010108
Xu Y, Shen Z, Gentekaki E, Xu J, Yi Z. Comparative Transcriptome Analyses during the Vegetative Cell Cycle in the Mono-Cellular Organism Pseudokeronopsis erythrina (Alveolata, Ciliophora). Microorganisms. 2020; 8(1):108. https://doi.org/10.3390/microorganisms8010108
Chicago/Turabian StyleXu, Yiwei, Zhuo Shen, Eleni Gentekaki, Jiahui Xu, and Zhenzhen Yi. 2020. "Comparative Transcriptome Analyses during the Vegetative Cell Cycle in the Mono-Cellular Organism Pseudokeronopsis erythrina (Alveolata, Ciliophora)" Microorganisms 8, no. 1: 108. https://doi.org/10.3390/microorganisms8010108
APA StyleXu, Y., Shen, Z., Gentekaki, E., Xu, J., & Yi, Z. (2020). Comparative Transcriptome Analyses during the Vegetative Cell Cycle in the Mono-Cellular Organism Pseudokeronopsis erythrina (Alveolata, Ciliophora). Microorganisms, 8(1), 108. https://doi.org/10.3390/microorganisms8010108






