Legacy Effects Overshadow Tree Diversity Effects on Soil Fungal Communities in Oil Palm-Enrichment Plantations
Abstract
:1. Introduction
2. Material and Methods
2.1. Research Site and Experimental Design
2.2. Sampling
2.3. Soil Nutrient Elements and pH
2.4. Vegetation Structure Complexity
2.5. Fungal Community
2.6. Sequence Data Deposition
2.7. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Sampling and Export Permission
References
- Carrasco, L.R.; Larrosa, C.; Milner-Gulland, E.J.; Edwards, D.P. A double-edged sword for tropical forests. Science 2014, 346, 38–40. [Google Scholar] [CrossRef]
- Sayer, J.; Ghazoul, J.; Nelson, P.; Klintuni Boedhihartono, A. Oil palm expansion transforms tropical landscapes and livelihoods. Glob. Food Secur. 2012, 1, 114–119. [Google Scholar] [CrossRef]
- Euler, M.; Krishna, V.; Schwarze, S.; Siregar, H.; Qaim, M. Oil palm adoption, household welfare, and nutrition among smallholder farmers in Indonesia. World Dev. 2017, 93, 219–235. [Google Scholar] [CrossRef] [Green Version]
- Euler, M.; Schwarze, S.; Siregar, H.; Qaim, M. Oil palm expansion among smallholder farmers in Sumatra, Indonesia. J. Agric. Econ. 2016, 67, 658–676. [Google Scholar] [CrossRef]
- Kubitza, C.; Krishna, V.V.; Alamsyah, Z.; Qaim, M. The economics behind an ecological crisis: Livelihood effects of oil palm expansion in Sumatra, Indonesia. Hum. Ecol. 2018, 46, 107–116. [Google Scholar] [CrossRef]
- Abood, S.A.; Lee, J.S.H.; Burivalova, Z.; Garcia-Ulloa, J.; Koh, L.P. Relative contributions of the logging, fiber, oil palm, and mining industries to forest loss in Indonesia. Conserv. Lett. 2015, 8, 58–67. [Google Scholar] [CrossRef] [Green Version]
- Gibbs, H.K.; Ruesch, A.S.; Achard, F.; Clayton, M.K.; Holmgren, P.; Ramankutty, N.; Foley, J.A. Tropical forests were the primary sources of new agricultural land in the 1980s and 1990s. Proc. Natl. Acad. Sci. USA 2010, 107, 16732–16737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miettinen, J.; Shi, C.; Liew, S.C. Deforestation rates in insular Southeast Asia between 2000 and 2010. Glob. Chang. Biol. 2011, 17, 2261–2270. [Google Scholar] [CrossRef]
- Vijay, V.; Pimm, S.L.; Jenkins, C.N.; Smith, S.J. The impacts of oil palm on recent deforestation and biodiversity loss. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Wilcove, D.S.; Koh, L.P. Addressing the threats to biodiversity from oil-palm agriculture. Biodivers. Conserv. 2010, 19, 999–1007. [Google Scholar] [CrossRef]
- Drescher, J.; Rembold, K.; Allen, K.; Beckschäfer, P.; Buchori, D.; Clough, Y.; Faust, H.; Fauzi, A.M.; Gunawan, D.; Hertel, D. Ecological and socio-economic functions across tropical land use systems after rainforest conversion. Phil. Trans. R. Soc. B 2016, 371, 20150275. [Google Scholar] [CrossRef] [PubMed]
- Fitzherbert, E.B.; Struebig, M.J.; Morel, A.; Danielsen, F.; Brühl, C.A.; Donald, P.F.; Phalan, B. How will oil palm expansion affect biodiversity? Trends Ecol. Evol. 2008, 23, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Rembold, K.; Mangopo, H.; Tjitrosoedirdjo, S.S.; Kreft, H. Plant diversity, forest dependency, and alien plant invasions in tropical agricultural landscapes. Biol. Conserv. 2017, 213, 234–242. [Google Scholar] [CrossRef]
- Barnes, A.D.; Allen, K.; Kreft, H.; Corre, M.D.; Jochum, M.; Veldkamp, E.; Clough, Y.; Daniel, R.; Darras, K.; Denmead, L.H.; et al. Direct and cascading impacts of tropical land-use change on multi-trophic biodiversity. Nat. Ecol. Evol. 2017, 1, 1511–1519. [Google Scholar] [CrossRef]
- Dislich, C.; Keyel, A.C.; Salecker, J.; Kisel, Y.; Meyer, K.M.; Auliya, M.; Barnes, A.D.; Corre, M.D.; Darras, K.; Faust, H.; et al. A review of the ecosystem functions in oil palm plantations, using forests as a reference system. Biol. Rev. 2017, 92, 1539–1569. [Google Scholar] [CrossRef]
- Grass, I.; Kubitza, C.; Krishna, V.V.; Corre, M.D.; Mußhoff, O.; Pütz, P.; Drescher, J.; Rembold, K.; Ariyanti, E.S.; Barnes, A.D.; et al. Trade-offs between multifunctionality and profit in tropical smallholder landscapes. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Foster, W.A.; Snaddon, J.L.; Turner, E.C.; Fayle, T.M.; Cockerill, T.D.; Ellwood, M.D.F.; Broad, G.R.; Chung, A.Y.C.; Eggleton, P.; Khen, C.V.; et al. Establishing the evidence base for maintaining biodiversity and ecosystem function in the oil palm landscapes of South East Asia. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2011, 366, 3277–3291. [Google Scholar] [CrossRef] [Green Version]
- Koh, L.P.; Levang, P.; Ghazoul, J. Designer landscapes for sustainable biofuels. Trends Ecol. Evol. 2009, 24, 431–438. [Google Scholar] [CrossRef]
- Bhagwat, S.A.; Willis, K.J.; Birks, H.J.B.; Whittaker, R.J. Agroforestry: A refuge for tropical biodiversity? Trends Ecol. Evol. 2008, 23, 261–267. [Google Scholar] [CrossRef]
- de Carvalho, W.R.; Vasconcelos, S.S.; Kato, O.R.; Capela, C.J.B.; Castellani, D.C. Short-term changes in the soil carbon stocks of young oil palm-based agroforestry systems in the eastern Amazon. Agrofor. Syst. 2014, 88, 357–368. [Google Scholar] [CrossRef]
- Ramos, H.M.N.; Vasconcelos, S.S.; Kato, O.R.; Castellani, D.C. Above- and belowground carbon stocks of two organic, agroforestry-based oil palm production systems in eastern Amazonia. Agrofor. Syst. 2018, 92, 221–237. [Google Scholar] [CrossRef]
- Schroth, G.; Izac, A.-M.N.; Vasconcelos, H.L.; Gascon, C.; da Fonseca, G.A.B.; Harvey, C.A. Agroforestry and Biodiversity Conservation in Tropical Landscapes; Island Press: Washington, DC, USA, 2004; ISBN 978-1-55963-357-4. [Google Scholar]
- Tscharntke, T.; Clough, Y.; Bhagwat, S.A.; Buchori, D.; Faust, H.; Hertel, D.; Hölscher, D.; Juhrbandt, J.; Kessler, M.; Perfecto, I.; et al. Multifunctional shade-tree management in tropical agroforestry landscapes—A review. J. Appl. Ecol. 2011, 619–629. [Google Scholar] [CrossRef] [Green Version]
- Frąc, M.; Hannula, S.E.; Bełka, M.; Jędryczka, M. Fungal biodiversity and their role in soil health. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Rillig, M.C. Arbuscular mycorrhizae and terrestrial ecosystem processes. Ecol. Lett. 2004, 7, 740–754. [Google Scholar] [CrossRef]
- van der Heijden, M.G.A.; Bardgett, R.D.; van Straalen, N.M. The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 2008, 11, 296–310. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.J.; Maldonado, C.; Frøslev, T.G.; Antonelli, A.; Rønsted, N. Unexpectedly high beta-diversity of root-associated fungal communities in the Bolivian Andes. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Brinkmann, N.; Schneider, D.; Sahner, J.; Ballauff, J.; Edy, N.; Barus, H.; Irawan, B.; Budi, S.W.; Qaim, M.; Daniel, R.; et al. Intensive tropical land use massively shifts soil fungal communities. Sci. Rep. 2019, 9, 3403. [Google Scholar] [CrossRef] [Green Version]
- Glassman, S.I.; Lubetkin, K.C.; Chung, J.A.; Bruns, T.D. The theory of island biogeography applies to ectomycorrhizal fungi in subalpine tree “islands” at a fine scale. Ecosphere 2017, 8, e01677. [Google Scholar] [CrossRef]
- Rodríguez-Echeverría, S.; Teixeira, H.; Correia, M.; Timóteo, S.; Heleno, R.; Öpik, M.; Moora, M. Arbuscular mycorrhizal fungi communities from tropical Africa reveal strong ecological structure. New Phytol. 2017, 213, 380–390. [Google Scholar] [CrossRef]
- Tedersoo, L.; Bahram, M.; Dickie, I.A. Does host plant richness explain diversity of ectomycorrhizal fungi? Re-evaluation of Gao et al. (2013) data sets reveals sampling effects. Mol. Ecol. 2014, 23, 992–995. [Google Scholar] [CrossRef]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Jakobsen, I.; Grønlund, M.; Smith, F.A. Roles of arbuscular mycorrhizas in plant phosphorus nutrition: Interactions between pathways of phosphorus uptake in arbuscular mycorrhizal roots have important implications for understanding and manipulating plant phosphorus acquisition. Plant Physiol. 2011, 156, 1050–1057. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Heijden, M.G.A.; Dombrowski, N.; Schlaeppi, K. Continuum of root–fungal symbioses for plant nutrition. Proc. Natl. Acad. Sci. USA 2017, 114, 11574–11576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buyer, J.S.; Baligar, V.C.; He, Z.; Arévalo-Gardini, E. Soil microbial communities under cacao agroforestry and cover crop systems in Peru. Appl. Soil Ecol. 2017, 120, 273–280. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, G.; Wu, Z.; Wen, X.; Zhong, H.; Zhong, Z.; Bian, F.; Gai, X. Agroforestry alters the rhizosphere soil bacterial and fungal communities of moso bamboo plantations in subtropical China. Appl. Soil Ecol. 2019, 143, 192–200. [Google Scholar] [CrossRef]
- Kerfahi, D.; Tripathi, B.M.; Lee, J.; Edwards, D.P.; Adams, J.M. The impact of selective-logging and forest clearance for oil palm on fungal communities in Borneo. PLoS ONE 2014, 9, e111525. [Google Scholar] [CrossRef]
- McGuire, K.L.; D’Angelo, H.; Brearley, F.Q.; Gedallovich, S.M.; Babar, N.; Yang, N.; Gillikin, C.M.; Gradoville, R.; Bateman, C.; Turner, B.L.; et al. Responses of soil fungi to logging and oil palm agriculture in Southeast Asian tropical forests. Microb. Ecol. 2015, 69, 733–747. [Google Scholar] [CrossRef] [Green Version]
- Sahner, J.; Budi, S.W.; Barus, H.; Edy, N.; Meyer, M.; Corre, M.D.; Polle, A. Degradation of root community traits as indicator for transformation of tropical lowland rain forests into oil palm and rubber plantations. PLoS ONE 2015, 10, e0138077. [Google Scholar] [CrossRef] [Green Version]
- Asmelash, F.; Bekele, T.; Birhane, E. The potential role of arbuscular mycorrhizal fungi in the restoration of degraded lands. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Kardol, P.; Wardle, D.A. How understanding aboveground–belowground linkages can assist restoration ecology. Trends Ecol. Evol. 2010, 25, 670–679. [Google Scholar] [CrossRef]
- Teuscher, M.; Gérard, A.; Brose, U.; Buchori, D.; Clough, Y.; Ehbrecht, M.; Hölscher, D.; Irawan, B.; Sundawati, L.; Wollni, M.; et al. Experimental biodiversity enrichment in oil-palm-dominated landscapes in Indonesia. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zemp, D.C.; Ehbrecht, M.; Seidel, D.; Ammer, C.; Craven, D.; Erkelenz, J.; Irawan, B.; Sundawati, L.; Hölscher, D.; Kreft, H. Mixed-species tree plantings enhance structural complexity in oil palm plantations. Agric. Ecosyst. Environ. 2019, 283, 106564. [Google Scholar] [CrossRef]
- Zemp, D.C.; Gérard, A.; Hölscher, D.; Ammer, C.; Irawan, B.; Sundawati, L.; Teuscher, M.; Kreft, H. Tree performance in a biodiversity enrichment experiment in an oil palm landscape. J. Appl. Ecol. 2019, 56, 2340–2352. [Google Scholar] [CrossRef]
- Allen, K.; Corre, M.D.; Tjoa, A.; Veldkamp, E. Soil nitrogen-cycling responses to conversion of lowland forests to oil palm and rubber plantations in Sumatra, Indonesia. PLoS ONE 2015, 10, e0133325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bell, T.; Lilley, A.K.; Hector, A.; Schmid, B.; King, L.; Newman, J.A. A linear model method for biodiversity–ecosystem functioning experiments. Am. Nat. 2009, 174, 836–849. [Google Scholar] [CrossRef] [PubMed]
- Gérard, A.; Wollni, M.; Hölscher, D.; Irawan, B.; Sundawati, L.; Teuscher, M.; Kreft, H. Oil-palm yields in diversified plantations: Initial results from a biodiversity enrichment experiment in Sumatra, Indonesia. Agric. Ecosyst. Environ. 2017, 240, 253–260. [Google Scholar] [CrossRef]
- Bray, R.; Kurtz, L. Determination of total, organic, and available forms of phosphorus in soils. Soil Sci. 1945, 59, 39–46. [Google Scholar] [CrossRef]
- Ehbrecht, M.; Schall, P.; Ammer, C.; Seidel, D. Quantifying stand structural complexity and its relationship with forest management, tree species diversity and microclimate. Agric. For. Meteorol. 2017, 242, 1–9. [Google Scholar] [CrossRef]
- Ehbrecht, M.; Schall, P.; Juchheim, J.; Ammer, C.; Seidel, D. Effective number of layers: A new measure for quantifying three-dimensional stand structure based on sampling with terrestrial LiDAR. For. Ecol. Manag. 2016, 380, 212–223. [Google Scholar] [CrossRef]
- Willim, K.; Stiers, M.; Annighöfer, P.; Ammer, C.; Ehbrecht, M.; Kabal, M.; Stillhard, J.; Seidel, D. Assessing understory complexity in beech-dominated forests (Fagus sylvatica L.) in Central Europe—From managed to primary forests. Sensors 2019, 19, 1684. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, D.Q.; Schneider, D.; Brinkmann, N.; Song, B.; Janz, D.; Schöning, I.; Daniel, R.; Pena, R.; Polle, A. Soil and root nutrient chemistry structure root-associated fungal assemblages in temperate forests. Environ. Microbiol. 2020, 22, 3081–3095. [Google Scholar] [CrossRef] [PubMed]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-coverage ITS primers for the DNA-based identification of Ascomycetes and Basidiomycetes in environmental samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols; Academic Press, Inc.: Cambridge, MA, USA, 1990; pp. 315–322. ISBN 978-0-12-372180-8. [Google Scholar]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina paired-end read merger. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughpt sequencing reads. Embnet. J. 2014, 17, 3. [Google Scholar]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.S.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. Bmc Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016, 44, D7–D19. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. 2019. Available online: https://www.researchgate.net/publication/313502495_Vegan_Community_Ecology_Package (accessed on 9 October 2020).
- Leinonen, R.; Sugawara, H.; Shumway, M.; Collaboration, on behalf of the I.N.S.D. The Sequence Read Archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Chao, A.; Gotelli, N.J.; Hsieh, T.C.; Sander, E.L.; Ma, K.H.; Colwell, R.K.; Ellison, A.M. Rarefaction and extrapolation with Hill numbers: A framework for sampling and estimation in species diversity studies. Ecol. Monogr. 2014, 84, 45–67. [Google Scholar] [CrossRef] [Green Version]
- Li, D. hillR: Taxonomic, functional, and phylogenetic diversity and similarity through Hill Numbers. J. Open Source Softw. 2018, 3, 1041. [Google Scholar] [CrossRef]
- Bachelot, B.; Uriarte, M.; Zimmerman, J.K.; Thompson, J.; Leff, J.W.; Asiaii, A.; Koshner, J.; McGuire, K. Long-lasting effects of land use history on soil fungal communities in second-growth tropical rain forests. Ecol. Appl. 2016, 26, 1881–1895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holste, E.K.; Holl, K.D.; Zahawi, R.A.; Kobe, R.K. Reduced aboveground tree growth associated with higher arbuscular mycorrhizal fungal diversity in tropical forest restoration. Ecol. Evol. 2016, 6, 7253–7262. [Google Scholar] [CrossRef]
- Kerfahi, D.; Tripathi, B.M.; Dong, K.; Go, R.; Adams, J.M. Rainforest Conversion to Rubber Plantation May Not Result in Lower Soil Diversity of Bacteria, Fungi, and Nematodes. Microb. Ecol. 2016, 72, 359–371. [Google Scholar] [CrossRef]
- de Vries, F.T.; Manning, P.; Tallowin, J.R.B.; Mortimer, S.R.; Pilgrim, E.S.; Harrison, K.A.; Hobbs, P.J.; Quirk, H.; Shipley, B.; Cornelissen, J.H.C.; et al. Abiotic drivers and plant traits explain landscape-scale patterns in soil microbial communities. Ecol. Lett. 2012, 15, 1230–1239. [Google Scholar] [CrossRef]
- Leff, J.W.; Bardgett, R.D.; Wilkinson, A.; Jackson, B.G.; Pritchard, W.J.; Long, J.R.D.; Oakley, S.; Mason, K.E.; Ostle, N.J.; Johnson, D.; et al. Predicting the structure of soil communities from plant community taxonomy, phylogeny, and traits. ISME J. 2018, 12, 1794–1805. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Tedersoo, L.; Soltis, P.S.; Soltis, D.E.; Gilbert, J.A.; Sun, M.; Shi, Y.; Wang, H.; Li, Y.; Zhang, J.; et al. Phylogenetic imprint of woody plants on the soil mycobiome in natural mountain forests of eastern China. ISME J. 2019, 13, 686–697. [Google Scholar] [CrossRef] [Green Version]
- Müller, K.; Kubsch, N.; Marhan, S.; Mayer-Gruner, P.; Nassal, P.; Schneider, D.; Daniel, R.; Piepho, H.-P.; Polle, A.; Kandeler, E. Saprotrophic and Ectomycorrhizal Fungi Contribute Differentially to Organic P Mobilization in Beech-Dominated Forest Ecosystems. Front. Glob. Chang. 2020, 3, 47. [Google Scholar] [CrossRef]
- Shigyo, N.; Umeki, K.; Hirao, T. Seasonal Dynamics of Soil Fungal and Bacterial Communities in Cool-Temperate Montane Forests. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kivlin, S.N.; Hawkes, C.V. Spatial and temporal turnover of soil microbial communities is not linked to function in a primary tropical forest. Ecology 2020, 101, e02985. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.; Cao, Y.; Yue, M.; Tian, T.; Yin, Q.; Dang, H.; Quan, J.; Zhang, R.; Wang, M. Soil abiotic properties and plant functional traits mediate associations between soil microbial and plant communities during a secondary forest succession on the Loess Plateau. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Moreira, C.C.; Celestino, D.; Sobrinho, T.G.; Cardoso, I.M.; Elliot, S.L. Agroforestry coffee soils increase the insect-suppressive potential offered by entomopathogenic fungi over full-sun soils: A case proposing a “bait survival technique”. Ecol. Evol. 2019, 9, 10777–10787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paquette, A.; Hector, A.; Castagneyrol, B.; Vanhellemont, M.; Koricheva, J.; Scherer-Lorenzen, M.; Verheyen, K. A million and more trees for science. Nat. Ecol. Evol. 2018, 2, 763–766. [Google Scholar] [CrossRef]
- Peay, K.G.; Russo, S.E.; McGuire, K.L.; Lim, Z.; Chan, J.P.; Tan, S.; Davies, S.J. Lack of host specificity leads to independent assortment of dipterocarps and ectomycorrhizal fungi across a soil fertility gradient. Ecol. Lett. 2015, 18, 807–816. [Google Scholar] [CrossRef] [Green Version]
- Schappe, T.; Albornoz, F.E.; Turner, B.L.; Neat, A.; Condit, R.; Jones, F.A. The role of soil chemistry and plant neighbourhoods in structuring fungal communities in three Panamanian rainforests. J. Ecol. 2017, 105, 569–579. [Google Scholar] [CrossRef] [Green Version]
- Bardgett, R.D.; van der Putten, W.H. Belowground biodiversity and ecosystem functioning. Nature 2014, 515, 505–511. [Google Scholar] [CrossRef]
- Schröter, K.; Wemheuer, B.; Pena, R.; Schöning, I.; Ehbrecht, M.; Schall, P.; Ammer, C.; Daniel, R.; Polle, A. Assembly processes of trophic guilds in the root mycobiome of temperate forests. Mol. Ecol. 2019, 28, 348–364. [Google Scholar] [CrossRef]
- Adams, R.I.; Miletto, M.; Taylor, J.W.; Bruns, T.D. Dispersal in microbes: Fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J. 2013, 7, 1262–1273. [Google Scholar] [CrossRef] [Green Version]
- Dumbrell, A.J.; Nelson, M.; Helgason, T.; Dytham, C.; Fitter, A.H. Relative roles of niche and neutral processes in structuring a soil microbial community. ISME J. 2010, 4, 337–345. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powell, J.R.; Karunaratne, S.; Campbell, C.D.; Yao, H.; Robinson, L.; Singh, B.K. Deterministic processes vary during community assembly for ecologically dissimilar taxa. Nat. Commun. 2015, 6, 8444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahram, M.; Kohout, P.; Anslan, S.; Harend, H.; Abarenkov, K.; Tedersoo, L. Stochastic distribution of small soil eukaryotes resulting from high dispersal and drift in a local environment. ISME J. 2016, 10, 885–896. [Google Scholar] [CrossRef] [Green Version]
- Kluber, L.A.; Carrino-Kyker, S.R.; Coyle, K.P.; DeForest, J.L.; Hewins, C.R.; Shaw, A.N.; Smemo, K.A.; Burke, D.J. Mycorrhizal response to experimental pH and P manipulation in acidic hardwood forests. PLoS ONE 2012, 7, e48946. [Google Scholar] [CrossRef] [Green Version]
- Sylvia, D.M. Principles and Applications of Soil Microbiology; Pearson Prentice Hall: Upper Saddle River, NJ, USA, 2005; ISBN 978-0-13-094117-6. [Google Scholar]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Rousk, J.; Bååth, E.; Brookes, P.C.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef] [PubMed]
- Lauber, C.L.; Strickland, M.S.; Bradford, M.A.; Fierer, N. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol. Biochem. 2008, 40, 2407–2415. [Google Scholar] [CrossRef]
- Vasco-Palacios, A.M.; Bahram, M.; Boekhout, T.; Tedersoo, L. Carbon content and pH as important drivers of fungal community structure in three Amazon forests. Plant Soil 2019. [Google Scholar] [CrossRef]
- Dalling, J.W.; Heineman, K.; Lopez, O.R.; Wright, S.J.; Turner, B.L. Nutrient Availability in tropical rain forests: The paradigm of phosphorus limitation. In Tropical Tree Physiology: Adaptations and Responses in a Changing Environment; Goldstein, G., Santiago, L.S., Eds.; Tree Physiology; Springer International Publishing: Cham, Switzerland, 2016; pp. 261–273. ISBN 978-3-319-27422-5. [Google Scholar]
- Turner, B.L.; Brenes-Arguedas, T.; Condit, R. Pervasive phosphorus limitation of tree species but not communities in tropical forests. Nature 2018, 555, 367–370. [Google Scholar] [CrossRef]
- McGee, K.M.; Eaton, W.D.; Shokralla, S.; Hajibabaei, M. Determinants of soil bacterial and fungal community composition toward carbon-use efficiency across primary and secondary forests in a Costa Rican conservation area. Microb. Ecol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Bahram, M.; Polme, S.; Koljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global diversity and geography of soil fungi. Science 2014, 346, 1256688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edy, N.; Yelianti, U.; Irawan, B.; Polle, A.; Pena, R. Differences in root nitrogen uptake between tropical lowland rainforests and oil palm plantations. Front. Plant Sci. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Danielsen, L.; Thürmer, A.; Meinicke, P.; Buée, M.; Morin, E.; Martin, F.; Pilate, G.; Daniel, R.; Polle, A.; Reich, M. Fungal soil communities in a young transgenic poplar plantation form a rich reservoir for fungal root communities. Ecol. Evol. 2012, 2, 1935–1948. [Google Scholar] [CrossRef]
- Moll, J.; Hoppe, B.; König, S.; Wubet, T.; Buscot, F.; Krüger, D. Spatial distribution of fungal communities in an arable soil. PLoS ONE 2016, 11, e0148130. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Hu, H.-W.; Guo, L.-D.; Anderson, I.C.; Powell, J.R. Dryland forest management alters fungal community composition and decouples assembly of root- and soil-associated fungal communities. Soil Biol. Biochem. 2017, 109, 14–22. [Google Scholar] [CrossRef]
- Bainard, L.D.; Koch, A.M.; Gordon, A.M.; Klironomos, J.N. Temporal and compositional differences of arbuscular mycorrhizal fungal communities in conventional monocropping and tree-based intercropping systems. Soil Biol. Biochem. 2012, 45, 172–180. [Google Scholar] [CrossRef]
- Chifflot, V.; Rivest, D.; Olivier, A.; Cogliastro, A.; Khasa, D. Molecular analysis of arbuscular mycorrhizal community structure and spores distribution in tree-based intercropping and forest systems. Agric. Ecosyst. Environ. 2009, 131, 32–39. [Google Scholar] [CrossRef]
- Furze, J.R.; Martin, A.R.; Nasielski, J.; Thevathasan, N.V.; Gordon, A.M.; Isaac, M.E. Resistance and resilience of root fungal communities to water limitation in a temperate agroecosystem. Ecol. Evol. 2017, 7, 3443–3454. [Google Scholar] [CrossRef] [Green Version]
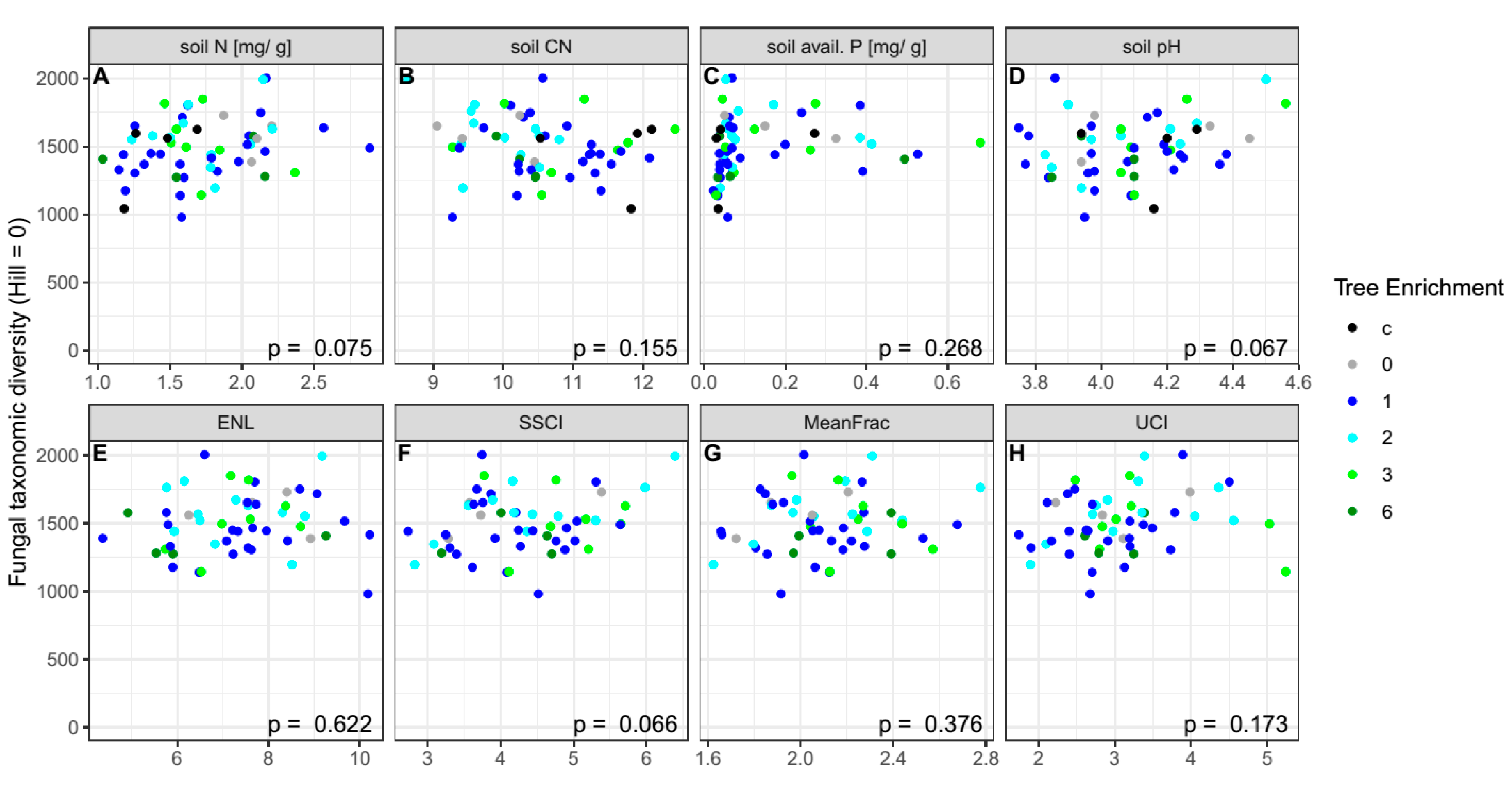
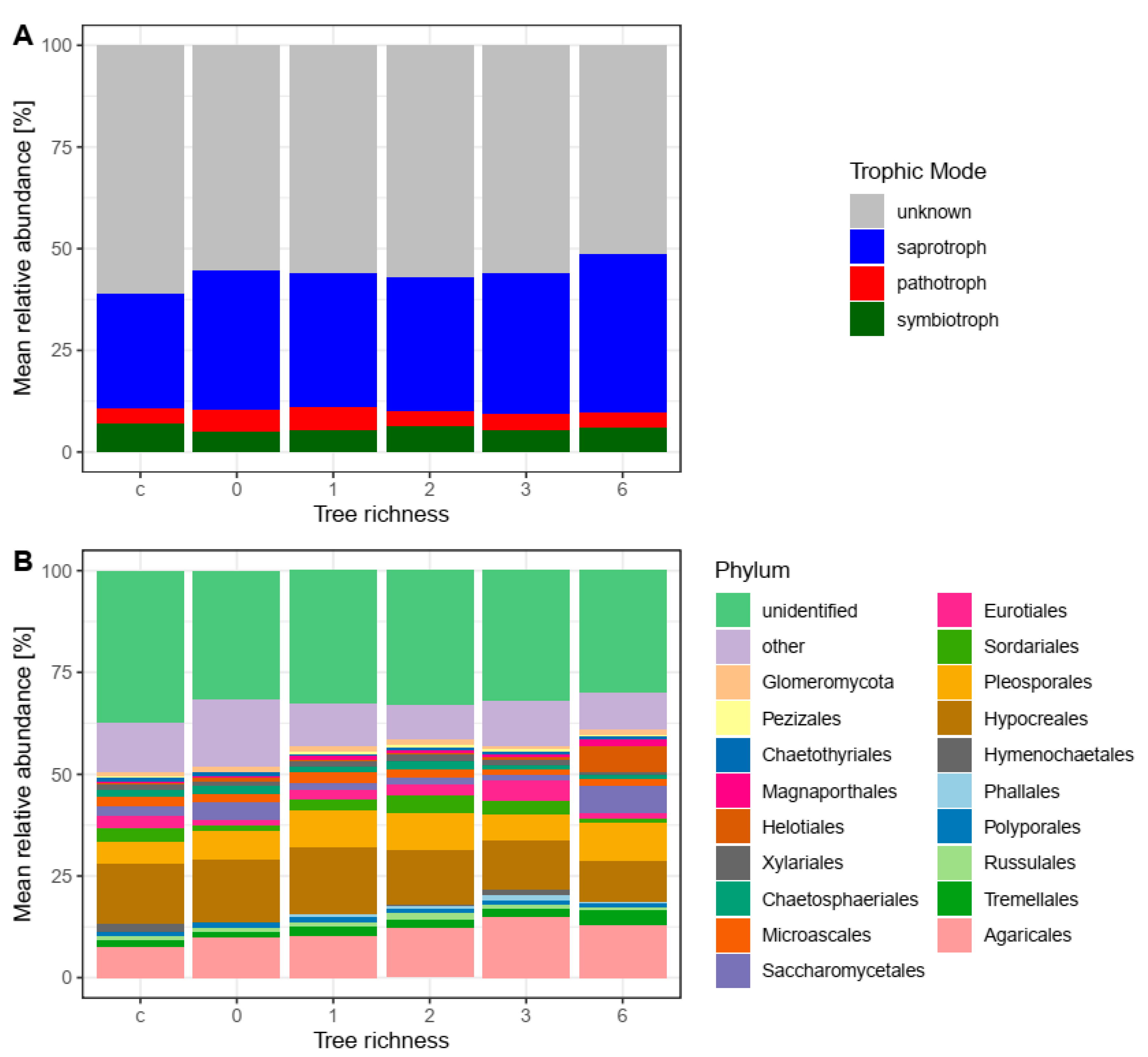
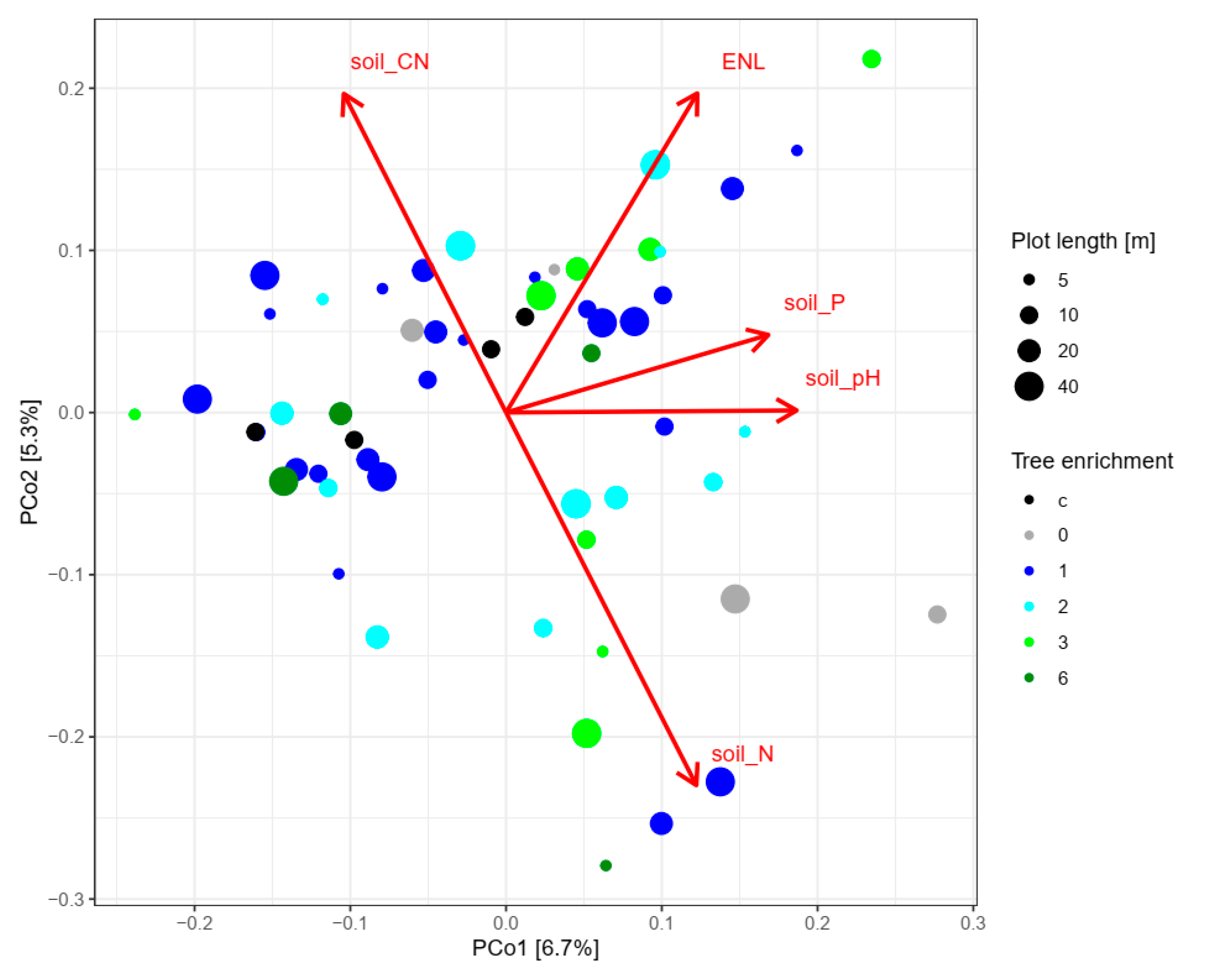
| Control | 0 | 1 | 2 | 3 | 6 | p-Value | |
|---|---|---|---|---|---|---|---|
| Fungal diversity | |||||||
| taxon. (0D) | 1459 (±139) | 1584 (±74) | 1460 (±46) | 1591 (±61) | 1532 (±84) | 1386 (±71) | 0.457 |
| taxon. (1D) | 162.7 (±26.3) | 172.6 (±25.5) | 178.9 (±9.5) | 174.3 (±17.1) | 180.7 (±9.8) | 134.0 (±35.2) | 0.681 |
| phylo. (0D) | 825 (±61) | 844 (±31) | 827 (±19) | 874 (±26) | 859 (±41) | 778 (±28) | 0.552 |
| phylo. (1D) | 39.6 (±2.4) | 38.4 (±3.5) | 43.2 (±1.5) | 42.0 (±2.9) | 45.1 (±2.0) | 37.5 (±5.3) | 0.552 |
| Soil variables | |||||||
| Soil pH | 4.15 (±0.07) | 4.18 (±0.13) | 4.11 (±0.07) | 4.42 (±0.24) | 4.31 (±0.13) | 4.00 (±0.06) | 0.454 |
| Soil nitrogen (mg·g−1) | 1.40 (±0.11) | 2.06 (±0.07) | 1.72 (±0.09) | 1.90 (±0.18) | 1.72 (±0.10) | 1.70 (±0.26) | 0.277 |
| Soil C/N | 11.60 (±0.36) | 9.79 (±0.33) | 10.73 (±0.15) | 9.90 (±0.18) | 10.95 (±0.36) | 10.26 (±0.08) | <0.001 *** |
| Soil phosphorus (mg·g−1) | 0.09 (±0.06) | 0.14 (±0.07) | 0.12 (±0.03) | 0.13 (±0.04) | 0.19 (±0.08) | 0.16 (±0.11) | 0.854 |
| Df | Sum Sq | Mean Sq | F-Value | p-Value | |
|---|---|---|---|---|---|
| Taxonomic diversity (0D) | |||||
| Soil pH | 1 | 226,996 | 226,996 | 5.1855 | 0.0290 * |
| Soil nitrogen (mg·g−1) | 1 | 98,642 | 98,642 | 2.2534 | 0.1423 |
| Soil C/N | 1 | 18,090 | 18,090 | 0.4132 | 0.5245 |
| Soil phosphorus (mg·g−1) | 1 | 11,620 | 11,620 | 0.2655 | 0.6096 |
| Linear tree richness | 1 | 12,349 | 12,349 | 0.2821 | 0.5987 |
| Tree species identity | 5 | 361,799 | 72,360 | 1.653 | 0.1719 |
| Nonlinear tree richness | 3 | 106,425 | 35,475 | 0.8104 | 0.4967 |
| Plot size | 3 | 56,147 | 18,716 | 0.4275 | 0.7345 |
| Residuals | 35 | 1,532,123 | 43,775 | ||
| Taxonomic diversity (1D) | |||||
| Soil pH | 1 | 14 | 13.6 | 0.0046 | 0.9462 |
| Soil nitrogen (mg·g−1) | 1 | 1302 | 1301.6 | 0.4414 | 0.5108 |
| Soil C/N | 1 | 1137 | 1136.7 | 0.3855 | 0.5387 |
| Soil phosphorus (mg·g−1) | 1 | 5841 | 5840.7 | 1.9808 | 0.1681 |
| Linear tree richness | 1 | 3681 | 3680.8 | 1.2483 | 0.2715 |
| Tree species identity | 5 | 1491 | 298.2 | 0.1011 | 0.9913 |
| Nonlinear tree richness | 3 | 2109 | 702.9 | 0.2384 | 0.869 |
| Plot size | 3 | 5329 | 1776.3 | 0.6024 | 0.6178 |
| Residuals | 35 | 103,205 | 2948.7 | ||
| Phylogenetic diversity (0D) | |||||
| Soil pH | 1 | 56,524 | 56,524 | 6.6913 | 0.01401 * |
| Soil nitrogen (mg·g−1) | 1 | 1987 | 1987 | 0.2353 | 0.63068 |
| Soil C/N | 1 | 553 | 553 | 0.0654 | 0.79963 |
| Soil phosphorus (mg·g−1) | 1 | 4 | 4 | 0.0004 | 0.98342 |
| Linear tree richness | 1 | 2657 | 2657 | 0.3145 | 0.57851 |
| Tree species identity | 5 | 60,943 | 12,189 | 1.4429 | 0.23345 |
| Nonlinear tree richness | 3 | 18,996 | 6332 | 0.7496 | 0.52997 |
| Plot size | 3 | 8775 | 2925 | 0.3463 | 0.79203 |
| Residuals | 35 | 295,657 | 8447 | ||
| Phylogenetic diversity (1D) | |||||
| Soil pH | 1 | 0.07 | 0.074 | 0.001 | 0.9752 |
| Soil nitrogen (mg·g−1) | 1 | 142.1 | 142.098 | 1.8859 | 0.1784 |
| Soil C/N | 1 | 69.96 | 69.96 | 0.9285 | 0.3419 |
| Soil phosphorus (mg·g−1) | 1 | 124.3 | 124.304 | 1.6498 | 0.2074 |
| Linear tree richness | 1 | 15.12 | 15.116 | 0.2006 | 0.657 |
| Tree species identity | 5 | 114.49 | 22.897 | 0.3039 | 0.9072 |
| Nonlinear tree richness | 3 | 118.95 | 39.65 | 0.5262 | 0.6672 |
| Plot size | 3 | 81.17 | 27.058 | 0.3591 | 0.7829 |
| Residuals | 35 | 2637.12 | 75.346 |
| Df | Sum Sq | F-Value | p-Value | |
|---|---|---|---|---|
| Soil pH | 1 | 0.2474 | 1.2965 | 0.017 * |
| Soil nitrogen (mg g−1) | 1 | 0.3727 | 1.9533 | 0.001 *** |
| Soil C/N | 1 | 0.2329 | 1.2204 | 0.063 |
| Soil phosphorus (mg g−1) | 1 | 0.2873 | 1.5054 | 0.003 ** |
| Linear tree richness | 1 | 0.1957 | 1.0255 | 0.381 |
| Tree species identity | 5 | 0.9462 | 0.9917 | 0.561 |
| Nonlinear tree richness | 3 | 0.5548 | 0.9692 | 0.657 |
| Plot size | 3 | 0.6293 | 1.0993 | 0.096 |
| Residuals | 35 | 6.6788 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ballauff, J.; Zemp, D.C.; Schneider, D.; Irawan, B.; Daniel, R.; Polle, A. Legacy Effects Overshadow Tree Diversity Effects on Soil Fungal Communities in Oil Palm-Enrichment Plantations. Microorganisms 2020, 8, 1577. https://doi.org/10.3390/microorganisms8101577
Ballauff J, Zemp DC, Schneider D, Irawan B, Daniel R, Polle A. Legacy Effects Overshadow Tree Diversity Effects on Soil Fungal Communities in Oil Palm-Enrichment Plantations. Microorganisms. 2020; 8(10):1577. https://doi.org/10.3390/microorganisms8101577
Chicago/Turabian StyleBallauff, Johannes, Delphine Clara Zemp, Dominik Schneider, Bambang Irawan, Rolf Daniel, and Andrea Polle. 2020. "Legacy Effects Overshadow Tree Diversity Effects on Soil Fungal Communities in Oil Palm-Enrichment Plantations" Microorganisms 8, no. 10: 1577. https://doi.org/10.3390/microorganisms8101577
APA StyleBallauff, J., Zemp, D. C., Schneider, D., Irawan, B., Daniel, R., & Polle, A. (2020). Legacy Effects Overshadow Tree Diversity Effects on Soil Fungal Communities in Oil Palm-Enrichment Plantations. Microorganisms, 8(10), 1577. https://doi.org/10.3390/microorganisms8101577





