The Untapped Australasian Diversity of Astaxanthin-Producing Yeasts with Biotechnological Potential—Phaffia australis sp. nov. and Phaffia tasmanica sp. nov.
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genome Sequencing and Assembly
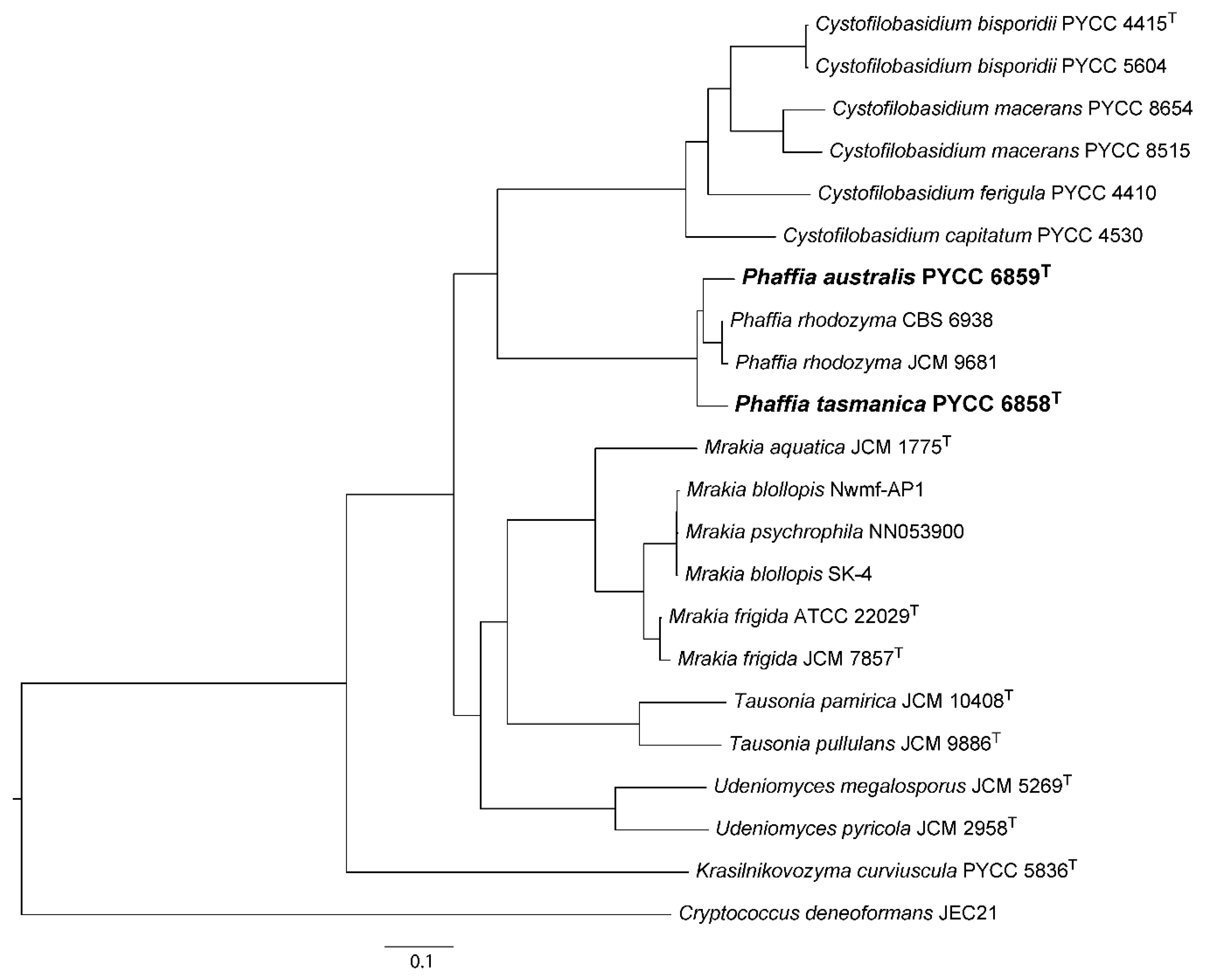
2.2. Orthology Mapping, Genomic Diversity, and Phylogenetic Analyses
2.3. Search for Relevant Genes
2.4. Phenotypic Characterization
3. Results
3.1. The Divergent Australasian Lineages Belong to the Genus Phaffia
3.2. Phenotypic Characterisation of P. australis sp. nov and P. tasmanica sp. nov.
3.3. Astaxanthin Production
3.4. Organization of MAT Loci
3.5. Taxonomy
3.5.1. Description of Phaffia australis sp. nov. M. David-Palma, D. Libkind, P. Gonçalves and J.P. Sampaio
3.5.2. Description of Phaffia tasmanica sp. nov. M. David-Palma, D. Libkind, P. Gonçalves and J.P. Sampaio
4. Concluding Remarks
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Miller, M.W.; Yoneyama, M.; Soneda, M. Phaffia, a new yeast genus in the Deuteromycotina (Blastomycetes). Int. J. Syst. Evol. Microbiol. 1976, 26, 286–291. [Google Scholar] [CrossRef] [Green Version]
- Golubev, W.I. Perfect state of Rhodomyces dendrorhous (Phaffia rhodozyma). Yeast 1995, 11, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Alcaíno, J.; Baeza, M.; Cifuentes, V. Carotenoid distribution in nature. In Carotenoids in Nature: Biosynthesis, Regulation and Function; Stange, C., Ed.; Springer International Publishing: Cham, Switzerland, 2016; pp. 3–33. ISBN 978-3-319-39126-7. [Google Scholar]
- Schmidt, I.; Schewe, H.; Gassel, S.; Jin, C.; Buckingham, J.; Humbelin, M.; Sandmann, G.; Schrader, J. Biotechnological production of astaxanthin with Phaffia rhodozyma/Xanthophyllomyces dendrorhous. Appl. Microbiol. Biotechnol. 2011, 89, 555–571. [Google Scholar] [CrossRef] [PubMed]
- Foss, P.; Storebakken, T.; Schiedt, K.; Liaaen-Jensen, S.; Austreng, E.; Streiff, K. Carotenoids in diets for salmonids. Aquaculture 1984, 41, 213–226. [Google Scholar] [CrossRef]
- Guerin, M.; Huntley, M.E.; Olaizola, M. Haematococcus astaxanthin: Applications for human health and nutrition. Trends Biotechnol. 2003, 21, 210–216. [Google Scholar] [CrossRef]
- Loto, I.; Gutierrez, M.S.; Barahona, S.; Sepulveda, D.; Martinez-Moya, P.; Baeza, M.; Cifuentes, V.; Alcaino, J. Enhancement of carotenoid production by disrupting the C22-sterol desaturase gene (CYP61) in Xanthophyllomyces dendrorhous. BMC Microbiol. 2012, 12, 235. [Google Scholar] [CrossRef] [Green Version]
- Gassel, S.; Breitenbach, J.; Sandmann, G. Genetic engineering of the complete carotenoid pathway towards enhanced astaxanthin formation in Xanthophyllomyces dendrorhous starting from a high-yield mutant. Appl. Microbiol. Biotechnol. 2014, 98, 345–350. [Google Scholar] [CrossRef]
- Ledetzky, N.; Osawa, A.; Iki, K.; Pollmann, H.; Gassel, S.; Breitenbach, J.; Shindo, K.; Sandmann, G. Multiple transformation with the crtYB gene of the limiting enzyme increased carotenoid synthesis and generated novel derivatives in Xanthophyllomyces dendrorhous. Arch. Biochem. Biophys. 2014, 545, 141–147. [Google Scholar] [CrossRef]
- Barredo, J.L.; García-Estrada, C.; Kosalkova, K.; Barreiro, C. Biosynthesis of astaxanthin as a main carotenoid in the heterobasidiomycetous yeast Xanthophyllomyces dendrorhous. J. Fungi 2017, 3, 44. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Sáiz, M.; De La Fuente, J.L.; Barredo, J.L. Xanthophyllomyces dendrorhous for the industrial production of astaxanthin. Appl. Microbiol. Biotechnol. 2010, 88, 645–658. [Google Scholar] [CrossRef]
- Libkind, D.; Ruffini, A.; Van Broock, M.; Alves, L.; Sampaio, J.P. Biogeography, host specificity, and molecular phylogeny of the basidiomycetous yeast Phaffia rhodozyma and its sexual form, Xanthophyllomyces dendrorhous. Appl. Environ. Microbiol. 2007, 73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- David-Palma, M.; Libkind, D.; Sampaio, J.P. Global distribution, diversity hot spots and niche transitions of an astaxanthin-producing eukaryotic microbe. Mol. Ecol. 2014, 23. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bushnell, B.; Rood, J.; Singer, E. BBMerge—Accurate paired shotgun read merging via overlap. PLoS ONE 2017, 12, e0185056. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Available online: https://academic.oup.com/bioinformatics/article/29/8/1072/228832 (accessed on 2 July 2020).
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.M.; Birol, I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef] [Green Version]
- Stanke, M.; Waack, S. Gene prediction with a hidden Markov model and a new intron submodel—PubMed. Bioinformatics 2003, 19 (Suppl. 2), ii215–ii225. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Stoeckert, C.J.; Roos, D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003, 13, 2178–2189. [Google Scholar] [CrossRef] [Green Version]
- Contreras-Moreira, B.; Vinuesa, P. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl. Environ. Microbiol. 2013, 79, 7696–7701. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Criscuolo, A.; Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010, 10, 210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haubold, B.; Pfaffelhuber, P.; Domazet-Los˘o, M.; Wiehe, T. Estimating mutation distances from unaligned genomes. J. Comput. Biol. 2009, 16, 1487–1500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gremme, G.; Steinbiss, S.; Kurtz, S. GenomeTools: A comprehensive software library for efficient processing of structured genome annotations. IEEE/ACM Trans. Comput. Biol. Bioinform. 2013, 10. [Google Scholar] [CrossRef]
- Bellora, N.; Moliné, M.; David-Palma, M.; Coelho, M.A.; Hittinger, C.T.; Sampaio, J.P.; Gonçalves, P.; Libkind, D. Comparative genomics provides new insights into the diversity, physiology, and sexuality of the only industrially exploited tremellomycete: Phaffia rhodozyma. BMC Genom. 2016, 17, 901. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Tusnady, G.E.; Simon, I. The HMMTOP transmembrane topology prediction server. Bioinformatics 2001, 17, 849–850. [Google Scholar] [CrossRef]
- Mitchell, A.; Chang, H.Y.; Daugherty, L.; Fraser, M.; Hunter, S.; Lopez, R.; McAnulla, C.; McMenamin, C.; Nuka, G.; Pesseat, S.; et al. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2015, 43, D213–D221. [Google Scholar] [CrossRef]
- Lin, J.; Hu, J. SeqNLS: Nuclear localization signal prediction based on frequent pattern mining and linear motif scoring. PLoS ONE 2013, 8, e76864. [Google Scholar] [CrossRef] [PubMed]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar] [CrossRef] [PubMed]
- Alva, V.; Nam, S.Z.; Soding, J.; Lupas, A.N. The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. 2016, 44, W410–W415. [Google Scholar] [CrossRef] [PubMed]
- Lupas, A.; Van Dyke, M.; Stock, J. Predicting coiled coils from protein sequences. Science 1991, 252, 1162–1164. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T.; Robert, V. Methods for isolation, phenotypic characterization and maintenance of yeasts. In The Yeasts, a Taxonomic Study; Kurtzman, C., Fell, J., Boekhout, T., Eds.; Elsevier: Amsterdam, The Netherlands, 2011; pp. 87–110. ISBN 978-0-444-52149-1. [Google Scholar]
- Libkind, D.; Moliné, M.; García, V.; Fontenla, S.; Broock, M. Characterization of a novel South American population of the astaxanthin producing yeast Xanthophyllomyces dendrorhous (Phaffia rhodozyma). Ind. Microbiol. Biotechnol. 2008, 35. [Google Scholar] [CrossRef]
- Libkind, D.; Moliné, M.; Tognetti, C. Isolation and selection of new astaxanthin producing strains of Xanthophyllomyces dendrorhous. In Microbial Carotenoids from Fungi: Methods and Protocols; Barredo, J.-L., Ed.; Humana Press: Totowa, NJ, USA, 2012; pp. 183–194. ISBN 978-1-61779-918-1. [Google Scholar]
- Liu, X.Z.; Wang, Q.M.; Göker, M.; Groenewald, M.; Kachalkin, A.V.; Lumbsch, H.T.; Millanes, A.M.; Wedin, M.; Yurkov, A.M.; Boekhout, T.; et al. Towards an integrated phylogenetic classification of the Tremellomycetes. Stud. Mycol. 2015, 81, 85–147. [Google Scholar] [CrossRef] [Green Version]
- Fell, J.W.; Boekhout, T.; Fonseca, A.; Scorzetti, G.; Statzell-Tallman, A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int. J. Syst. Evol. Microbiol. 2000, 50 Pt 3, 1351–1371. [Google Scholar] [CrossRef]
- Hibbett, D.S.; Binder, M.; Bischoff, J.F.; Blackwell, M.; Cannon, P.F.; Eriksson, O.E.; Huhndorf, S.; James, T.; Kirk, P.M.; Lücking, R.; et al. A higher-level phylogenetic classification of the Fungi. Mycol. Res. 2007, 111. [Google Scholar] [CrossRef]
- Libkind, D.; Čadež, N.; Opulente, D.A.; Langdon, Q.K.; Rosa, C.A.; Sampaio, J.P.; Gonçalves, P.; Hittinger, C.T.; Lachance, M.A. Towards yeast taxogenomics: Lessons from novel species descriptions based on complete genome sequences. FEMS Yeast Res. 2020, 20, foaa042. [Google Scholar] [CrossRef]
- Rokas, A.; Mead, M.E.; Steenwyk, J.L.; Raja, H.A.; Oberlies, N.H. Biosynthetic gene clusters and the evolution of fungal chemodiversity. Nat. Prod. Rep. 2020, 37, 868–878. [Google Scholar] [CrossRef] [PubMed]
- David-Palma, M.; Sampaio, J.P.; Gonçalves, P. Genetic dissection of sexual reproduction in a primary homothallic basidiomycete. PLoS Genet. 2016, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coelho, M.A.; Bakkeren, G.; Sun, S.; Hood, M.E.; Giraud, T. Fungal Sex: The Basidiomycota. In The Fungal Kingdom; Heitman, J., Howlett, B.J., Crous, P.W., Stukenbrock, E.H., James, T., Gow, N.A.R., Eds.; American Society of Microbiology: Washington, DC, USA, 2017; pp. 147–175. [Google Scholar]
- Devier, B.; Aguileta, G.; Hood, M.E.; Giraud, T. Ancient trans-specific polymorphism at pheromone receptor genes in basidiomycetes. Genetics 2009, 181, 209–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNeill, J.; Barrie, F.; Buck, W.; Demoulin, V.; Greuter, W.; Al, E. International Code of Nomenclature for Algae, Fungi, and Plants (Melbourne Code). Regnum Vegetabile 154; A.R.G. Gantner Verlag KG: Ruggell, Liechtenstein, 2012; ISBN 978-3-87429-425-6. [Google Scholar]




| Species | Sequenced Strain | Other Collections | Genome Data (BioProject) | Origin |
|---|---|---|---|---|
| Cryptococcus deneoformans | JEC21 | PRJNA13856 | NCBI | |
| Cystofilobasidium bisporidii | PYCC 5604 | CBS 6347 | PRJNA371778 | This study |
| Cystofilobasidium bisporidii | PYCC 4415T | CBS 6346T | PRJNA371780 | This study |
| Cystofilobasidium capitatum | PYCC 4530 | CBS 7420 | PRJNA371774 | This study |
| Cystofilobasidium ferigula | PYCC 4410 | CBS 7201 | PRJNA371786 | This study |
| Cystofilobasidium macerans | PYCC 8515 | CBS 6532 | PRJNA371809 | This study |
| Cystofilobasidium macerans | PYCC 8654 | CBS 2425 | PRJNA371814 | This study |
| Krasilnikovozyma curviuscula | PYCC 5836T | PRJNA371818 | This study | |
| Mrakia aquatica | JCM 1775T | PRJDB3647 | RIKEN BioResource Center | |
| Mrakia blollopis | SK-4 | PRJDB3253 | NCBI | |
| Mrakia blollopis | Nwmf-AP1 | RRJNA268263 | NCBI | |
| Mrakia frigida | JCM 7857T | PRJDB3713 | RIKEN BioResource Center | |
| Mrakia frigida | ATCC 22029T | PRJNA334195 | JGI | |
| Mrakia psychrophila | NN053900 | PRJNA304674 | NCBI | |
| Phaffia australis | PYCC 6859T | CBS 14095T | PRJNA371751 | This study |
| Phaffia rhodozyma | CBS 6938 | PYCC 6916 | PRJEB6925 | NCBI |
| Phaffia rhodozyma | JCM 9681 | PYCC 6917 | PRJDB3716 | RIKEN BioResource Center |
| Phaffia tasmanica | PYCC 6858T | CBS 14096T | PRJNA371754 | This study |
| Tausonia pamirica | JCM 10408T | PRJDB3689 | RIKEN BioResource Center | |
| Tausonia pullulans | JCM 9886T | PRJDB3678 | RIKEN BioResource Center | |
| Udeniomyces megalosporus | JCM 5269T | PRJDB3720 | RIKEN BioResource Center | |
| Udeniomyces pyricola | JCM 2958T | PRJDB3672 | RIKEN BioResource Center |
| Fermentation | P. australis | P. tasmanica | P. rhodozyma |
|---|---|---|---|
| D-Glucose | + | + | + |
| D-Galactose | - | - | - |
| D-Xylose | - | - | - |
| Sucrose | +, W | + | D, - |
| Maltose | - | - | D, - |
| α,α-Trehalose | - | - | D, - |
| Melibiose | - | - | - |
| Lactose | - | - | - |
| Methyl-α-D-glucoside | - | - | - |
| Cellobiose | - | - | D, - |
| Melezitose | - | + | D, - |
| Raffinose | - | W | - |
| Inulin | - | - | - |
| Soluble Starch | - | - | - |
| Assimilation of carbon compounds | |||
| D-Glucose | + | + | + |
| D-Galactose | - | D, W | - |
| L-Sorbose | - | - | D, - |
| D-Glucosamine | - | - | - |
| D-Ribose | - | - | D, - |
| D-Xylose | + | - | + |
| L-Arabinose | + | + | + |
| D-Arabinose | - | - | D, - |
| L-Rhamnose | - | - | D, - |
| Sucrose | + | + | + |
| Maltose | + | + | + |
| α,α-Trehalose | + | + | + |
| Methyl-α-D-glucoside | + | + | D, - |
| Cellobiose | + | + | + |
| Salicin | + | + | + |
| Melibiose | + | + | - |
| Lactose | - | - | - |
| Raffinose | + | + | + |
| Melezitose | + | + | + |
| Inulin | - | - | - |
| Soluble Starch | + | + | + |
| Glycerol | + | + | + |
| Erythritol | - | - | - |
| Ribitol | D | - | D, - |
| Xylitol | + | - | - |
| D-Glucitol | + | - | D |
| D-Mannitol | + | + | + |
| Galactitol | - | - | - |
| Inositol | - | - | - |
| Glucono-δ-lactone | + | + | + |
| D-Gluconic acid | + | + | + |
| D-Glucuronic acid | + | w | +, D |
| D,L-Lactic acid | - | - | V |
| Succinic acid | + | + | + |
| Citric acid | + | + | + |
| D-Tartaric acid | - | - | - |
| m-Tartaric acid | - | - | - |
| Saccharic acid | - | - | - |
| Mucic acid | - | - | - |
| Methanol | - | - | - |
| Ethanol | + | + | + |
| Assimilation of nitrogen compounds | |||
| Nitrate | - | - | - |
| Nitrite | - | - | - |
| Ethylamine | + | - | - |
| L-Lysine | + | + | + |
| Cadaverine | W | W | + |
| Creatine | - | - | - |
| Creatinine | - | - | - |
| Other tests | |||
| Growth in vitamin-free medium | - | - | - |
| Growth in the presence of 0.01% cycloheximide | - | - | - |
| Growth in the presence of 0.1% cycloheximide | - | - | - |
| Growth at 25 °C | + | + | + |
| Growth at 30 °C | - | - | - |
| Formation of starch-like compounds | + | + | + |
| Hydrolysis of urea | + | + | + |
| Colour reaction with Diazonium Blue B | + | + | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
David-Palma, M.; Libkind, D.; Brito, P.H.; Silva, M.; Bellora, N.; Coelho, M.A.; Heitman, J.; Gonçalves, P.; Sampaio, J.P. The Untapped Australasian Diversity of Astaxanthin-Producing Yeasts with Biotechnological Potential—Phaffia australis sp. nov. and Phaffia tasmanica sp. nov. Microorganisms 2020, 8, 1651. https://doi.org/10.3390/microorganisms8111651
David-Palma M, Libkind D, Brito PH, Silva M, Bellora N, Coelho MA, Heitman J, Gonçalves P, Sampaio JP. The Untapped Australasian Diversity of Astaxanthin-Producing Yeasts with Biotechnological Potential—Phaffia australis sp. nov. and Phaffia tasmanica sp. nov. Microorganisms. 2020; 8(11):1651. https://doi.org/10.3390/microorganisms8111651
Chicago/Turabian StyleDavid-Palma, Márcia, Diego Libkind, Patrícia H. Brito, Margarida Silva, Nicolás Bellora, Marco A. Coelho, Joseph Heitman, Paula Gonçalves, and José Paulo Sampaio. 2020. "The Untapped Australasian Diversity of Astaxanthin-Producing Yeasts with Biotechnological Potential—Phaffia australis sp. nov. and Phaffia tasmanica sp. nov." Microorganisms 8, no. 11: 1651. https://doi.org/10.3390/microorganisms8111651
APA StyleDavid-Palma, M., Libkind, D., Brito, P. H., Silva, M., Bellora, N., Coelho, M. A., Heitman, J., Gonçalves, P., & Sampaio, J. P. (2020). The Untapped Australasian Diversity of Astaxanthin-Producing Yeasts with Biotechnological Potential—Phaffia australis sp. nov. and Phaffia tasmanica sp. nov. Microorganisms, 8(11), 1651. https://doi.org/10.3390/microorganisms8111651






