Abstract
Bacterial collections are invaluable tools for microbiologists. However, their practical use is compromised by imprecise taxonomical assignation of bacterial strains. This is particularly true for soft rotting plant pathogens of the Pectobacterium genus. We analysed the taxonomic status of 265 Pectobacterium strains deposited at CIRM-CFBP collection from 1944 to 2020. This collection gathered Pectobacterium strains isolated in 27 countries from 32 plant species representing 17 botanical families or from nonhost environments. The MLSA approach completed by genomic analysis of 15 strains was performed to update the taxonomic status of these 265 strains. The results showed that the CIRM-CFBP Pectobacterium collection harboured at least one strain of each species, with the exception of P. polonicum. Yet, seven strains could not be assigned to any of the described species and may represent at least two new species. Surprisingly, P. versatile, recently described in 2019, is the most prevalent species among CIRM-CFBP strains. An analysis of P. versatile strains revealed that this species is pandemic and isolated from various host plants and environments. At the opposite, other species gathered strains isolated from only one botanical family or exclusively from a freshwater environment. Our work also revealed new host plants for several Pectobacterium spp.
1. Introduction
Bacterial collections are invaluable tools for microbiologists, as they host many strains isolated at different times on different hosts or environments and on different countries and continents. As such, they summarise the collective sampling and research efforts performed by bacteriologists from all over the world on many different bacterial species. This collective treasure is nevertheless often underexploited for several reasons, the main one being the poor taxonomical assignation of many deposited strains to current taxonomical standard. This situation results from the fact that many strains are ancient strains that were deposited in collection before the precise taxonomic delineation of species through genome analysis were performed. Even if most collections are doing great efforts to improve this situation, a lot of work is still necessary. Therefore, currently, collections harbour many strains with old names no longer reflecting their actual taxonomical status. Such ancient strains are nevertheless important to understand the epidemiology of a given species, when and where a particular species was first isolated in the world and what is its historical prevalence all over the world.
Soft rot plant pathogenic bacteria of the Pectobacterium genus represent an archetype of this situation. They are characterised by their ability to degrade plant cell walls through the secretion of a cocktail of plant cell wall degrading enzymes (PCWDEs) [1,2]. Pectobacterium spp. are a major cause of harvest loss of potatoes both on the field and during potato storage. However, strains of this genus have also been collected on a large number of host plants and are thus known as large host range plant pathogens, although the extent of the host range varies between species [3,4]. Pectobacterium spp. were previously regrouped in the Erwinia genus founded in 1917 to unite all Gram-negative, fermentative, nonsporulating and peritrichous flagellated plant pathogenic bacteria [5]. Early taxonomy recognised three taxa within these soft rot bacteria: Erwinia carotovora subsp. carotovora, Erwinia carotovora subsp. atroseptica and Erwinia chrysanthemi [6,7] that were included in the Approved Lists of Bacterial Names in 1980 either under the species named Erwinia or Pectobacterium [8], and the Pectobacterium genus was formally described in 1998 [9]. In 2005, on the basis of the 16S RNA sequence, P. chrysanthemi was reclassified within the new Dickeya genus [10]. Furthermore, in addition to Pectobacterium carotovorum subsp. carotovorum and Pectobacterium carotovorum subsp. atrosepticum, several new subspecies were progressively described for P. carotovorum: Pectobacterium carotovorum subsp. brasiliense [11,12], Pectobacterium carotovorum subsp. wasabiae, Pectobacterium carotovorum subsp. betavasculorum, Pectobacterium carotovorum subsp. odoriferum [9] and P. carotovorum subsp. actinidiae [13]. All these subspecies were latter elevated to species level [14,15] following genomic analysis [16]. In addition, new Pectobacterium species were progressively described, most of them recently on the basis of whole-genome sequence analysis. Today, the Pectobacterium genus encompasses 17 recognised species: Pectobacterium actinidiae [15], Pectobacterium aquaticum [17], Pectobacterium aroidearum [18], Pectobacterium atrosepticum [14], Pectobacterium betavasculorum [14], Pectobacterium brasiliense [15], Pectobacterium cacticida [9,19], Pectobacterium carotovorum [8,15,20], Pectobacterium fontis [21], Pectobacterium odoriferum [15], Pectobacterium parmentieri [22], Pectobacterium parvum [23], Pectobacterium polaris [24], Pectobacterium polonicum [25], Pectobacterium punjabense [26], Pectobacterium versatile [15] and Pectobacterium wasabiae [14] and two proposed species not yet validated by ad hoc committees: “Pectobacterium peruviense” and “Pectobacterium zantedeschiae” [27,28]. Given the taxonomic evolution in the past ten years, the ecological importance, repartition and habitat of most species need to be evaluated. Bacterial collections are interesting tools to reach that goal.
The CIRM-CFBP, French Collection for Plant-associated Bacteria (DOI: 10.15454/1.5103266699001077E12) located in France at INRAE, hosts many strains of the Pectobacterium genus isolated from 1944 to 2019. However, the taxonomical status of most of these strains is unclear. Many strains were deposited as Pectobacterium spp., which indicates they belong to this genus. Furthermore, other strains were deposited as P. carotovorum. Since, historically, P. carotovorum has gathered seven subspecies that are now elevated to species level, it is currently difficult to know to which species these strains belong. Finally, many ancient strains were likely characterised solely on the basis of phenotypic tests, and this could have led to incorrect taxonomic assignation. As a result, it is currently impossible to have a synthetic view of the Pectobacterium collection hosted at the CIRM-CFBP.
The aim of this work was to clarify and update the taxonomic status of 265 Pectobacterium strains deposited at the CIRM-CFBP collection and to gain insight of the frequency and isolation habitat of the 19 described Pectobacterium species within the CIRM-CFBP collection. To do so, we performed a phylogenetic analysis based on the partial sequences or dnaX, leuS and recA housekeeping genes that allocated strains to specific clades. To understand how clades were related to delineate species, this analysis was completed with the genome sequencing of 15 strains. This allowed determining the frequency of each species within the CIRM-CFBP collection. Some species appeared to be pandemic Pectobacterium species found all over the world on various host plants and environments, while others, at the opposite, gathered strains isolated from only one botanical family or one specific environment.
2. Materials and Methods
2.1. Bacterial Strains, Culture Conditions and DNA Extraction
The 265 strains used in this study are provided Table S1. For housekeeping genes amplification, PCRs were conducted directly on colonies grown overnight on solid King B medium (2-g protease peptone, 15-g agar, 10-mL glycerol, 1.5-g KH2PO4 and 1.5-g MgSO4-7H2O per one litre of medium) and boiled at 100 °C for 10 min. For the preparation of genomic DNA, the strains were first grown overnight at 28 °C on solid LB medium (10-g tryptone, 5-g yeast extract, 10-g NaCl and 15-g agar per one litre of medium). A single colony was then picked up and grown overnight in 2 mL of liquid LB medium at 28 °C agitated at 120 rpm. After centrifugation of the culture broth (5 min at 12,000 rpm), DNA was extracted with the wizard® genomic DNA extraction kit (Promega, Madison, WI, USA) following the supplier’s instructions. DNA was suspended in 100 μL of sterile distilled water, and the quantity and quality of DNA was assessed by NanoDrop measurement, spectrophotometry analysis and agarose gel electrophoresis on 1% agarose gels.
2.2. dnaX-leuS-recA Phylogeny
Housekeeping genes dnaX leuS and recA were amplified and sequenced for the 261 strains. PCR protocols and primers were described in Portier et al. [15]. PCR products sequencing was performed by Genoscreen (Lille, France). The consensus sequences for each gene for each strain were extracted from forward and reverse sequence assemblies using Geneious Pro version 9.1.8 (www.geneious.com). The sequences were then aligned and trimmed using BioEdit version 5.0.6. All the obtained sequences were deposited in public databases, and Table S1 summarises the data. A phylogenetic tree was constructed with concatenated alignments of all genes with MEGA 7.0.26 using the neighbour-joining method with 1000 bootstrap replicates, and the evolutionary distances were computed by using the Kimura two-parameter method. All the Pectobacterium type strains were included in the phylogenetic tree. When type strains were not present at CIRM-CFBP (P. polonicum DPMP 315T, P. actinidiae KKH3T, P. zantedeschiae 9MT and P. peruviense UGC32T), dnaX, leuS and recA sequences were retrieved from the genome sequences available at NCBI. In addition, to help species delineation on the phylogenetic tree, dnaX, leuS and recA sequences retrieved of other NCBI genomes recently reclassified in their correct assignation species by Portier et al. [15] were also included. To root the phylogenetic tree, dnaX, leuS and recA sequences were retrieved from the genome of Dickeya solani CFBP 7704 (RNS 08.23). Twelve strains were not included in the phylogenetic analysis provided in Figure S1, because at least one of the three sequences of dnaX, leuS or recA was not correctly amplified. However, for these 12 strains, the remaining amplified sequences (data not shown) allowed their assignation to known species without ambiguities. Finally, 4 strains (CFBP 8719, CFBP 8720, CFBP 8723 and CFBP 8724) deposited at the CIRM-CFBP in 2019 were assigned to their species following amplification and sequencing of the gapA housekeeping gene as described by Cigna et al. [29]. All the sequences used for the phylogenetic tree were deposited at NCBI, and the accession numbers are listed in Table S1.
2.3. Genome Analysis
Genome sequencing was performed at the next-generation sequencing core facilities of the Institute for Integrative Biology of the Cell (91190 Gif-sur-Yvette, Avenue de la Terrasse, France). Nextera DNA libraries were prepared from 50 ng of high-quality genomic DNA. Paired-end 2 × 75-pb sequencing was performed on an Illumina NextSeq 500 apparatus, with a High Output 150 cycle kit. CLC Genomics Workbench (Version 9.5.2, Qiagen Bioinformatics) was used to assemble reads. Final sequencing coverage was between 118× and 216× (Table 1). Coding sequences were predicted using the RAST server [30] with the Glimmer 3 prediction tool [31]. Statistics of the 15 newly sequenced draft genomes are presented in Table 1.

Table 1.
General information for the 15 sequenced Pectobacterium genomes.
Pairwise comparison of the genomes was computed using the average nucleotide identity (ANI) Pyani python module (https://github.com/widdowquinn/pyani) [32] with the blast algorithm (ANIb). The species threshold was set at 96%. Digital DNA-DNA hybridisation (dDDH) values were calculated between each sequenced genome and reference species genomes using a dedicated pipeline (http://ggdc.dsmz.de/) from the formula 2 (sum of all identities found in high-scoring segment pairs (HSPs) divided by overall HSP length); this measure is normalised to the genome length and, therefore, is still robust when incomplete draft genomes are analysed. The species threshold was set to 70%.
A phylogenetic tree was constructed from concatenated sequences of 1053 homologous genes retrieved from the 15 newly sequenced genomes and 18 genomes of type strains or reference strains for each species. MLSA analysis was performed as described in Portier et al. [15]. The genome of D. solani strain RNS_08.23 was used to root the tree. The species P. cacticida was not included in this analysis, as no reference genome has yet been published.
3. Results and Discussion
3.1. The CIRM-CFBP Studied Strains
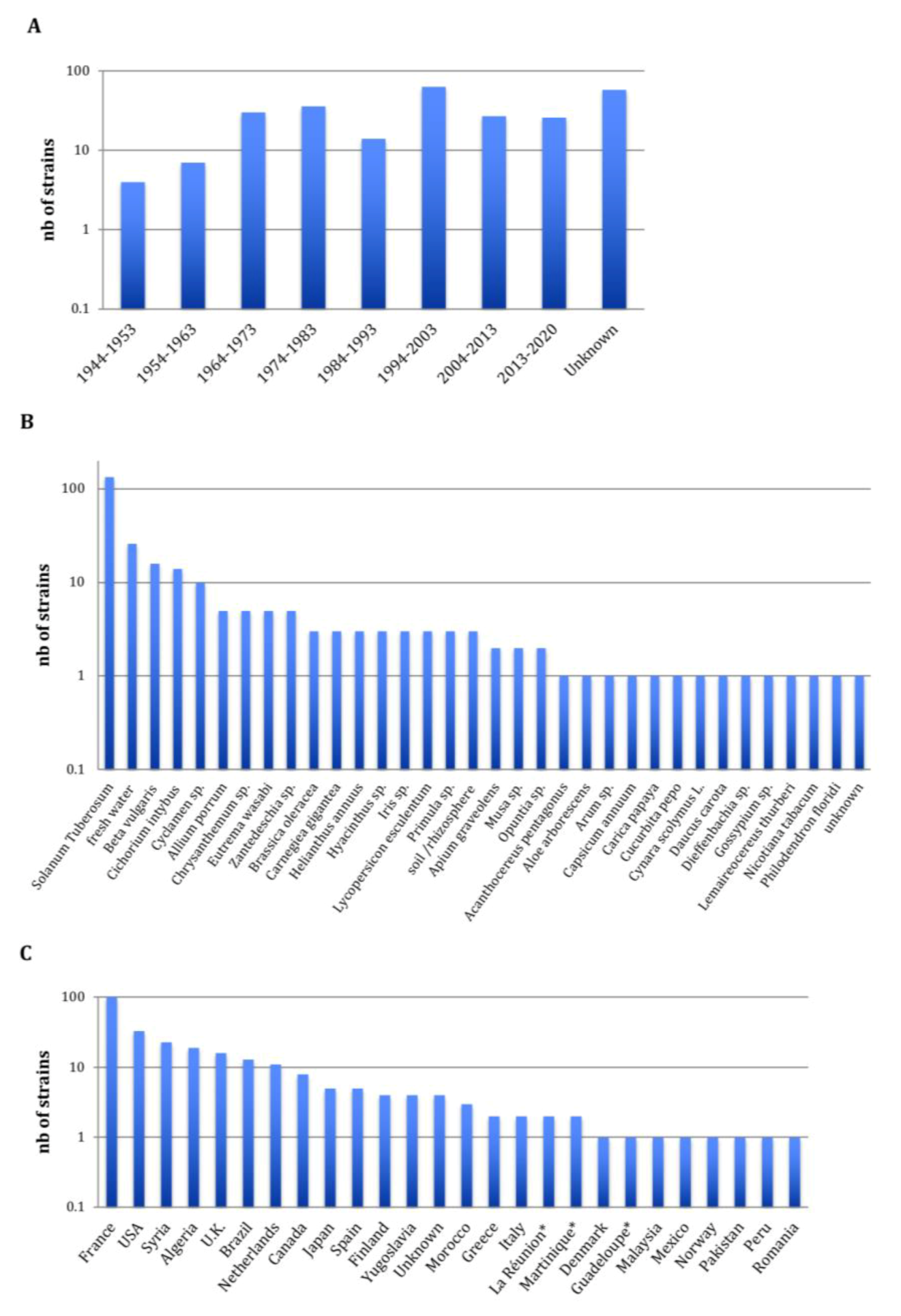
The 265 Pectobacterium CIRM-CFBP-studied strains are presented in Figure 1. They were isolated from 1944 to 2019 covering, therefore, a 75-year period. For 56 strains, the year of isolation was not reported; however, for these strains the year of the deposit at the CIRM-CFBP indicated that 22 strains were isolated at least 46 years ago (deposited from 1970 to 1974), 1 strain was isolated at least 38 years ago (deposited in 1982), 6 strains were isolated at least 28 years ago (deposited from 1991 to 1992) and 28 strains were isolated at least 19 years ago (deposited in 2001) (Figure 1A). Concerning the host plants or environments from which these 265 strains were isolated, a large majority of 136 strains were isolated from potato tubers or potato plants, accounting for the threat provoked by Pectobacterium spp. on this economically important crop plant (Figure 1B) [4]. In addition, the studied collection gathered 100 strains isolated from 31 host plants covering 17 botanical families, accounting for the broad host range of Pectobacterium spp. [3], as well as 29 strains isolated from freshwater, soil or rhizosphere. As the CIRM-CFBP collection is a French collection, it is not surprising that a large majority of 100 strains originated from this country (Figure 1C). Fours strains originating from overseas French territories (Martinique, Guadeloupe and La Réunion) were considered apart, as these territories are located respectively on the North American continent (Martinique and Guadeloupe) and African continent (La Réunion). Overall, the collection gathers strains originating from 27 countries in Europe, Africa, North America, South America, Asia and Indonesia. Finally, 94 strains were deposited at the CIRM-CFBP as Pectobacterium spp., without any indication of the species they belong. Furthermore, many strains were deposited under names that no longer exist or under the P. carotovorum name, which could be misleading, since it previously gathered several subspecies. In summary, although this collection gathers many strains sampled at different times over different countries and environments, its practical use is hampered by the poor taxonomic designation of the deposited strains.
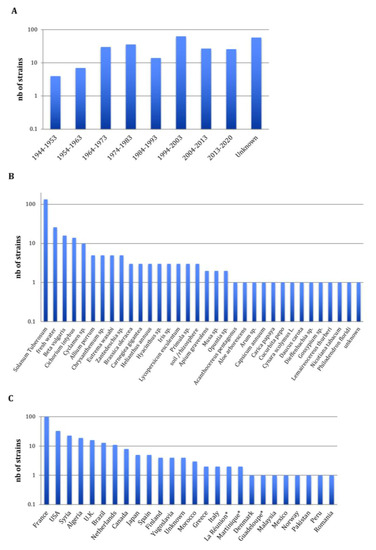
Figure 1.
Overview of the 265 CIRM-CFBP Pectobacterium strains at the beginning of our study. (A) Decade of isolation, (B) host or environment from which the strains were isolated and (C) country from which the strains were isolated; the stars indicate French overseas territories. Complete details for strains are available in Table S1.
3.2. dnaX-leuS-recA Phylogeny of the Collection
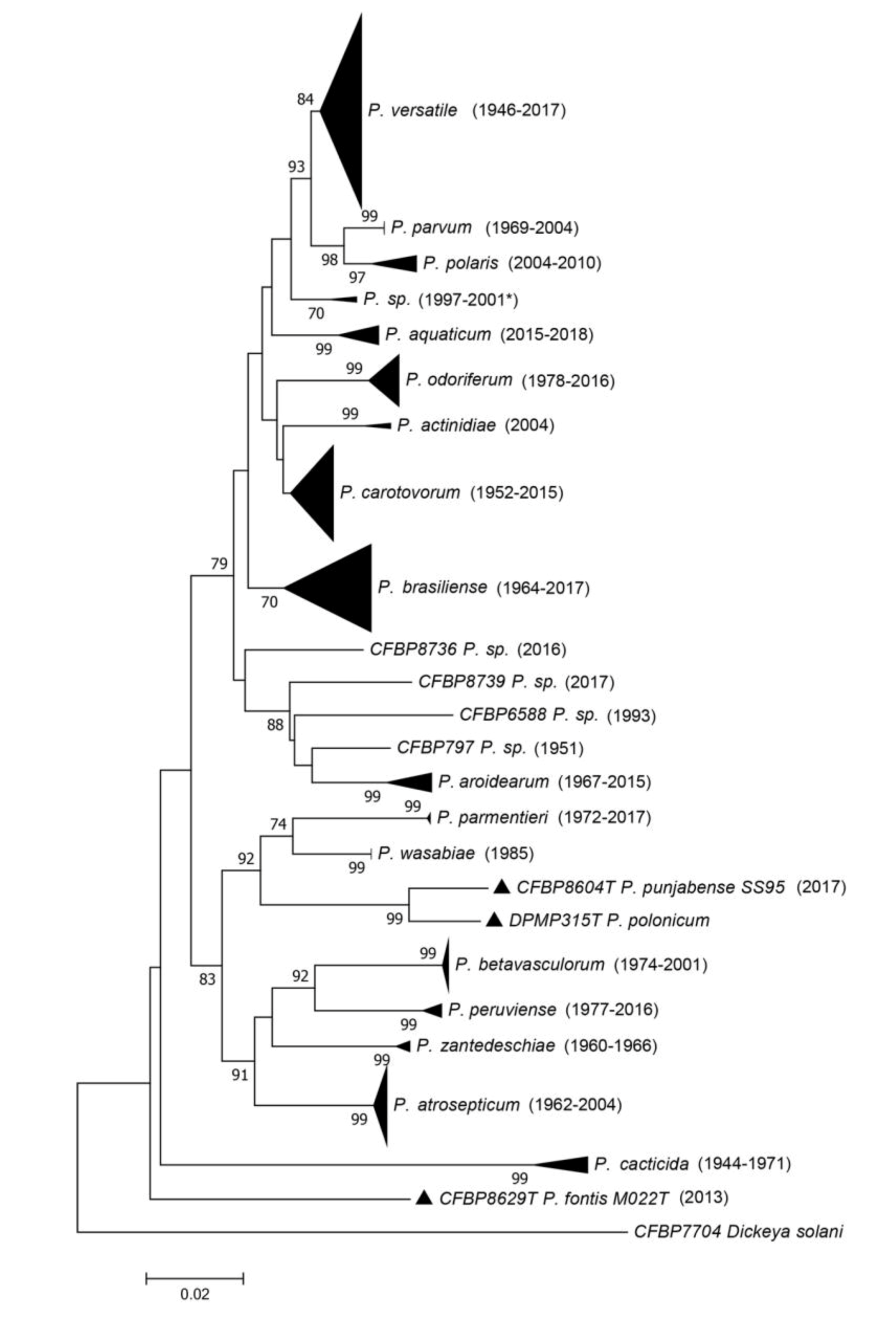
The dnaX-leuS-recA phylogeny (Figure 2 and Figure S1) revealed that most strains spread out in clades that are separated from each other’s and supported by usually high bootstrap values. As most of these clades included the type strain of a given species, this allowed assigning most of the strains of CIRM-CFBP to 18 of the 19 described species within Pectobacterium, except P. polonicum.
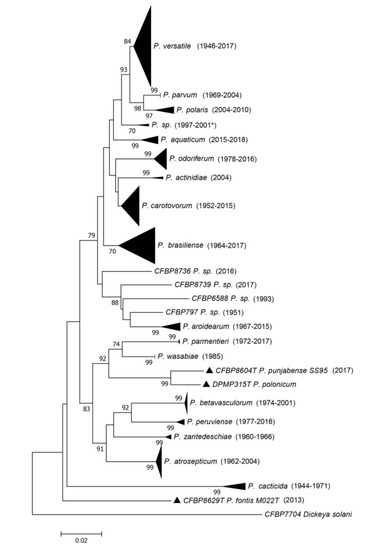
Figure 2.
Phylogenetic tree reconstructed from concatenated partial sequences from dnaX, leuS and recA housekeeping genes. The phylogenetic tree was reconstructed with concatenated alignments of all genes with MEGA 7.0.26 using the neighbour-joining method with 1000 bootstrap replicates, and the evolutionary distances were computed by using the Kimura two-parameter method. Bootstrap values are shown when over 70. Between parentheses are indicated the earliest and latest isolation years for each species. When there is only one strain in the clade, the year of isolation for that strain is indicated. For one clade, the latest year of isolation is unknown, and the latest year of deposit is indicated as follows: 2001*. Full view of this tree and the accession numbers of the sequences are available in Figure S1 and Table S1, respectively.
The relative position of the P. caticida species inside the Pectobacterium genus remains questionable, since no genome of this species has been sequenced, and, at the time of the description in 1991 by Alcorn et al. [19], most of the Pectobacterium species were not yet described. Interestingly, the leuS, recA and dnaX phylogeny performed here grouped all the P. cacticida strains in the same clade as a deep branching species within the Pectobacterium genus (Figure 2).
For seven strains (CFBP 797, CFBP 5380, CFBP 6067, CFBP 6168, CFBP 6588, CFBP 8736 and CFBP 8739), the assignation to an already described species was either not possible or their position on the phylogenetic tree was ambiguous (Figure 2). These seven strains may belong to yet undescribed species; however, the dnaX-leuS-recA phylogeny performed here is not adequate to delineate new species. Three of these strains grouped together in the same dnaX-leuS-recA clade and may represent a single species (two are displayed in the phylogenetic tree in Figure S1), while the four remaining strains each represent a different clade. We sequenced the genomes of two strains out of these seven strains. Furthermore, in order to check that large clades revealed by the dnaX-leuS-recA phylogeny indeed represented a single species, we sequenced 13 genomes to complete this analysis.
3.3. Whole-Genome Strains Analysis
A phylogenetic tree constructed from concatenated sequences of 1053 homologous genes retrieved from the 15 analysed genomes and the genomes of type strain or representative species is presented in Figure 3. Pairwise ANIb and dDDH were performed between the genomes of these 15 strains and the genomes of type strains or representative species. The results of ANIb and dDDH (Table S2) allowed to classify all the newly sequenced strains but two to already described species: four of the sequenced strains were classified as P. carotovorum, four as P. versatile, three as P. brasiliense, one as P. odoriferum and one as P. aroidearum.
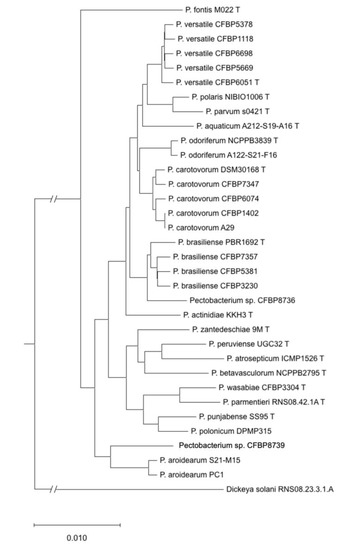
Figure 3.
Phylogenetic tree reconstructed from the concatenated sequences of 1053 homologous gene sequences retrieved from complete genome sequences for the 15 sequenced strains and type strains or reference strains of other species.
Strain CFBP 8736 displays pairwise ANI and DDH values below the species threshold with the P. brasiliense type strain (ANI: 94.7%; DDH 61.1%) (Table 2). However, the pairwise ANI/DDH values are higher (95.5% to 95.7% and 64.7% to 65.3%, respectively) between CFBP 8736 and the other P. brasiliense genomes (Table 2). The P. brasiliense species shows a relatively high level of divergence between its strains and is probably ongoing a diversification process [15]. Our results show that strain CFBP 8736 belongs to a separate species but is very close to P. brasiliense.

Table 2.
Pairwise average nucleotide identity (ANI) (below diagonal) and digital DNA-DNA hybridisation (dDDH) (above diagonals) for 6 of the analysed genomes.
The remaining genome, corresponding to strain CFBP 8739, could not be assigned to known or proposed species. Analysis of the pairwise ANI/dDDH values obtained for this latter genome with type strains or reference strains of other species indicated that its closest species was P. aroidearum, with a pairwise ANI value of 91.1% and dDDH value of 43.2%, well below the cut-off values of the species limit (Table 2). Strain CFBP 8739 therefore belongs to a still-uncharacterised species. Its position in the dnaX-leuS-recA phylogenetic tree was distinct but close to two other strains, CFBP 6588 and CFBP 797, which could not be assigned to any known species (Figure 2 and Figure S1). However, it remains ambiguous to decipher if CFBP 6588 and CFBP 797 could be grouped either with P. aroidearum or with this new species represented by strain CFBP 8739 or if each strain represents a new species. Interestingly, these three strains were isolated from various environments: a monocot (musa sp. for CFBP6588), a dicot (nicotiana tabacum for CFBP 797) and freshwater (for CFBP 8739).
3.4. Comparison of the Updated Taxonomy with the Former One
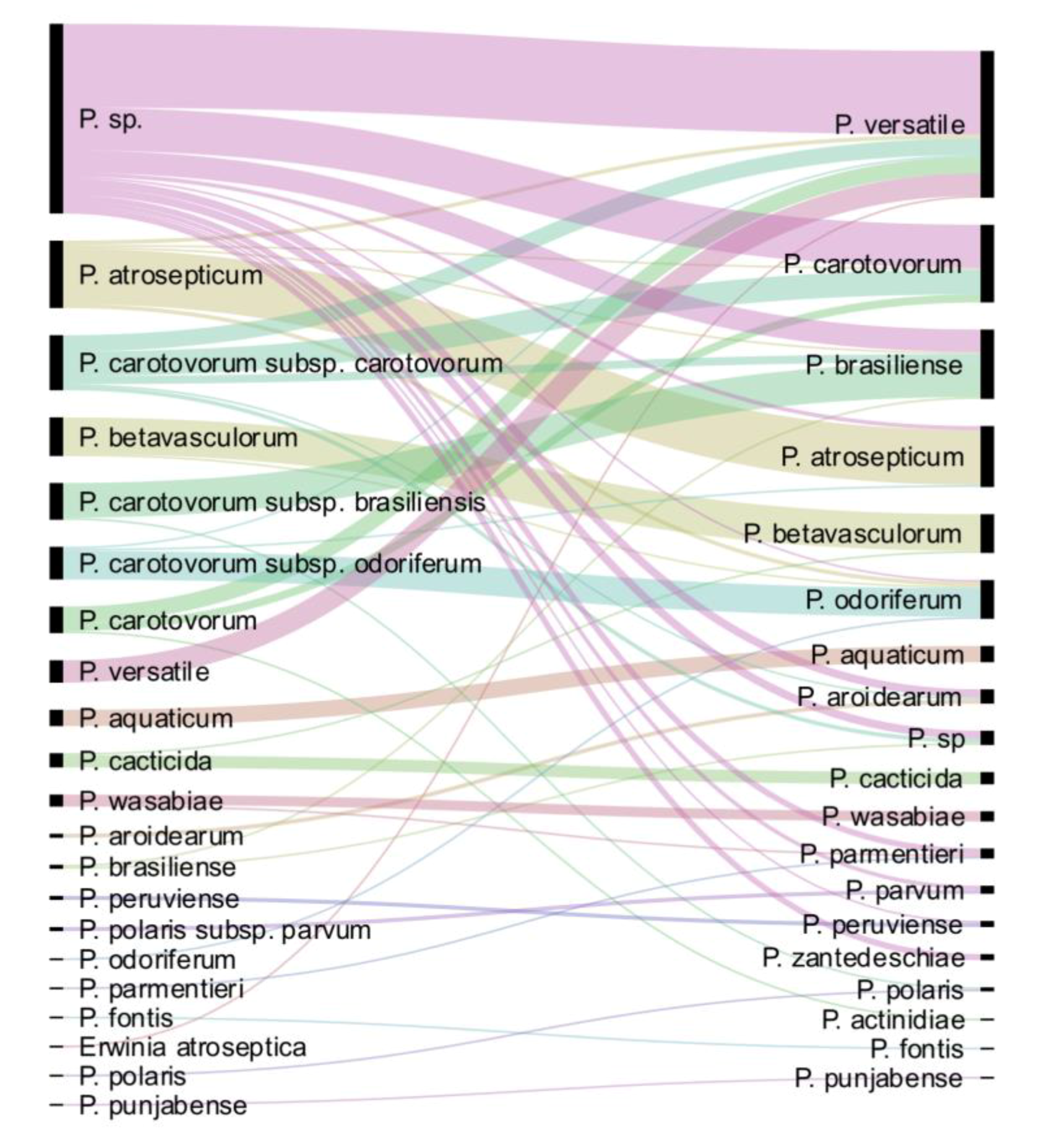
Following dnaX-leuS-recA phylogeny and genome analysis, out of the 265 strains analysed, 157 strains could be assigned to 18 known or proposed species. The only species not represented in the CIRM-CFBP collection is P. polonicum. We performed a comparison of the updated taxonomy with the former one (Figure 4). Among the 94 formerly unassigned Pectobacterium sp. strains, 90 were taxonomically assigned to 10 different species. The more frequently assigned species were P. versatile (41/94 strains), P. carotovorum (21/94 strains) and P. brasiliense (11/94). Interestingly, three strains of formerly unassigned Pectobacterium sp. were finally assigned to P. zantedeschiae, a species without known representative in the CIRM-CFBP collection before our work. Most of the 27 former P. carotovorum subsp. carotovorum strains were split between P. carotovorum (12/27) and P. versatile (8/27). As well, the 13 strains previously classified as P. carotovorum were mostly split between P. versatile (8/13) and P. carotovorum (4/13). This highlights the close proximity between P. versatile and P. carotovorum, as already noted [15]. Conversely, strains assigned to the former P. carotovorum subsp. brasiliense and P. carotovorum subsp. odoriferum were mostly assigned to their cognate species P. brasiliense (16/17) and P. odoriferum (14/16) (Figure 4). As well, strains assigned to the species P. atrosepticum (27/33), P. betavasculorum (18/19), P. aquaticum (8/8), P. cacticida (6/7) and P. wasabiae (5/6) remained mostly associated to the same species, indicating former good taxonomic resolution of these groups. Finally, while the former classification assigned only two strains to the P. aroidearum species, this species was enriched of five strains following our taxonomical update. Out of these five strains, four were previously designated as Pectobacterium sp. and one as P. carotovorum.
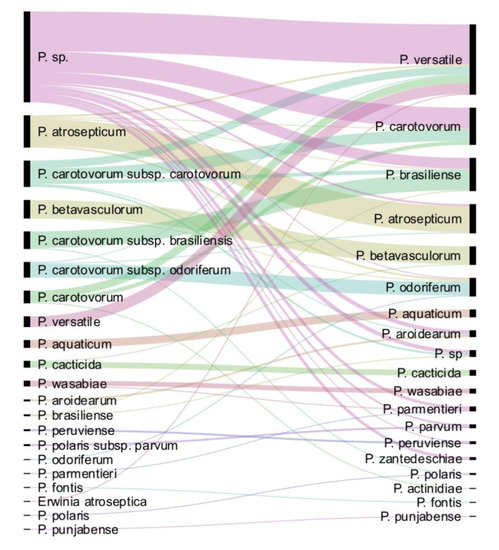
Figure 4.
Comparison of the updated taxonomy with the former one. On the left are displayed the taxonomic names under which the strains were deposited; on the right, the updated taxonomy for the 265 strains is displayed.
Interestingly, seven strains could not be assigned to any known or proposed species. Three strains (CFBP 5380, CFBP 6067 and CFBP 6068, the latter not displayed in the phylogenetic tree; Figure S1) could probably be gathered in the same new species, since they are closely clustered in the same clade following dnaX-leuS-recA analysis. It remains unclear how many different species could be described with the four remaining strains. Among these, CFBP 8736 and CFBP 8739, whose genomes were sequenced, represent potentially two new species close to P. brasiliense and P. aroidearum, respectively. These two strains were isolated from river water in France in 2016 and 2017.
3.5. Analysis of Strains Isolated on Potato Plants
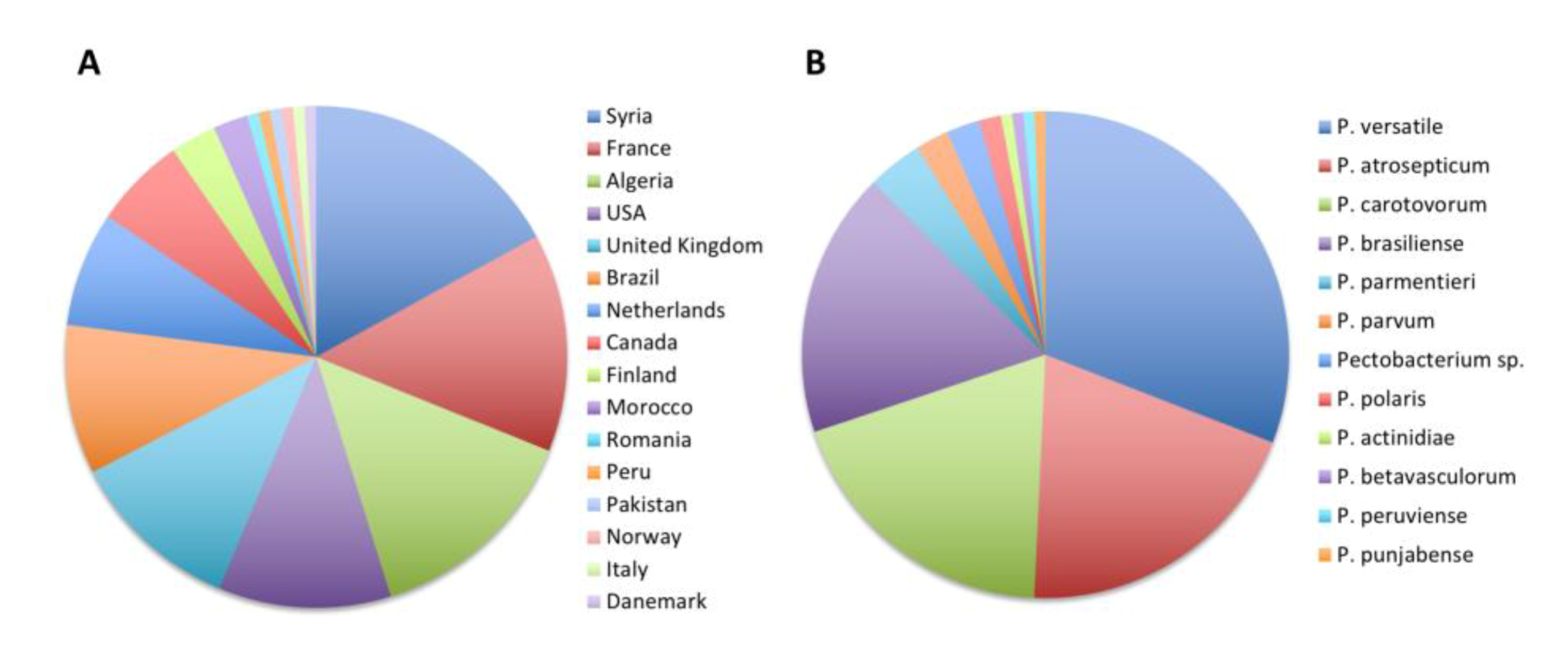
As already stated, a large majority of 136 strains were isolated from potato tubers or potato plants. These 136 strains were isolated in 16 countries covering four continents (Figure 5). We were therefore interested in understanding which species were isolated from this economically important crop. We found that strains isolated from potatoes belonged to 11 different already described Pectobacterium species (Figure 5). The most frequently deposited species was the recently described species P. versatile (42/136), followed by P. atrosepticum (27/136), P. carotovorum (26/136) and P. brasiliense (24/136). Less frequently, other recently described species known to infect potatoes, such as P. parmentieri, P. polaris, P. parvum, P. peruviense and P. punjabense, were also deposited to the collection (Figure 5 and Table 3). Surprisingly, our taxonomical update also identified one strain of P. actinidiae and one strain of P. betavasculorum that were isolated from potatoes in Syria in 2004 and Romania in 1992, respectively. Among the nonclassified strains, the three strains that grouped into a putative new species (CFBP 5380, CFBP 6067 and CFBP 6068) were isolated from potato plants and may represent a new pathogen species on this host plant.
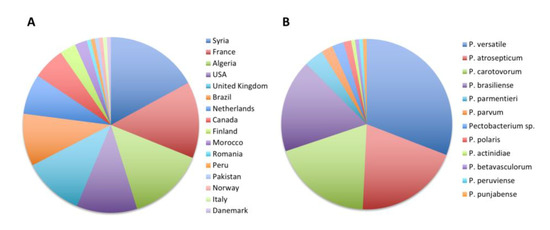
Figure 5.
Updated taxonomy of the 136 strains isolated from potatoes. (A) Country of isolation and (B) proportion of strain isolated in each Pectobacterium species.

Table 3.
Species isolated from potato plants and reported symptoms.
On potatoes, soft rot is the name usually used for tuber rotting, while the blackleg disease refers to the spread of the pathogen to the base of the potato stem, where it causes darkening and the decay of the aerial part [4]. Not all Pectobacterium spp. cause blackleg, but we could infer that a strain isolated from the stem or aerial part of the potato plant was isolated from blackleg disease symptoms. Out of the 136 strains isolated from potatoes, 31 were isolated from the stem or leaf and 33 from the tuber, and, unfortunately, it was not clearly documented for the remaining 71 strains (Table 3). The 31 strains isolated from the stem or leaf belonged to six species: five of which—P. brasiliense, P. atrosepticum, P. parvum, P. parmentieri and P. punjabense—are well-known species triggering blackleg disease [4,23,26]. Interestingly, the newly described P. versatile species was also isolated from potato stems or leaves in the UK, Morocco and France from 1972 to 2016. Whether P. versatile could be responsible for blackleg outbreak, or whether it is associated with blackleg symptoms as a secondary invader, remain to be determined.
3.6. Analysis of Species Host Range and Geographic Distribution
The update of taxonomical assignation prompted us to check the host range and geographic distribution of species deposited at the CIRM-CFBP collection (Table 4).

Table 4.
Host plant, environment and country of isolation for each of the Pectobacterium species deposited at the CIRM-CFBP.
The P. versatile species is by far the most represented species of the CIRM-CFBP collection. The 72 P. versatile strains were isolated from 12 host plants representing nine botanical families and from water. P. versatile strains were isolated from 11 countries on three continents. This highlights the broad ecological and geographical distributions of this species. The most ancient P. versatile strain deposited in CIRM-CFBP was isolated in 1946. This contrasts with the recent description of this species in 2019. The reason why this species was so long-neglected probably comes from its close genomic proximity with P. carotovorum [15], and the taxonomical update performed here confirms that these two species were often mixed up by bacteriologists (Figure 4). P. carotovorum is the second-most represented species deposited in CIRM-CFBP, and, as P. versatile, it also has a broad ecological and geographical distribution (Table 4), also explaining why these two species were often mixed up. Another species with broad ecological and geographical distribution is P. brasiliense (Table 4). This is in contrast with P. atrosepticum strains, which were isolated from only one botanical family, Solanaceae, with 27 strains isolated from potatoes, 2 from tomatoes and 1 from the soil environment, certainly indicating a narrower ecological niche. The host range of Pectobacterium spp. was reviewed by Ma et al. in 2007 and updated by Charkowski in 2018 [3,4]. Our taxonomical update of the CIRM-CFBP extends the number of plant hosts from which Pectobacterium strains were isolated (Table 4, new plant hosts in bold). For example, we found that P. brasiliense and P. versatile could be isolated from Chrysanthemum sp., an ornamental plant previously described as infected by Dickeya spp. and not by Pectobacterium spp. [3]. As well, P. brasiliense was isolated from Musa sp., a plant also previously reported to be infected only by Dickeya sp. [3].
The recently described “P. zantedeschiae” [27] and P. parvum species [23] had only a few representatives in CIRM-CFBP (Table 4). Nevertheless, some of the strains were isolated more than 50 years ago. Indeed, the first P. parvum strain was isolated in 1969, and the three “P. zandeteschiae” strains were respectively isolated in 1960, 1964 and 1966. This indicates that both species were already present on their respective host plants well before their description. Interestingly, although most of the P. parvum strains were isolated from potato plants, the P. parvum strain of 1969 was isolated from sunflowers enlarging the host range of this species. For “P. zantedeschiae”, the three strains hosted at CIRM-CFBP were all isolated from Araceae plants, as the other strains described for this species [27]. Concerning the recently described freshwater environmental species P. aquaticum [17], P. fontis [21] and P. polonicum [25] and the strain CFBP 8739, we were surprised that these species had either no representative in the CIRM-CFBP or only strains deposited following the recent description of the species. The fact that no strain representative of these species was isolated from crop plants and ornamentals along the 75-year period that covered our analysis strongly suggests that these species are not an important threat for crop plants and ornamentals. Whether these species evolved toward saprophytism and are no longer able to damage living plants or whether they infect living plants with no economic value remains to be determined.
4. Conclusions
Bacterial culture collections hold resources that are diverse and can help scientists to better understand, in the case of pathogens, the history of the epidemics, the emergence of diseases and the host range of pathogenic bacteria. However, even if the situation improved a lot during the last decade, the quality of associated data to deposited resources is often scarce for old resources and can hamper the benefit that could be gained from bacterial collections.
By this publication, we wanted to demonstrate how efforts made by the collections to cure their resources are beneficial to the whole scientist community and bring a better understanding of the dynamic of the taxa considered. We strongly encourage culture collections to initiate or continue their efforts to update the identity of their resources.
We also encourage scientists to deposit their resources in public culture collections, where they will be made available long-term for the benefit of the whole scientific community. The quality of the resources being dependent on the quality of the associated data, the depositors are also strongly encouraged to transmit the most accurate and complete data, even if these data do not appear to be important at the time of deposit. Indeed, while the plant species or environment from which strains have been isolated is generally indicated, the type of symptoms is often missing, and for environmental strains isolated from water, the water temperature is often not indicated. As well, the year of isolation is important data that is sometimes neglected.
In the case of Pectobacterium spp., our work permitted a better overview of the extent of the diversity in the collection, uncovering potential new species and giving insights into the epidemiology and ecology of this genus.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/9/1441/s1: Two supplementary tables (Table S1 and Table S2) and one supplementary figure (Figure S1) are available with the online version of this article.
Author Contributions
M.-A.B.: conceptualisation, funding acquisition, writing—original draft; P.P. and M.-A.B.: supervision and project administration; P.P. and J.P.: writing—review and editing and formal analysis; P.P., J.P., and M.-A.B.: validation; P.P., J.P., M.-A.B. and G.T.: visualisation; P.P., J.P. and G.T.: data curation and methodology; P.P., M.-A.B., G.T. and C.D.: investigation and C.D.: resources. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the ANR Project SPREE ANR-17-CE32-0004.
Acknowledgments
We acknowledge the help of Claire Bertrand for the extraction of genomic DNA prior to sequencing; Emma Caullireau for dnaX-leuS-recA PCRs; Audrey Lathus for strain conservation at CIRM-CFBP and Nicole Hugouvieux-Cotte-Pattat for providing gapA sequences for strains CFBP 8719, CFBP 8720, CFBP 8723 and CFBP 8724. This work benefited from the expertise of the High-Throughput Sequencing Platform of I2BC, Gif sur Yvette, France.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
References
- Hugouvieux-Cotte-Pattat, N.; Condemine, G.; Shevchik, V.E. Bacterial pectate lyases, structural and functional diversity: Bacterial pectate lyases. Environ. Microbiol. Rep. 2014, 6, 427–440. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, Y.; Liang, S.; Tian, Y.; Yin, S.; Xie, S.; Xie, H. Comparative genomics of 84 Pectobacterium genomes reveals the variations related to a pathogenic lifestyle. Bmc Genom. 2018, 19, 889. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Hibbing, M.E.; Kim, H.-S.; Reedy, R.M.; Yedidia, I.; Breuer, J.; Breuer, J.; Glasner, J.D.; Perna, N.T.; Kelman, A.; et al. Host range and molecular phylogenies of the soft rot enterobacterial genera pectobacterium and dickeya. Phytopathol. 2007, 97, 1150–1163. [Google Scholar] [CrossRef] [PubMed]
- Charkowski, A.O. The changing face of bacterial soft-rot diseases. Annu. Rev. Phytopathol. 2018, 56, 269–288. [Google Scholar] [CrossRef]
- Winslow, C.E.; Broadhurst, J.; Buchanan, R.E.; Krumwiede, C.; Rogers, L.A.; Smith, G.H. The families and genera of the bacteria: Preliminary report of the committee of the society of American bacteriologists on characterization and classification of bacterial types. J. Bacteriol. 1917, 2, 505–566. [Google Scholar] [CrossRef]
- Waldee, E.L. Comparative studies of some peritrichous phytopathogenic bacteria. Iowa State Coll. J. Sci. 1945, 19, 435–484. [Google Scholar]
- Brenner, D.J.; Fanning, G.R.; Steigerwalt, A.G. Deoxyribonucleic acid relatedness among Erwiniae and other Enterobacteriaceae: The gall, wilt and dry-necrosis organisms (Genus Erwinia WINSLOW et al. sensu stricto). Int. J. Syst. Bacteriol. 1973, 24, 197–204. [Google Scholar] [CrossRef]
- Skerman, V.B.D.; McGowan, V.; Sneath, P.H.A. Approved lists of bacterial names. Int. J. Syst. Bacteriol. 1980, 30, 225–420. [Google Scholar] [CrossRef]
- Hauben, L.; Moore, E.R.B.; Vauterin, L.; Steenackers, M.; Mergaert, J.; Verdonck, L.; Swings, J. Phylogenetic position of phytopathogens within the Enterobacteriaceae. Syst. Appl. Microbiol. 1998, 21, 384–397. [Google Scholar] [CrossRef]
- Samson, R.; Legendre, J.B.; Christen, R.; Fischer-Le Saux, M.; Achouak, W.; Gardan, L. Transfer of Pectobacterium chrysanthemi (Burkholder et al. 1953) Brenner et al. 1973 and Brenneria paradisiaca to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dickeya dadantii sp. nov., Dickeya dianthicola sp. nov., Dickeya dieffenbachiae sp. nov. and Dickeya zeae sp. nov. Int. J. Syst. Evol. Microbiol. 2005, 55, 1415–1427. [Google Scholar]
- Thomson, S.V.; Hildebrand, D.C.; Schroth, M.N. Identification and nutritional differentiation of the Erwinia sugarbeet pathogen from members of Erwinia carotovora and Erwinia Chrysanthemi. Phytopathology. 1981, 71, 1037–1042. [Google Scholar] [CrossRef]
- Nabhan, S.; De Boer, S.H.; Maiss, E.; Wydra, K. Taxonomic relatedness between Pectobacterium carotovorum subsp. carotovorum, Pectobacterium carotovorum subsp. odoriferum and Pectobacterium carotovorum subsp. brasiliense subsp. nov. J. Appl. Microbiol. 2012, 113, 904–913. [Google Scholar] [CrossRef] [PubMed]
- Koh, Y.; Kim, G.; Lee, Y.; Sohn, S.; Koh, H.; Kwon, S.; Heu, S.; Jung, J. Pectobacterium carotovorum subsp. actinidiae subsp. nov., a new bacterial pathogen causing canker-like symptoms in yellow kiwifruit, Actinidia chinensis. N. Z. J. Crop. Hortic. Sci. 2012, 40, 269–279. [Google Scholar] [CrossRef][Green Version]
- Gardan, L.; Cécile, G.; Christen, R.; Samson, R. Elevation of three subspecies of Pectobacterium carotovorum to species level: Pectobacterium atrosepticum sp. nov., Pectobacterium betavasculorum sp. nov. and Pectobacterium wasabiae sp. nov. Int. J. Syst. Evol. Microbiol. 2003, 53, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Portier, P.; Pédron, J.; Taghouti, G.; Fischer-Le Saux, M.; Caullireau, E.; Bertrand, C.; Laurent, A.; Chawki, K.; Oulgazi, S.; Moumni, M.; et al. Elevation of Pectobacterium carotovorum subsp. odoriferum to species level as Pectobacterium odoriferum sp. nov., proposal of Pectobacterium brasiliense sp. nov. and Pectobacterium actinidiae sp. nov., emended description of Pectobacterium carotovorum and description of Pectobacterium versatile sp. nov., isolated from streams and symptoms on diverse plants. Int. J. Syst. Evol. Microbiol. 2019, 69, 3207–3216. [Google Scholar] [CrossRef]
- Zhang, Y.; Fan, Q.; Loria, R. A re-evaluation of the taxonomy of phytopathogenic genera Dickeya and Pectobacterium using whole-genome sequencing data. Syst. Appl. Microbiol. 2016, 39, 252–259. [Google Scholar] [CrossRef]
- Pédron, J.; Bertrand, C.; Taghouti, G.; Portier, P.; Barny, M.-A. Pectobacterium aquaticum sp. nov., isolated from waterways. Int. J. Syst. Evol. Microbiol. 2019, 69, 745–751. [Google Scholar] [CrossRef]
- Nabhan, S.; De Boer, S.H.; Maiss, E.; Wydra, K. Pectobacterium aroidearum sp. nov., a soft rot pathogen with preference for monocotyledonous plants. Int. J. Syst. Evol. Microbiol. 2013, 63, 2520–2525. [Google Scholar] [CrossRef]
- Alcorn, S.M.; Orum, T.V.; Steigerwalt, A.G.; Foster, J.L.; Fogelman, J.C.; Brenner, D.J. Taxonomy and pathogenicity of Erwinia cacticida sp. nov. Int. J. Syst. Bacteriol. 1991, 41, 197–212. [Google Scholar] [CrossRef]
- Gallois, A.; Samson, R.; Ageron, E.; Grimont, P. Erwinia carotovora subsp. odorifera subsp. nov., associated with odorous soft rot of chicory (Cichorium intybus L.). Int. J. Syst. Bacteriol. 1992, 42, 582–588. [Google Scholar] [CrossRef]
- Oulghazi, S.; Cigna, J.; Lau, Y.Y.; Moumni, M.; Chan, K.G.; Faure, D. Transfer of the waterfall source isolate Pectobacterium carotovorum M022 to Pectobacterium fontis sp. nov., a deep-branching species within the genus Pectobacterium. Int. J. Syst. Evol. Microbiol. 2019, 69, 470–475. [Google Scholar] [CrossRef]
- Khayi, S.; Cigna, J.; Chong, T.; Quêtu-Laurent, A.; Chan, K.; Helias, V.; Faure, D. Transfer of the potato plant isolates of Pectobacterium wasabiae to Pectobacterium parmentieri sp. nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 5379–5383. [Google Scholar] [CrossRef] [PubMed]
- Pasanen, M.; Waleron, M.; Schott, T.; Cleenwerck, I.; Misztak, A.; Waleron, K.; Pritchard, L.; Bakr, R.; Degefu, Y.; van der Wolf, J.; et al. Pectobacterium parvum sp. nov., having a salmonella SPI-1-like Type III secretion system and low virulence. Int. J. Syst. Evol. Microbiol. 2020, 70, 2440–2448. [Google Scholar] [CrossRef] [PubMed]
- Dees, M.W.; Lysøe, E.; Rossmann, S.; Perminow, J.; Brurberg, M.B. Pectobacterium polaris sp. nov., isolated from potato (Solanum tuberosum). Int. J. Syst. Evol. Microbiol. 2017, 67, 5222–5229. [Google Scholar] [CrossRef] [PubMed]
- Waleron, M.; Misztak, A.; Waleron, M.; Jonca, J.; Furmaniak, M.; Waleron, K. Pectobacterium polonicum sp. nov. isolated from vegetable fields. Int. J. Syst. Evol. Microbiol. 2019, 69, 1751–1759. [Google Scholar] [CrossRef]
- Sarfraz, S.; Riaz, K.; Oulghazi, S.; Cigna, J.; Sahi, S.T.; Khan, S.H.; Faure, D. Pectobacterium punjabense sp. nov., isolated from blackleg symptoms of potato plants in Pakistan. Int. J. Syst. Evol. Microbiol. 2018, 68, 3551–3556. [Google Scholar] [CrossRef]
- Waleron, M.; Misztak, A.; Waleron, M.; Franczuk, M.; Wielgomas, B.; Waleron, K. Transfer of Pectobacterium carotovorum subsp. carotovorum strains isolated from potatoes grown at high altitudes to Pectobacterium peruviense sp. nov. Syst. Appl. Microbiol. 2018, 41, 85–93. [Google Scholar] [CrossRef]
- Waleron, M.; Misztak, A.; Waleron, M.; Franczuk, M.; Jońca, J.; Wielgomas, B.; Mikiciński, A.; Popović, T.; Waleron, K. Pectobacterium zantedeschiae sp. nov. a new species of a soft rot pathogen isolated from Calla lily (Zantedeschia spp.). Syst. Appl. Microbiol. 2019, 42, 275–283. [Google Scholar] [CrossRef]
- Cigna, J.; Dewaegeneire, P.; Beury, A.; Gobert, V.; Faure, D. A gapA PCR-sequencing assay for identifying the Dickeya and Pectobacterium potato pathogens. Plant. Dis. 2017, 101, 1278–1282. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Delcher, A.L.; Harmon, D.; Kasif, S.; White, O.; Salzberg, S.L. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999, 27, 4636–4641. [Google Scholar] [CrossRef]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).