Diversity and Activity of Sulfate-Reducing Prokaryotes in Kamchatka Hot Springs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Analytical Techniques
2.2. Radiotracing Experiments
2.3. DNA Extraction, 16S rRNA and dsrB Genes Amplification, Sequencing and Analyses
2.4. Bioinformatics Processing and Data Analyses
2.5. Cultivation and Identification of SRP
3. Results
3.1. Characteristics of the Hot Springs Studied
3.2. Rates of Sulfate Reduction
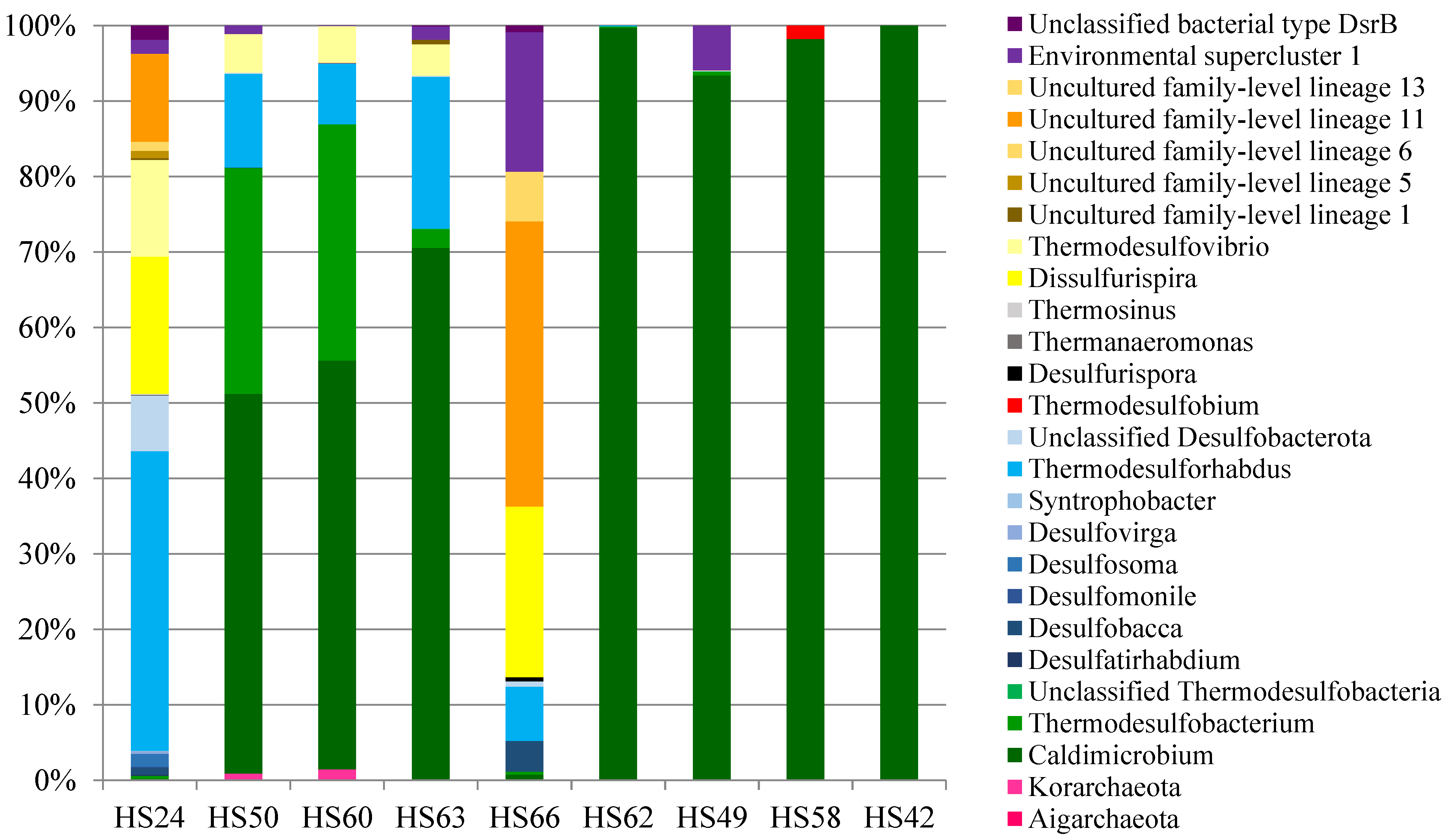
3.3. Diversity of dsrB-Carrying Prokaryotes in Kamchatka
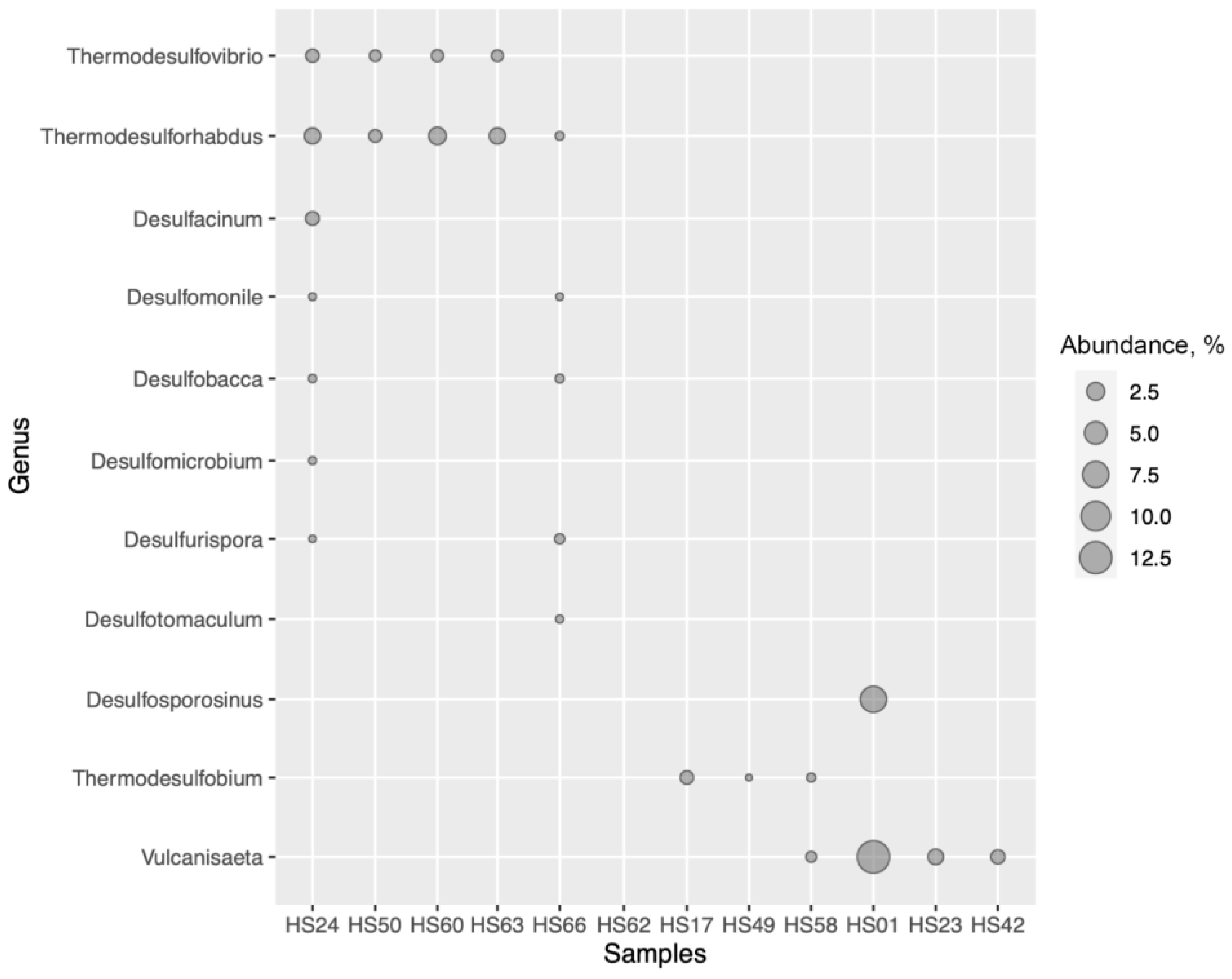
3.4. Sulfate-Reducing Prokaryotes Detected by 16S rRNA Gene-Based Analyses
3.5. Cultivation of Thermophilic Sulfate-Reducing Prokaryotes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muyzer, G.; Stams, A.J.M. The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 2008, 6, 441–454. [Google Scholar] [CrossRef]
- Rabus, R.; Venceslau, S.S.; Wöhlbrand, L.; Voordouw, G.; Wall, J.D.; Pereira, I.A. A post-genomic view of the ecophysiology, catabolism and biotechnological relevance of sulphate-reducing prokaryotes. Adv. Microb. Physiol. 2015, 66, 55–321. [Google Scholar] [CrossRef]
- Waite, D.W.; Chuvochina, M.; Pelikan, C.; Parks, D.H.; Yilmaz, P.; Wagner, M.; Loy, A.; Naganuma, T.; Nakai, R.; Whitman, W.B.; et al. Proposal to reclassify the proteobacterial classes Deltaproteobacteria and Oligoflexia, and the phylum Thermodesulfobacteria into four phyla reflecting major functional capabilities. Int. J. Syst. Evol. Microbiol. 2020, 70, 5972–6016. [Google Scholar] [CrossRef] [PubMed]
- Thiel, V.; Garcia Costas, A.M.; Fortney, N.W.; Martinez, J.N.; Tank, M.; Roden, E.E.; Boyd, E.S.; Ward, D.M.; Hanada, S.; Bryant, D.A. “Candidatus Thermonerobacter thiotrophicus”, a non-phototrophic member of the Bacteroidetes/Chlorobi with dissimilatory sulfur metabolism in hot spring mat communities. Front. Microbiol. 2019, 9, 3159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chernyh, N.A.; Neukirchen, S.; Frolov, E.N.; Sousa, F.L.; Miroshnichenko, M.L.; Merkel, A.Y.; Pimenov, N.V.; Sorokin, D.Y.; Ciordia, S.; Mena, M.C.; et al. Dissimilatory sulfate reduction in the archaeon “Candidatus Vulcanisaeta moutnovskia” sheds light on the evolution of sulfur metabolism. Nat. Microbiol. 2020, 5, 1428–1438. [Google Scholar] [CrossRef] [PubMed]
- Zeikus, J.G.; Dawson, N.A.; Thompson, T.E.; Ingvorsen, K.; Hatchikian, E.C. Microbial ecology of volcanic sulphidogenesis: Isolation and characterization of Thermodesulfobacterium commune gen. nov. and sp. nov. J. Gen. Microbiol. 1982, 129, 1159–1169. [Google Scholar] [CrossRef] [Green Version]
- Jeanthon, C.; L’Haridon, S.; Cueff, V.; Banta, A.; Reysenbach, A.-L.; Prieur, D. Thermodesulfobacterium hydrogeniphilum sp. nov., a thermophilic, chemolithoautotrophic, sulfate-reducing bacterium isolated from a deep-sea hydrothermal vent at Guaymas Basin, and emendation of the genus Thermodesulfobacterium. Int. J. Syst. Evol. Microbiol. 2002, 52, 765–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sekiguchi, Y.; Muramatsu, M.; Imachi, H.; Narihiro, T.; Ohashi, A.; Harada, H.; Hanada, S.; Kamagata, Y. Thermodesulfovibrio aggregans sp. nov. and Thermodesulfovibrio thiophilus sp. nov., anaerobic, thermophilic, sulfate-reducing bacteria isolated from thermophilic and methanogenic sludge, and emended description of the genus Thermodesulfovibrio. Int. J. Syst. Evol. Microbiol. 2008, 58, 2541–2548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beeder, J.; Torsvik, T.; Lien, T. Thermodesulforhabdus norvegicus gen. nov., sp. nov., a novel thermophilic sulfate-reducing bacterium from oil field water. Arch. Microbiol. 1995, 164, 331–336. [Google Scholar] [CrossRef]
- Kaksonen, A.H.; Spring, S.; Schumann, P.; Kroppenstedt, R.M.; Puhakka, J.A. Desulfurispora thermophila gen. nov., sp. nov., a thermophilic, spore-forming sulfate-reducer isolated from a sulfidogenic fluidized-bed reactor. Int. J. Syst. Evol. Microbiol. 2007, 57, 1089–1094. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Kojima, H.; Fukui, M. Desulfotomaculum intricatum sp. nov., a sulfate reducer isolated from freshwater lake sediment. Int. J. Syst. Evol. Microbiol. 2013, 63, 3574–3578. [Google Scholar] [CrossRef]
- Bonch-Osmolovskaya, E.A. Studies of thermophilic microorganisms at the Institute of Microbiology, Russian Academy of Sciences. Microbiology 2004, 73, 644–658. [Google Scholar] [CrossRef]
- Mardanov, A.V.; Gumerov, V.M.; Beletsky, A.V.; Perevalova, A.A.; Karpov, G.A.; Bonch-Osmolovskaya, E.A.; Ravin, N.A.; Skryabin, K.G. Uncultured archaea dominate in the thermal groundwater of Uzon Caldera, Kamchatka. Extremophiles 2011, 15, 365–372. [Google Scholar] [CrossRef]
- Merkel, A.Y.; Pimenov, N.V.; Rusanov, I.I.; Slobodkin, A.I.; Slobodkina, G.B.; Tarnovetckii, I.Y.; Frolov, E.N.; Dubin, A.V.; Perevalova, A.A.; Bonch-Osmolovskaya, E.A. Microbial diversity and autotrophic activity in Kamchatka hot spring. Extremophiles 2017, 21, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Miroshnichenko, M.L.; Tourova, T.P.; Kolganova, T.P.; Kostrikina, N.A.; Bonch-Osmolovskaya, E.A. Ammonifex thiophilus sp. nov., a hyperthermophilic anaerobic bacterium from a Kamchatka hot spring. Int. J. Syst. Evol. Microbiol. 2008, 58, 2935–2938. [Google Scholar] [CrossRef] [Green Version]
- Frolov, E.N.; Kublanov, I.V.; Toshchakov, S.; Bonch-Osmolovskaya, E.A.; Novikov, A.A.; Chernyh, N.A. Thermodesulfobium acidiphilum sp. nov., a new thermoacidophilic, sulfate-reducing, chemoautotrophic bacterium from a thermal site. Int. J. Syst. Evol. Microbiol. 2017, 67, 1482–1485. [Google Scholar] [CrossRef] [PubMed]
- Frolov, E.N.; Zayulina, K.S.; Kopitsyn, D.S.; Kublanov, I.V.; Bonch-Osmolovskaya, E.A.; Chernyh, N.A. Desulfothermobacter acidiphilus gen. nov., sp. nov., a thermoacidophilic sulfate-reducing bacterium isolated from a terrestrial hot spring. Int. J. Syst. Evol. Microbiol. 2018, 68, 871–875. [Google Scholar] [CrossRef] [PubMed]
- Prokofeva, M.I.; Kublanov, I.V.; Nercessian, O.; Tourova, T.P.; Kolganova, T.V.; Lebedinsky, A.V.; Bonch-Osmolovskaya, E.A.; Spring, S. Cultivated anaerobic acidophilic/acidotolerant thermophiles from terrestrial and deep-sea hydrothermal habitats. Extremophiles 2005, 9, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Gumerov, V.M.; Mardanov, A.V.; Beletsky, A.V.; Prokofeva, M.I.; Bonch-Osmolovskaya, E.A.; Ravin, N.V.; Skryabin, K.G. Complete genome sequence of “Vulcanisaeta moutnovskia” strain 768-28, a novel member of the hyperthermophilic crenarchaeal genus Vulcanisaeta. J. Bacterol. 2011, 193, 2355–2356. [Google Scholar] [CrossRef] [Green Version]
- Bonch-Osmolovskaya, E.A.; Gorlenko, V.M.; Karpov, G.A.; Starynin, D.A. Anaerobic degradation of organic matter of microbial mats in the Termofil’nyi spring (Uzon caldera, Kamchatka). Microbiology 1987, 56, 1022–1028. [Google Scholar]
- Chernyh, N.A.; Mardanov, A.V.; Gumerov, V.M.; Miroshnichenko, M.L.; Lebedinsky, A.V.; Merkel, A.Y.; Crowe, D.; Pimenov, N.V.; Rusanov, I.I.; Ravin, N.V.; et al. Microbial life in Bourlyashchy, the hottest thermal pool of Uzon Caldera, Kamchatka. Extremophiles 2015, 19, 1157–1171. [Google Scholar] [CrossRef]
- Frolov, E.N.; Merkel, A.Y.; Pimenov, N.V.; Kvashchevskaya, A.A.; Bonch-Osmolovskaya, E.A.; Chernyh, N.A. Sulfate reduction and inorganic carbon assimilation in acidic thermal springs of the Kamchatka Peninsula. Microbiology 2016, 85, 471–480. [Google Scholar] [CrossRef]
- Gumerov, V.M.; Mardanov, A.V.; Beletsky, A.V.; Bonch-Osmolovskaya, E.A.; Ravin, N.V. Molecular analysis of microbial diversity in Zavarzin Spring, the Uzon Caldera, Kamchatka. Microbiology 2011, 80, 244–251. [Google Scholar] [CrossRef]
- Mardanov, A.V.; Gumerov, V.M.; Beletsky, A.V.; Bonch-Osmolovskaya, E.A.; Ravin, N.V.; Skryabin, K.G. Characteristic of biodiversity of thermophilic microbial community by parallel pyrosequencing method. Dokl. Biochem. Biophys. 2010, 432, 110–113. [Google Scholar] [CrossRef]
- Burgess, E.A.; Unrine, J.M.; Mills, G.L.; Romanek, C.S.; Wiegel, J. Comparative geochemical and microbiological characteristization of two thermal pools in Uzon Caldera, Kamchatka, Russia. Microb. Ecol. 2012, 63, 471–489. [Google Scholar] [CrossRef]
- Wemheuer, B.; Taube, R.; Akyol, P.; Wemheuer, F.; Daniel, R. Microbial diversity and biochemical potential encoded by thermal spring metagenomes derived from the Kamchatka Peninsula. Archaea 2013, 2013, 136714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozanov, A.S.; Bryanskaya, A.V.; Malup, T.K.; Lazareva, E.V.; Taran, O.P.; Ivanisenko, T.V.; Zhmodik, S.M.; Kolchanov, N.A.; Peltek, S.E. Molecular analysis of the benthos microbial community in Zavarzin thermal spring (Uzon Caldera, Kamchatka, Russia). BMC Genom. 2014, 15, S12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobretsov, N.I.; Lazareva, E.V.; Zhmodik, S.M.; Bryanskaya, A.V.; Morozova, V.V.; Tikunova, N.V.; Peltek, S.E.; Karpov, G.A.; Taran, O.P.; Ogorodmikova, O.L.; et al. Geological, hydrochemical and microbiological characteristics of the Oil site of the Uzon Caldera (Kamchatka). Russ. Geol. Geophys. 2015, 56, 39–63. [Google Scholar] [CrossRef]
- Menzel, P.; Gudbergsdottir, S.R.; Rike, A.G.; Lin, L.; Zhang, Q.; Contursi, P.; Moracci, M.; Krostjansson, J.K.; Bolduc, B.; Gavrilov, S.; et al. Comparative metagenomic of eight geographically remote terrestrial hot springs. Microb. Ecol. 2015, 70, 411–424. [Google Scholar] [CrossRef]
- Wagner, M.; Loy, A.; Klein, M.; Lee, N.; Ramsing, N.B.; Stahl, D.A.; Friedrich, M.W. Functional marker genes for identification of sulfate-reducing prokaryotes. Methods Enzymol. 2005, 397, 469–489. [Google Scholar] [CrossRef]
- Lücker, S.; Steger, D.; Kjeldsen, K.U.; MacGregor, B.J.; Wagner, M.; Loy, A. Improved 16S rRNA-targeted probe set for analysis of sulfate-reducing bacteria by fluorescence in situ hybridization. J. Microbiol. Methods 2007, 69, 523–528. [Google Scholar] [CrossRef]
- Müller, A.L.; Kjeldsen, K.U.; Rattei, T.; Pester, M.; Loy, A. Phylogenetic and environmental diversity of DsrAB-type dissimilatory (bi)sulfite reductases. ISME J. 2015, 9, 1152–1165. [Google Scholar] [CrossRef] [Green Version]
- Pelikan, C.; Herbold, C.W.; Hausmann, B.; Müller, A.L.; Pester, M.; Loy, A. Diversity analysis of sulfite- and sulfate-reducing microorganisms by multiplex dsrA and dsrB amplicon sequencing using new primers and mock community-optimized bioinformatics. Environ. Microbiol. 2016, 18, 2994–3009. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.; Friedrich, M.; Roger, A.J.; Hugenholtz, P.; Fishbain, S.; Abicht, H.; Blackall, L.L.; Stahl, D.A.; Wagner, M. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 2001, 183, 6028–6035. [Google Scholar] [CrossRef] [Green Version]
- Zverlov, V.; Klein, M.; Lücker, S.; Friedrich, M.W.; Kellermann, J.; Stahl, D.A.; Loy, A.; Wagner, M. Lateral gene transfer of dissimilatory (bi)sulfite reductase revisited. J. Bacteriol. 2005, 187, 2203–2208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loy, A.; Duller, S.; Baranyi, C.; Mussmann, M.; Ott, J.; Sharon, I.; Béjà, O.; Le Paslier, D.; Dahl, C.; Wagner, M. Reverse dissimilatory sulfite reductase as phylogenetic marker for a subgroup of sulfur-oxidizing prokaryotes. Environ. Microbiol. 2009, 11, 289–299. [Google Scholar] [CrossRef] [Green Version]
- Moreau, J.W.; Zierenberg, R.A.; Banfield, J.F. Diversity of dissimilatory sulfite reductase genes (dsrAB) in a salt marsh impacted by long-term acid mine drainage. Appl. Environ. Microbiol. 2010, 76, 4819–4828. [Google Scholar] [CrossRef] [Green Version]
- Mori, Y.; Purdy, K.J.; Oakley, B.B.; Kondo, R. Comprehensive detection of phototrophic sulfur bacteria using PCR primers that target reverse dissimilatory sulfite reductase gene. Microbes Environ. 2010, 25, 190–196. [Google Scholar] [CrossRef] [Green Version]
- Pester, M.; Bittner, N.; Deevong, P.; Wagner, M.; Loy, A. A “rare biosphere” microorganism contributes to sulfate reduction in a peatland. ISME J. 2010, 4, 1591–1602. [Google Scholar] [CrossRef] [PubMed]
- Lenk, S.; Arnds, J.; Zerjatke, K.; Musat, N.; Amann, R.; Mussmann, M. Novel groups of Gamma-proteobacteria catalyse sulfur oxidation and carbonbfixation in a coastal, intertidal sediment. Environ. Microbiol. 2011, 13, 758–774. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Soto, M.F.; Cerqueda-Garcia, D.; Alcantara-Hernandez, R.J.; Falcon, L.I.; Pech, D.; Arcega-Cabrera, F.; Aguirre-Macedo, M.L.; Garcia-Maldonado, J.Q. Assessing the diversity of benthic sulfate-reducing microorganisms in northwestern Gulf of Mexico by Illumina Sequencing of dsrB gene. Microb. Ecol. 2021, 81, 908–921. [Google Scholar] [CrossRef]
- Trüper, H.G.; Schlegel, H.G. Sulfur metabolism in Thiorodaceae. I. Quantitative measurements in growing cells of Chromatium okenii. Antonie Leeuwennhoek 1964, 30, 225–238. [Google Scholar] [CrossRef]
- Pimenov, N.V.; Bonch-Osmolovskaya, E.A. In Situ activity studies in thermal environments. Methods Microbiol. 2006, 35, 29–53. [Google Scholar]
- Lever, M.A.; Torti, A.; Eickenbusch, P.; Michaud, A.B.; Šantl-Temkiv, T.; Jørgensen, B.B. A modular method for the extraction of DNA and RNA, and the separation of DNA pools from diverse environmental sample types. Front. Microbiol. 2015, 6, 476. [Google Scholar] [CrossRef] [Green Version]
- Fadrosh, D.W.; Ma, B.; Gajer, P.; Sengamalay, N.; Ott, S.; Brotman, R.M.; Ravel, J. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2014, 2, 6. [Google Scholar] [CrossRef] [Green Version]
- Frey, B.; Rime, T.; Phillips, M.; Stierli, B.; Hajdas, I.; Widmer, F.; Hartmann, M. Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol. Ecol. 2016, 92, fiw018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merkel, A.Y.; Tarnovetskii, I.Y.; Podosokorskaya, O.A.; Toshchakov, S.V. Analysis of 16S rRNA primer systems for profiling of thermophilic microbial communities. Microbiology 2019, 88, 671–680. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Renaud, G.; Stenzel, U.; Maricic, T.; Wiebe, V.; Kelso, J. deML: Robust demultiplexing of Illumina sequences using a likelihood-based approach. Bioinformatics 2015, 31, 770–772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ansorge, R.; Birolo, G.; James, S.A.; Telatin, A. Dadaist2: A Toolkit to Automate and Simplify Statistical Analysis and Plotting of Metabarcoding Experiments. Int. J. Mol. Sci. 2021, 22, 5309. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [Green Version]
- Valero-Mora, P.M. ggplot2: Elegant Graphics for Data Analysis. J. Stat. Softw. 2010, 35, 1–3. [Google Scholar] [CrossRef] [Green Version]
- Kevbrin, V.; Zavarzin, G. Effect of sulfur compounds on the growth of the halophilic homoacetogenic bacterium Acetohalobium arabaticum. Microbiology 1992, 61, 563–567. [Google Scholar]
- Wolin, E.A.; Wolin, M.J.; Wolfe, R.S. Formation of methane by bacterial extracts. J. Biol. Chem. 1963, 238, 2882–2888. [Google Scholar] [CrossRef]
- Perevalova, A.A.; Kublanov, I.V.; Baslerov, R.V.; Zhang, G.; Bonch-Osmolovskaya, E.A. Brockia lithotrophica gen. nov., sp. nov., an anaerobic thermophilic bacterium from a terrestrial hot spring. Int. J. Syst. Evol. Microbiol. 2013, 63, 479–483. [Google Scholar] [CrossRef]
- Benson, D.A.; Boguski, M.S.; Lipman, D.J.; Ostell, J.; Ouellette, B.F.; Rapp, B.A.; Wheeler, D.L. GenBank. Nucleic Acids Res. 1999, 27, 12–17. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Fishbain, S.; Dillon, J.G.; Gough, H.L.; Stahl, D.A. Linkage of high rates of sulfate reduction in Yellowstone hot springs to unique sequence types in the dissimilatory sulfate respiration pathway. Appl. Environ. Microbiol. 2003, 69, 3663–3667. [Google Scholar] [CrossRef] [Green Version]
- Ferris, M.J.; Magnuson, T.S.; Fagg, J.A.; Thar, R.; Kühl, M.; Sheehan, K.B.; Henson, J.M. Microbially mediated sulphide production in a thermal, acidic algal mat community in Yellowstone National Park. Environ. Microbiol. 2003, 5, 954–9604. [Google Scholar] [CrossRef] [Green Version]
- Roychoudhury, A.N. Sulfate respiration in extreme enviroments: A kinetic study. Geomicrobiol. J. 2004, 21, 33–43. [Google Scholar] [CrossRef]
- Dillon, J.G.; Fishbain, S.; Miller, S.R.; Bebout, B.M.; Habicht, K.S.; Stahl, D.A. High rates of sulfate reduction in a low-sulfate hot spring microbial mat are driven by a low level of diversity of sulfate-respiring microorganisms. Appl. Environ. Microbiol. 2007, 73, 5218–5226. [Google Scholar] [CrossRef] [Green Version]
- Pimenov, N.V. Radioisotopic studies of microbial activity in the hot springs of the Uzon caldera (Kamchatka). Proc. Winogradsky Inst. Microbiol. 2011, 16, 144–159. [Google Scholar]
- Hugenholtz, P.; Pitulle, C.; Hershberger, K.L.; Pace, N.R. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 1998, 180, 366–376. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, T.; Hanada, S.; Maruyama, A.; Marumo, K.; Urabe, T.; Fukui, M. Distribution and diversity of thermophilic sulfate-reducing bacteria within a Cu-Pb-Zn mine (Toyoha, Japan). FEMS Microbiol. Ecol. 2002, 41, 199–209. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sogabe, M.; Okumura, K.; Kurane, R. A highly selective direct method of detecting sulphate-reducing bacteria in crude oil. Lett. Appl. Microbiol. 2002, 35, 242–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhillon, A.; Teske, A.; Dillon, J.; Stahl, D.A.; Sogin, M.L. Molecular characterization of sulfate-reducing bacteria in the Guaymas Basin. Appl. Environ. Microbiol. 2003, 69, 2765–2772. [Google Scholar] [CrossRef] [Green Version]
- Guan, J.; Zhang, B.L.; Mbadinga, S.M.; Liu, J.F.; Gu, J.D.; Mu, B.Z. Functional genes (dsr) approach reveals similar sulphidogenic prokaryotes diversity but different structure in saline waters from corroding high temperature petroleum reservoirs. Appl. Microbiol. Biotechnol. 2014, 98, 1871–1882. [Google Scholar] [CrossRef] [PubMed]
- Henry, E.A.; Devereux, R.; Maki, J.S.; Gilmour, C.C.; Woese, C.R.; Mandelco, L.; Schauder, R.; Remsen, C.C.; Mitchell, R. Characterization of a new thermophilic sulfate-reducing bacterium Thermodesulfovibrio yellowstonii, gen. nov. and sp. nov.: Its phylogenetic relationship to Thermodesulfobacterium commune and their origins deep within the bacterial domain. Arch. Microbiol. 1994, 161, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Sonne-Hansen, J.; Ahring, B.K. Thermodesulfobacterium hveragerdense sp. nov., and Thermodesulfovibrio islandicus sp. nov., two thermophilic sulfate reducing bacteria isolated from a Icelandic hot spring. Syst. Appl. Microbiol. 1999, 22, 559–564. [Google Scholar] [CrossRef]
- Haouari, O.; Fardeau, M.L.; Cayol, J.L.; Fauque, G.; Casiot, C.; ElbazPoulichet, F.; Hamdi, M.; Ollivier, B. Thermodesulfovibrio hydrogeniphilus sp. nov., a new thermophilic sulphate-reducing bacterium isolated from a Tunisian hot spring. Syst. Appl. Microbiol. 2008, 31, 38–42. [Google Scholar] [CrossRef] [PubMed]
- Beana, S.; Perdomo, N.; Carvajal, C.; Diaz, C.; Patel, B.K. Desulfosoma caldarium gen. nov., sp. nov., a thermophilic sulfate-reducing bacterium from a terrestrial hot spring. Int. J. Syst. Evol. Microbiol. 2011, 61, 732–736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gregoire, P.; Ferdeau, M.L.; Guasco, S.; Lagiere, J.; Cambar, J.; Michotey, V.; Bonin, P.; Olivier, B. Desulfosoma profundi sp. nov., a thermophilic sulfate-reducing bacterium isolated from a deep terrestrial geothermal spring in France. Antonie Leeuwenhoek 2012, 101, 595–602. [Google Scholar] [CrossRef] [PubMed]
- Rozanova, E.P.; Khudiakova, A.I. A new non-spore forming thermophilic organism, reducing sulfates, Desulfovibrio thermophilus nov. sp. Microbiology 1974, 43, 1069–1075. [Google Scholar] [PubMed]
- Hamilton-Brehm, S.D.; Gibson, R.A.; Green, S.J.; Hopmans, E.C.; Schouten, S.; van der Meer, M.T.; Shields, J.P.; Damsté, J.S.; Elkins, J.G. Thermodesulfobacterium geofontis sp. nov., a hyperthermophilic, sulfate-reducing bacterium isolated from Obsidian Pool, Yellowstone National Park. Extremophiles 2013, 17, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Rees, G.N.; Grassia, G.S.; Sheehy, A.J.; Dwivedi, P.P.; Patel, B.K.C. Desulfacinum infernum gen. nov., sp. nov., a thermophilic sulfate-reducing bacterium from a petroleum reservoir. Int. J. Syst. Bacteriol. 1995, 45, 85–89. [Google Scholar] [CrossRef] [Green Version]
- Sievert, S.M.; Kuever, J. Desulfacinum hydrothermale sp. nov., a thermophilic, sulfate-reducing bacterium from geothermally heated sediments near Milos Island (Greece). Int. J. Syst. Evol. Microbiol. 2000, 50, 1239–1246. [Google Scholar] [CrossRef]
- Rozanova, E.P.; Turova, T.P.; Kolganova, T.V.; Lysenko, A.M.; Mitiushina, L.L.; Iusupov, S.K.; Beliaev, S.S. Desulfacinum subterraneum sp. nov. a new thermophilic sulfate-reducing bacterium isolated from a high temperature oil field. Microbiology 2001, 70, 536–542. [Google Scholar]
- Rubiano-Labrador, C.; Díaz-Cárdenas, C.; López, G.; Gómez, J.; Baena, S. Colombian Andean thermal springs: Reservoir of thermophilic anaerobic bacteria producing hydrolytic enzymes. Extremophiles 2019, 23, 793–808. [Google Scholar] [CrossRef]
- Zavarzina, D.G.; Kochetkova, T.V.; Chistyakova, N.I.; Gracheva, M.A.; Antonova, A.V.; Merkel, A.Y.; Perevalova, A.A.; Chernov, M.S.; Koksharov, Y.A.; Bonch-Osmolovskaya, E.A.; et al. Siderite-based anaerobic iron cycle driven by autotrophic thermophilic microbial consortium. Sci. Rep. 2020, 10, 21661. [Google Scholar] [CrossRef]
- Baker, B.J.; Moser, D.P.; MacGregor, B.J.; Fishbain, S.; Wagner, M.; Fry, N.K.; Jackson, B.; Speolstra, N.; Loos, S.; Takai, K.; et al. Related assemblages of sulphate-reducing bacteria associated with ultradeep gold mines of South Africa and deep basalt aquifers of Washington State. Environ. Microbiol. 2003, 5, 267–277. [Google Scholar] [CrossRef]
- Trimarco, E.; Balkwill, D.; Davidson, M.; Onstott, T.C. In Situ enrichment of a diverse community of bacteria from a 4–5 km deep fault zone in South Africa. Geomicrobiol. J. 2006, 23, 463–473. [Google Scholar] [CrossRef]
- Kaksonen, A.H.; Spring, S.; Schumann, P.; Kroppenstedt, R.M.; Puhakka, J.A. Desulfotomaculum thermosubterraneum sp. nov., a thermophilic sulfate-reducer isolated from an underground mine located in a geothermally active area. Int. J. Syst. Evol. Microbiol. 2006, 56, 2603–2608. [Google Scholar] [CrossRef] [Green Version]
- Giese, R.; Henninges, J.; Lüth, S.; Morozova, D.; Schmidt-Hattenberger, C.; Würdemann, H.; Zimmer, M.; Cosma, C.; Juhlin, C. Monitoring at the CO2 SINK site: A concept integrating geophysics, geochemistry and microbiology. Energy Procedia 2009, 1, 2251–2259. [Google Scholar] [CrossRef] [Green Version]
- Aüllo, T.; Berlendis, S.; Lascourrèges, J.F.; Dessort, D.; Duclerc, D.; Saint-Laurent, S.; Schraauwers, B.; Mas, J.; Patriarche, D.; Boesinger, C.; et al. New bio-indicators for long term natural attenuation of monoaromatic compounds in deep terrestrial Aquifers. Front. Microbiol. 2016, 7, 122. [Google Scholar] [CrossRef] [PubMed]
- Nazina, T.N.; Rozanova, E.P. Thermophilic sulfate-reducing bacteria from oil strata. Microbiology 1978, 47, 113–118. [Google Scholar]
- Cha, I.T.; Roh, S.W.; Kim, S.J.; Hong, H.J.; Lee, H.W.; Lim, W.T.; Rhee, S.K. Desulfotomaculum tongense sp. nov., a moderately thermophilic sulfate-reducing bacterium isolated from a hydrothermal vent sediment collected from the Tofua Arc in the Tonga Trench. Antonie Leeuwenhoek 2013, 104, 1185–1192. [Google Scholar] [CrossRef] [PubMed]
- Aüllo, T.; Ranchou-Peyruse, A.; Ollivier, B.; Magot, M. Desulfotomaculum spp. and related gram-positive sulfate-reducing bacteria in deep subsurface environments. Front. Microbiol. 2013, 4, 362. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.W.; Lu, Z. Phylogenomic evaluation of members above the species level within the phylum Firmicutes based on conserved proteins. Environ. Microbiol. Rep. 2015, 7, 273–281. [Google Scholar] [CrossRef]
- Kunisawa, T. Evolutionary relationship of completely sequenced Clostridia species and close relatives. Int. J. Syst. Evol. Microbiol. 2015, 65, 4276–4283. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.A.; Hugenholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 10362–10366. [Google Scholar] [CrossRef] [PubMed]
- Frolov, E.N.; Kublanov, I.V.; Toshchakov, S.V.; Lunev, E.A.; Pimenov, N.V.; Bonch-Osmolovskaya, E.A.; Lebedinsky, A.V.; Chernyh, N.A. Form III RubisCO-mediated transaldolase variant of the Calvin cycle in a chemolithoautotrophic bacterium. Proc. Natl. Acad. Sci. USA 2019, 116, 18638–18646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, K.; Kim, H.; Kakegawa, T.; Hanada, S. A novel lineage of sulfate-reducing microorganisms: Thermodesulfobiaceae fam. nov., Thermodesulfobium narugense, gen. nov., sp. nov., a new thermophilic isolate from a hot spring. Extremophiles 2003, 7, 283–290. [Google Scholar] [CrossRef] [PubMed]
- Itoh, T.; Suzuki, K.; Sanchez, P.C.; Nakase, T. Caldivirga maquilingensis gen. nov., sp. nov., a new genus of rod-shaped crenarchaeote isolated from a hot spring in the Philippines. Int. J. Syst. Bacteriol. 1999, 49, 1157–1163. [Google Scholar] [CrossRef] [Green Version]
- Siebers, B.; Zaparty, M.; Raddatz, G.; Tjaden, B.; Albers, S.V.; Bell, S.D.; Blombach, F.; Kletzin, A.; Kyrpides, N.; Lanz, C.; et al. The complete genome sequence of Thermoproteus tenax: A physiologically versatile member of the Crenarchaeota. PLoS ONE 2011, 6, e24222. [Google Scholar] [CrossRef] [Green Version]
- June Yim, K.; Seon Song, H.; Choi, J.S.; Woon Roh, S. Thermoproteus thermophilus sp. nov., a hyperthermophilic crenarchaeon isolated from solfataric soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 2507–2510. [Google Scholar] [CrossRef] [Green Version]
- Umezawa, K.; Kojima, H.; Kato, Y.; Fukui, M. Disproportionation of inorganic sulfur compounds by a novel autotrophic bacterium belonging to Nitrospirota. Syst. Appl. Microbiol. 2020, 43, 126110. [Google Scholar] [CrossRef]
- Miroshnichenko, M.L.; Lebedinsky, A.V.; Chernyh, N.A.; Tourova, T.P.; Kolganova, T.V.; Spring, S.; Bonch-Osmolovskaya, E.A. Caldimicrobium rimae gen. nov., sp. nov., an extremely thermophilic, facultatively lithoautotrophic, anaerobic bacterium from the Uzon Caldera, Kamchatka. Int. J. Syst. Evol. Microbiol. 2009, 59, 1040–1044. [Google Scholar] [CrossRef] [Green Version]
- Kojima, H.; Umezawa, K.; Fukui, M. Caldimicrobium thiodismutans sp. nov., a sulfur-disproportionating bacterium isolated from a hot spring, and emended description of the genus Caldimicrobium. Int. J. Syst. Evol. Microbiol. 2016, 66, 1828–1831. [Google Scholar] [CrossRef]
- Umezawa, K.; Kojima, H.; Kato, Y.; Fukui, M. Dissulfurispira thermophila gen. nov., sp. nov., a thermophilic chemolithoautotroph growing by sulfur disproportionation, and proposal of novel taxa in the phylum Nitrospirota to reclassify the genus Thermodesulfovibrio. Syst. Appl. Microbiol. 2021, 44, 126184. [Google Scholar] [CrossRef] [PubMed]
- McKay, L.J.; Dlakić, M.; Fields, M.W.; Delmont, T.O.; Eren, A.M.; Jay, Z.J.; Klingelsmith, K.B.; Rusch, D.B.; Inskeep, W.P. Co-occurring genomic capacity for anaerobic methane and dissimilatory sulfur metabolisms discovered in the Korarchaeota. Nat. Microbiol. 2019, 4, 614–622. [Google Scholar] [CrossRef]
| Sample Name | Spring 1, Coordinates | Sample Description | T (°C) | pH | SO42−, mM | Approach Applied 2 |
|---|---|---|---|---|---|---|
| Foot of Mutnovskii volcano: | ||||||
| HS01 | Unnamed; 52°31.809′ N 158°11.472′ E, 823 m | Yellow to gray deposit | 90 | 3.5 | 3.2 | HTS, RT |
| HS08 | Unnamed; 52°32.087′ N 158°11.851′ E, 794 m | Black deposit | 60 | 5.6 | 1.1 | RT |
| Uzon Caldera: | ||||||
| HS12 | Unnamed, OTF; 54°30.413′ N 160°00.043′ E, 659 m | Gray deposit | 82 | 2.5 | 9.9 | RT |
| HS17 | Unnamed, CTF; 54°30.008′ N 160°00.322′ E, 657 m | Yellow/orange deposit | 53 | 5.0 | 1.2 | HTS |
| HS23 | Unnamed, WTF; 54°30.009′ N 159°56.983′ E, 707 m | Gray deposit | 72 | 5.0 | 2.7 | HTS, RT |
| HS24 | Solnechny, CTF; 54°29.941′ N 159°59.530′ E, 657 m | Black deposit | 52 | 6.1 | 0.6 | HTS, I, RT |
| HS27 | Oil Site, CTF; 54°30.023′ N 160°00.088′ E, 654 m | Black deposit | 61 | 4.2 | 2.2 | RT, I |
| HS42 | Sery, ETF; 54°29.882′ N 160°00.862′ E, 662 m | Short gray filaments around the margins | 80 | 6.1 | 1.9 | HTS |
| HS49 | Unnamed, ETF; 54°29.892′ N 160°00.870′ E, 664 m | Short white filaments around the margins | 63 | 6.3 | 0.5 | HTS |
| HS50 | Unnamed, ETF; 54°29.892′ N 160°00.870′ E, 664 m | Short gray filaments around the margins | 72 | 6.6 | 0.2 | HTS |
| HS58 | Arkashin Shurf, CTF; 54°30.000′ N 160°00.337′ E, 660 m | Yellow/orange deposit | 64 | 5.0 | 0.8 | HTS, I |
| HS60 | Unnamed, CTF; 54°30.013′ N 160°00.441′ E, 657 m | White filaments | 68 | 6.1 | 1.7 | HTS |
| HS62 | Unnamed, ETF; 54°29.993′ N 160°00.810′ E, 659 m | Gray deposit | 72 | 5.1 | 1.5 | HTS, I |
| HS63 | Vertoletny, ETF; 54°30.005′ N 160°00.732′ E, 664 m | Gray deposit | 55 | 5.6 | 0.6 | HTS |
| Valley of Geysers: | ||||||
| HS66 | Unnamed; 54°26.299′ N 160°08.375′ E, 451 m | Black deposit | 62 | 5.3 | 1.2 | HTS, I |
| Substrate | Sulfate Reduction Rates (nmol/cm3d) 1 | |||||
|---|---|---|---|---|---|---|
| HS24 | HS08 | HS27 | HS01 | HS23 | HS12 | |
| In situ | 2.4 ± 0.5 | 3.1 ± 1.2 | 13.0 ± 3.6 | 8.7 ± 3.0 | ND | ND |
| Hydrogen | 24.2 ± 7.6 | 34.2 ± 4.4 | 136.6 ± 2.2 | ND | ND | ND |
| Acetate | 33.5 ± 1.5 | 5.9 ± 0.1 | 7.0 ± 3.4 | ND | ND | ND |
| Lactate | 20.5 ± 0.6 | 18.2 ± 1.2 | 12.9 ± 4.8 | ND | 6.2 ± 3.1 | ND |
| Ethanol | 41.7 ± 3.8 | 83.4 ± 8.1 | 19.9 ± 0.1 | ND | ND | ND |
| Methanol | 8.7 ± 0.9 | 2.4 ± 0.4 | ND 2 | ND | ND | ND |
| Yeast extract | 45.6 ± 8.2 | 25.3 ± 4.8 | 98.3 ± 8.4 | ND | ND | ND |
| Sample | Isolate | Growth, T °C | Growth, pH | Electron Donor | Carbon Source | Closest Relative | 16S rRNA Gene Identity, % |
|---|---|---|---|---|---|---|---|
| HS62 | 3462-1 | 65 | 5.5 | H2 | Acetate | Thermodesulfovibrio aggregans | 97.6 |
| HS27 | 3427-1 | 55 | 4.7 | H2 | CO2 | Thermodesulfobium acidiphilum | 99.9 |
| HS58 | 3458-1 | 55 | 5.0 | H2 | CO2 | Thermodesulfobium acidiphilum | 99.9 |
| HS24 | Microbial association | 50 | 6.0 | Lactate | Lactate | Thermodesulforhabdus (74%) 1 Thermoanaerobacter (23%) 1 | 96.0 2 100.0 2 |
| HS66 | Microbial association | 60 | 5.0 | Ethanol | Ethanol | Desulfotomaculum (97%) 1 Thermoanaerobacterium (2%) 1 | 98.0 2 99.0 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frolov, E.N.; Gololobova, A.V.; Klyukina, A.A.; Bonch-Osmolovskaya, E.A.; Pimenov, N.V.; Chernyh, N.A.; Merkel, A.Y. Diversity and Activity of Sulfate-Reducing Prokaryotes in Kamchatka Hot Springs. Microorganisms 2021, 9, 2072. https://doi.org/10.3390/microorganisms9102072
Frolov EN, Gololobova AV, Klyukina AA, Bonch-Osmolovskaya EA, Pimenov NV, Chernyh NA, Merkel AY. Diversity and Activity of Sulfate-Reducing Prokaryotes in Kamchatka Hot Springs. Microorganisms. 2021; 9(10):2072. https://doi.org/10.3390/microorganisms9102072
Chicago/Turabian StyleFrolov, Evgenii N., Alexandra V. Gololobova, Alexandra A. Klyukina, Elizaveta A. Bonch-Osmolovskaya, Nikolay V. Pimenov, Nikolay A. Chernyh, and Alexander Y. Merkel. 2021. "Diversity and Activity of Sulfate-Reducing Prokaryotes in Kamchatka Hot Springs" Microorganisms 9, no. 10: 2072. https://doi.org/10.3390/microorganisms9102072
APA StyleFrolov, E. N., Gololobova, A. V., Klyukina, A. A., Bonch-Osmolovskaya, E. A., Pimenov, N. V., Chernyh, N. A., & Merkel, A. Y. (2021). Diversity and Activity of Sulfate-Reducing Prokaryotes in Kamchatka Hot Springs. Microorganisms, 9(10), 2072. https://doi.org/10.3390/microorganisms9102072






