Abstract
The formation of biofilms results from a multicellular mode of growth, in which bacteria remain enwrapped by an extracellular matrix of their own production. Many different bacteria form biofilms, but among the most studied species are those that belong to the Pseudomonas genus due to the metabolic versatility, ubiquity, and ecological significance of members of this group of microorganisms. Within the Pseudomonas genus, biofilm studies have mainly focused on the opportunistic human pathogen Pseudomonas aeruginosa due to its clinical importance. The extracellular matrix of P. aeruginosa is mainly composed of exopolysaccharides, which have been shown to be important for the biofilm architecture and pathogenic features of this bacterium. Notably, some of the exopolysaccharides recurrently used by P. aeruginosa during biofilm formation, such as the alginate and polysaccharide synthesis loci (Psl) polysaccharides, are also used by pathogenic and beneficial plant-associated Pseudomonas during their interaction with plants. Interestingly, their functions are multifaceted and seem to be highly dependent on the bacterial lifestyle and genetic context of production. This paper reviews the functions and significance of the exopolysaccharides produced by plant-associated Pseudomonas, particularly the alginate, Psl, and cellulose polysaccharides, focusing on their equivalents produced in P. aeruginosa within the context of pathogenic and beneficial interactions.
1. Introduction
Biofilms are matrix-enclosed bacterial populations that are adherent to each other and to surfaces and/or interfaces and are mainly composed of polysaccharides, proteins, lipids, and extracellular DNA [1,2,3]. During biofilm formation, the cells transit from a motile to a sessile lifestyle by interacting with a surface and starting to produce an extracellular matrix that holds them together and attaches them to the surface [2]. Therefore, the cells forming biofilms are referred to as sessile cells, which differ from their non-encased free-swimming counterparts, the planktonic cells [4]. Recent studies indicate that biofilms represent the main mechanism of active bacterial life due to their dominance in all habitats throughout the world [5,6]. Compared to the planktonic lifestyle, the biofilm lifestyle confers several benefits to the integrating cells, such as protection against antimicrobial agents and predators, tolerance towards changing environmental conditions, and colonization aptitudes [3,7,8].
Bacteria form biofilms in artificial and natural environments, including the soil, internal and external tissues of all living organisms, rocks, and water, among others [5]. Many different bacteria form biofilms, but the Pseudomonas genus is among the most studied for several reasons: (1) it harbors species with the ability to colonize a wide variety of environments due to the high metabolic and physiologic versatility found in this group of microorganisms, (2) it has ecological relevance due to its interactions with living organisms, and (3) it has potential biotechnological applications [9]. The Pseudomonas aeruginosa species, a ubiquitous bacterium that can also act as an opportunistic human pathogen, has long been used as a model bacterium within the Pseudomonas genus for the study of biofilm formation and pathogenesis due to its relevance in the clinical environment [10]. The extracellular matrix of P. aeruginosa has been studied in-depth and, to date, is known to contain three exopolysaccharides: alginate, polysaccharide synthesis loci (Psl), and pellicle loci (Pel) [11]. The role of these exopolysaccharides in the biofilm architecture of P. aeruginosa and the impact of their production in the clinical setting, such as protection against antibiotic treatments and host defenses, have been explored in several studies [12,13,14,15,16,17,18,19,20]. Although P. aeruginosa produces infections in humans, there are also some examples in which this bacterium can act as a pathogen for plants [21,22]. However, the biological significance of alginate, Psl, and Pel exopolysaccharides in a nonclinical context has not been studied.
Bacteria belonging to the Pseudomonas genus are common inhabitants of plant surfaces [23,24]. The role played by Pseudomonas in the agricultural industry is remarkable as several economically important activities are derived from their interaction with plants. Among these activities, there are harmful diseases that involve severe economic losses and beneficial activities such as plant growth stimulation, the promotion of plant health and nutrient availability in soils, and induction of plant immune defenses [25,26]. Pathogenic plant-associated Pseudomonas are predominantly present on the phyllosphere. The phyllosphere is an extreme and unstable habitat as it is exposed to highly variable nutrient and water availability, temperatures, and ultraviolet (UV) radiation. Therefore, the microbial populations associated with the phyllosphere must be adapted to these continuously fluctuating conditions [23,27,28]. The extracellular matrix of epiphytic bacteria contributes to the fitness [29,30,31], protection [8,32], and hydration of the cells [33], allowing cells to cope with these ever-changing conditions. Conversely, beneficial plant-associated Pseudomonas usually prevail in the rhizosphere. Compared to the phyllosphere, the environmental fluctuations that take place on the rhizosphere are weak and buffered [34]. Nevertheless, the rhizosphere is not considered a uniform and stable environment as the conditions can change abruptly in extremely short distance ranges [35,36]. Biofilm formation by beneficial plant-associated Pseudomonas plays advantageous roles for both the plant and bacteria [27,37]. On the one hand, they can increase plant yield by improving mineral uptake and phytohormone production, inducing the competitive suppression of pathogens and triggering plant-induced systemic resistance [38]. On the other hand, these biofilms allow the attachment of the cells to a nutrient source and confer protection against plant defenses and environmental fluctuations [27,37]. Furthermore, the biofilms produced by rhizospheric bacteria enhance soil aggregation, which improves the water-holding capacity, fertility, and porosity of the soils, leading to an increase in agricultural productivity [39,40,41,42].
Some of the biofilm components, mainly exopolysaccharides, that are required for biofilm formation and pathogenesis in P. aeruginosa find their equivalents in pathogenic and beneficial plant-interacting Pseudomonas. In this review, we shed light on the extracellular matrix exopolysaccharides of plant-associated Pseudomonas within the context of pathogenic and beneficial interactions.
2. Ecological Significance of Biofilm Formation by Plant-Interacting Bacteria
Plant-associated bacteria develop a biofilm lifestyle during their interactions with plants [27,37]. Depending on whether biofilms are formed by pathogenic or beneficial individuals, the ecological outcome resulting from the interaction can be completely different. In the context of pathogenic plant-associated bacteria, the role of different components involved in biofilm formation has been studied. For instance, the biofilms formed by Erwinia amylovora, the causal agent of fire blight disease in different plant species of the Rosaceae family, and specifically the amylovoran and levan exopolysaccharides, physically blocked the vascular system of plants [43,44,45]. A mutant of Ralstonia solanacearum, the causal agent of bacterial wilt disease, in the lecM gene, which encodes a lectin, showed reduced biofilm formation in vitro and colonization of the intercellular spaces of tomato leaves and was impaired in virulence [46]. The gumB mutant of Xanthomonas citri, which produces canker disease in citrus plants, was unable to produce the polysaccharide xantan and exhibited reduced biofilm formation, survival and symptom development on lemon leaves [30]. Similarly, Xylella fastidiosa, which causes economically important diseases in several host plants, produced exopolysaccharides that played roles in the virulence of this bacterium, as these are required for bacterial movement within plants and plant-to plant transmission through insects [47,48].
Notably, the Pseudomonas syringae complex harbors most of the phytopathogens within the Pseudomonas genus [49,50]. In particular, the species P. syringae is one of the most ubiquitous bacterial participants of the phyllosphere [51]. This ubiquity, together with the fact that it can infect almost all important agricultural crops [25,52], has made it a model for the study of plant–bacteria interactions. P. syringae possesses a great diversity of virulence factors that engage in plant infection, as well as adaptation mechanisms that improve bacterial survival over the plant surface. Generally, P. syringae produces a type III secretion system (T3SS), effector proteins, motility appendages, phytotoxins, multidrug efflux pumps, extracellular polysaccharides, cell wall-degrading enzymes, and ice nucleation activity [53]. Copper- and UV radiation-resistance genes, as well as exopolysaccharide production, play fundamental roles in P. syringae fitness and survival [31,50,54,55,56,57,58]. In P. syringae pv. syringae (Pss), biofilm formation has been proven to influence the transition between pathogenic and epiphytic lifestyles in plants [29,31,59].
In the context of beneficial plant-associated bacteria, Bacillus subtilis, a Gram-positive bacterium that acts as a biocontrol agent of several plant pathogens, requires the production of extracellular matrix components involved in biofilm formation, such as those encoded by tapA-sipW-tasA and epsA-O operons, for the colonization of the plant roots and for conferring plant protection [60]. Pseudomonas fluorescens, an important rhizobacterium that promotes plant health and nutrition, requires biofilm formation for the colonization of plant surfaces [61]. A cellulose exopolysaccharide mutant in the P. fluorescens SBW25 strain was compromised in the colonization of the rhizosphere and the phyllosphere of sugar beet compared to the wild-type strain [61]. In general, the P. fluorescens species and some closely related species that belong to the P. fluorescens complex are among the most studied bacteria within soil communities, because they frequently show agricultural, biotechnological, and ecological interest, mostly due to their beneficial plant features [62]. In particular, the P. fluorescens and Pseudomonas chlororaphis species stand out because of their potential use as biocontrol agents as they frequently contribute to plant health by exerting antagonist activities against pathogens [63,64,65]. Phenotypes linked to biofilm formation have also been observed to favor bacteria–plant root interactions and biocontrol activity of P. chlororaphis and Pseudomonas putida species [66,67,68,69,70]. Usually, biocontrol agents can form biofilms, and increasing evidence strongly suggests that biofilm-forming ability should be considered in assessing their potential beneficial performance [71].
3. Main Exopolysaccharides Produced by Plant-Associated Pseudomonas
Among all the exopolysaccharides that are produced by plant-associated Pseudomonas [72], those that have been mainly studied are alginate, cellulose, and Psl (Table 1). A description of their functions in biofilm formation and architecture and their ecological significance during pathogenic and beneficial plant–bacteria interactions are listed below.

Table 1.
Main exopolysaccharides produced by different Pseudomonas spp. strains that are involved in biofilm formation.
3.1. Alginate Exopolysaccharide
Alginate is a copolymer made of O-acetylated D-mannuronic and L-glucuronic acid residues joined by β-1,4 linkages [74]. In PAO1, the alginate polysaccharide is encoded on a twelve gene operon that corresponds to the PA3540-PA3551 genomic region [13]. During infections in cystic fibrosis (CF) patients, P. aeruginosa undergoes a switch into a mucoid phenotype characterized by alginate overproduction [75,76,77]. Alginate overexpression increases the resistance of P. aeruginosa to antimicrobial treatments, predators, and host defenses [12,78]. The high frequency in which this conversion occurs, and the protective capacities described for alginate, suggests that alginate is the main exopolysaccharide of the P. aeruginosa extracellular matrix. However, studies performed on nonmucoid P. aeruginosa strains (e.g., PAO1 and PA14), the truly predominant phenotype and the one responsible for the colonization of the lungs of CF patients [13], have shown that, although it is not critical for biofilm constitution, this polysaccharide is a component of the P. aeruginosa extracellular matrix and can influence its biofilm architecture [13,14,79,80].
Studies performed on alginate in some plant-associated Pseudomonas have revealed that this polysaccharide plays minor structural roles in their biofilms, including the bacterial phytopathogen P. syringae and the plant-beneficial bacteria P. fluorescens, P. chlororaphis, and P. putida [59,70,73,81,82]. The alginate-deficient derivative of the P. syringae pv. glycinea PG4180.muc strain formed biofilms to the same extent as the wild-type strain in flow-cell chambers [81]. However, the biofilm architecture of the PssUMAF0158 ∆alg8 strain, which does not produce alginate, showed slightly but significantly lower surface coverage and volume than the wild-type strain [59], as was previously described in P. aeruginosa [80]. Alginate is overproduced in some strains of P. syringae upon exposure to copper bactericides, which are usually applied to reduce the disease incidence caused by some plant pathogens [32]. This could be explained because exopolysaccharide production has been generally associated with a higher tolerance against toxic compounds [2,83]. Previous works have indicated that alginate polysaccharides are involved in the pathogenic interaction of P. syringae with plants [29,84,85]. For instance, the alginate mutant of the P. syringae pv. syringae 3525 strain, the causal agent of bacterial brown spot on bean, is significantly impaired in the colonization of bean (host) and tomato (non-host) leaves, and although it retains the ability to generate symptoms, the symptoms are less severe than those induced by the wild-type [29]. However, these results have not been observed in other P. syringae strains [59,86,87,88]. For example, in P. syringae pv. glycinea PG4180 strain, the causal agent of bacterial blight of soybean, the expression of the AlgT regulator protein, but not alginate production per se, promotes survival and symptom development in plants [88]. Similarly, the PssUMAF0158 ∆alg8 mutant strain is not altered in the induction of symptoms in tomato compared to the wild-type strain [59].
The structural functions displayed by alginate in the biofilms of plant-pathogenic Pseudomonas are in line with those observed in the plant-beneficial Pseudomonas. The alginate mutant of P. fluorescens SBW25 still forms biofilms in flow-cell chamber experiments, but they are thinner than those formed by the wild-type strain [82]. This result is consistent with the flow-cell chamber phenotypes of the PssUMAF0158 and PAO1 alginate mutant derivative strains [59,80]. However, the alginate mutant of the biocontrol agent P. chlororaphis PCL1606 (PcPCL1606 ∆alg8) forms biofilms to the same extent as the wild-type in flow-cell chamber experiments and is not impaired in initial surface attachment, showing nonsignificant differences in surface coverage and volume values with respect to the wild-type [70]. Alginate has been described as the primary polysaccharide that promotes hydration under desiccating stress in P. putida [89,90]. In fact, alginate slightly contributes to the biofilm architecture of P. putida under water-limiting conditions [90]. The functions performed by alginate polysaccharide in both P. fluorescens and P. putida strains in vivo seem to be more relevant than those in vitro. For instance, the CHA211 and CHA213M mucoid variants of the P. fluorescens CHA0 strain, which overproduce alginate, enhance their biofilm formation abilities on carrot roots compared to the wild-type strain [91]. The genomic region located upstream of the algD gene of P. putida KT2440 is active during the colonization of maize root, which suggests that this polysaccharide could be a fitness determinant for the rhizosphere colonization ability of this bacterium [92]. Overall, these studies indicate that alginate is not a critical component for biofilm formation in vitro in plant-associated Pseudomonas and that its role seems to be more prominent in vivo, facilitating colonization and providing protection against stressors.
3.2. Cellulose Exopolysaccharide
Cellulose is a polymer made of D-glucose residues joined by β-1,4 glycosidic linkages and is considered a relevant biofilm matrix molecule in many environmental Pseudomonas species [93,94]. Several biosynthesis and regulation mechanisms have been described for bacterial cellulose, but a common role of this component is to facilitate the establishment of efficient host-bacteria interactions [95]. Previous studies reported that several plant-associated Pseudomonas species can produce cellulose, including the plant-associated pathogenic bacteria P. syringae, P. asplenii, P. marginalis, P. corrugate, and P. savastanoi and beneficial bacteria, such as P. fluorescens and P. putida [72,93,94]. Within the Pseudomonas genus, P. fluorescens SBW25 (SBW25) is traditionally used as the model strain for the study of bacterial cellulose. In SBW25, cellulose polysaccharide is encoded on a ten-gene operon (wssA-J) that corresponds to the PFLU0300-PFLU0309 genomic region [96]. This exopolysaccharide is involved in the formation of floating biofilms, also called pellicles, in many strains of the species mentioned above, including the SBW25 [31,93,96,97,98,99]. The P. aeruginosa species does not contain the cellulose operon [72]. In particular, P. aeruginosa PAO1 and PA14 strains, which have been traditionally used as model strains for conducting biofilm studies within the Pseudomonas genus, were tested for cellulose production, but in line with in silico observations [72], they were not found to produce this exopolysaccharide [94]. However, the PAO1 and PA14 strains contain a seven-gene operon that encodes Pel, which is a polymer composed of partially acetylated 1→4 glycosidic linkages of N-acetylgalactosamine and N-acetylglucosamine [100]. The pel operon is poorly conserved among environmental Pseudomonas [72,100,101]. Interestingly, Pel promotes the formation of pellicle biofilms, as has also been described for cellulose [102].
Among all plant-pathogenic Pseudomonas that have been reported to produce cellulose, studies regarding its structural roles within biofilms and biological significance have essentially been conducted on P. syringae. The P. syringae pv. syringae (Pss) UMAF0158 (PssUMAF0158) strain, the causal agent of bacterial apical necrosis (BAN) on mango trees, and P. syringae pv. tomato DC3000 (PtoDC3000), responsible for bacterial speck disease on tomato plants, produce cellulose as the main exopolysaccharide of their biofilms [31,59,103]. The biofilm structures formed by PssUMAF0158 and PtoDC3000 in micro-well plates are highly similar, consisting of pellicles with wrinkles on the surface that are weakly attached to the walls of the culture vessels [31,103]. Despite the structural similarities found in vitro, the biological performance of cellulose seems to differ in both strains. Cellulose allows PssUMAF0158 to adhere to mango leaves, and its production intimately affects the epiphytic and pathogenic stages of this strain over the plant surface [31]. Hence, the incidence and severity of necrotic symptoms developed by PssUMAF0158 on tomato leaflets are lower in the wild-type than in cellulose mutants (∆wssB and ∆wssE mutants) and practically nonexistent in the cellulose-overproducing strain [31]. These results, together with the fact that the highest BAN symptoms coincide with cool and wet periods [104], support the proposed lifecycle of Pss strains over the mango tree, in which biofilm formation would be mainly needed during the epiphytic phase (spring/summer) when the bacteria are more exposed to the external environment, and protection against its challenging conditions becomes crucial for survival [50]. Interestingly, the link observed between cellulose production and PssUMAF0158 transition through epiphytic and pathogenic stages over the mango plant surface has not been reported in PtoDC3000. The disease symptoms developed in tomato by the PtoDC3000 wild-type strain were not different from those of its ∆wssBC-derived mutant [105]. Furthermore, in disagreement with what has been observed in PssUMAF0158, cellulose overproduction in PtoDC3000 does not lead to a significant impact on virulence [103]. However, the PtoDC3000 armZ gene mutant, which does not produce alginate and does overproduce cellulose, has a reduced virulence compared to the wild-type strain [105]. Although PssUMAF0158 and PtoDC3000 are categorized as P. syringae species and belong to the P. syringae complex, this complex is comprised of a hodgepodge that, in effect, includes many other taxonomically related species [49]. A previous study revealed that the phylogenetic relationship between P. syringae pv. syringae B728a strain, closely related to PssUMAF0158, and PtoDC3000, is not very proximate. In fact, PtoDC3000, together with other strains of the tomato pathovar, seems to form a new species Pseudomonas tomato, pending a deeper taxonomic analysis [49]. This evidence, together with the fact that the infection assays were performed using different tomato cultivars and inoculation approaches, could all eventually account for different results.
Regarding beneficial plant-interacting Pseudomonas, studies on bacterial cellulose have been mainly conducted on P. fluorescens and P. putida species. Biofilm experiments on SBW25 determined that the gradients occurring within a static microcosm immediately select for the emergence of variants that occupy different niches [106]. Among those variants, the air-liquid (A–L) interface is colonized by wrinkly spreader (WS) pellicles, an SBW25-derived mutant that overproduces cellulose compared to the wild-type equivalent [107]. In P. putida mt2 and its plasmid-free derivative KT2440 [108] strains, cellulose plays minor roles in biofilm formation in vitro [73,89], while two additional exopolysaccharide gene clusters, putida exopolysaccharide A (Pea) and putida exopolysaccharide B (Peb), are essential for biofilm formation in this species [73,89]. Instead, the role of cellulose exopolysaccharide in P. putida seems to be directed more towards conferring protection, as water-limiting conditions and increasing osmolarity highly induce cellulose expression of P. putida mt2 [89,109]. In addition, the cellulose mutant of P. putida mt2 strain accumulates significantly more reactive oxygen species (ROS) than the wild-type strain upon exposure to matric and copper stressors [109]. During plant–bacteria interactions, the cellulose exopolysaccharide of SBW25 contributes to the ecological performance of this strain in the rhizosphere and phyllosphere of sugar beet [61]. Thus, a cellulose-defective mutant of SBW25 (SM13) was compared against the wild-type in the rhizosphere, phyllosphere, and bulk soil surrounding the rhizosphere of sugar beet seedlings, and the results showed no significant differences between the fitness of SM13 relative to the wild-type in bulk soil, but significant differences were found in the rhizosphere and phyllosphere, especially in the phyllosphere [61]. Something similar has been reported in the P. putida mt2 strain in which the cellulose mutant is impaired in the colonization of the maize rhizosphere during competition with the wild-type equivalent [73]. These studies indicate that, while the cellulose operon does not seem to be critical for biofilm formation under laboratory conditions in P. fluorescens and P. putida, their roles in these species seem to be more pronounced in vivo.
3.3. Psl Exopolysaccharide
The Psl polysaccharide was first described in P. aeruginosa [102,110,111], and its structural analysis determined that it consists of a repeating pentasaccharide subunit of D-mannose, D-glucose, and L-rhamnose in a 3:1:1 ratio [112]. In PAO1, Psl was formerly described to be encoded by the 15-gene operon psl (pslA-O), which corresponds to the PA2231-PA2245 genomic region [102,110,111]. However, later works revealed that the last three genes of the operon (pslMNO) constitute an independent transcriptional unit [113,114,115] and are not truly required to produce Psl [112]. Except for the case of the P. aeruginosa PA14 strain, which does not produce Psl due to the absence of pslA-D genes [102], the psl gene cluster is present in multiple strains of P. aeruginosa [72,80], where it plays key roles in their biofilm lifestyles [80]. Several studies have proven the involvement of Psl in adhesion to biotic and abiotic surfaces, biofilm architecture, motility, and protection against stressors [16,19,116,117,118,119]. Although research on Psl polysaccharides has been mostly conducted in P. aeruginosa, the existence of a psl-like gene cluster has been reported in some environmental nonaeruginosa Pseudomonas [59,70,72,101]. Generally, the psl-like gene clusters found in nonaeruginosa Pseudomonas either lack orthologues to pslMNO genes or are found scattered in the genome outside the cluster. The bacterial phytopathogen PssUMAF0158 contains a psl-like gene cluster that does not include orthologues to the pslCLMNO genes and encodes a putative acetyltransferase between the pslJ-and pslK-like genes that might perform a related function to that of acyltransferase PslL [59]. Interestingly, the psl-like gene cluster of PssUMAF0158 seems to be highly conserved among the plant-associated phylogroups belonging to the P. syringae complex [59]. The biocontrol agent PcPCL1606 also contains a psl-like gene cluster, which lacks the pslLMNO genes and encodes a putative acetyltransferase between the pslJ- and pslK-like genes, similar to that of PssUMAF0158 [70]. However, the psl-like gene cluster of PcPCL1606 is not present in some phylogroups of the P. fluorescens complex and is partially present in others, according to the strains included in a previous study [70]. It is completely absent in the corrugata, jessenii, and koreensis phylogroups; only present in Pseudomonas GM21 strain of the mandelii phylogroup; and is partially present within the P. fluorescens phylogroup. Interestingly, a psl-like gene cluster is found in all the strains of the P. chlororaphis phylogroup that have been assessed [70], which suggests that this polysaccharide could be relevant for biofilm formation in this species.
The first study regarding Psl composition in P. aeruginosa PAO1 determined that this polysaccharide was a galactose- and mannose-rich exopolysaccharide [120]. Support for this information came from three pieces of evidence. First, a chemical composition analysis of exopolysaccharide preparations of WFPA801, a PAO1-derived Psl-inducible strain, determined the presence of galactose, mannose, and glucose, as well as trace amounts of xylose, rhamnose, and N-acetylglucosamine. Second, staining of planktonically grown WFPA801 cells with FITC-HHA lectin, which binds to some mannosyl units, and FITC-MOA lectin, which binds to some galactosyl units, revealed green fluorescent signals on the WFPA801 surface. Ultimately, mutants of the pslH gene, which encodes a putative galactosyltransferase, and the pslI gene, which encodes a putative mannosyltransferase, were deficient in attachment, yielding a similar phenotype to that of the WFPA800 null Psl-producing strain [120]. Two years later, the structural analysis of Psl was published, indicating that it likely consisted of a pentasaccharide repeating unit of mannose, glucose, and rhamnose in approximate ratios of 3:1:1 [112]. Interestingly, galactose, which was reported as the major component of Psl in the first study [120], was not detected as a component of Psl in the structural analysis [112]. The authors stated that different growth conditions were used in both studies, which could account for some variations in composition, as described previously [121]. Therefore, there is some thought that different forms of Psl might be produced even in the same strain depending on the growth conditions. Be that as it may, mannose seems to be a key component of the Psl structure in P. aeruginosa. An analysis of the composition and structure of the putative Psl polysaccharide produced by PssUMAF0158 and PcPCL1606 has not yet been conducted, but some hints exist regarding the existence of a polysaccharide that resembles Psl in P. syringae and P. fluorescens. Thus, it was reported that, in addition to alginate and levan, P. syringae PG4180 produced a third exopolysaccharide (EPS) that consisted of a fibrous structure in its biofilms and bound to Naja mossambica lectin (NML) [81]. Interestingly, the monosaccharide specificity of NML is mannose [122]. Furthermore, two P. fluorescens strains isolated from rotted bell pepper, PF-05-2 and PM-LB-1, produced a novel exopolysaccharide composed of mannose, rhamnose, and glucose (1:1:1 molar ratio) substituted with pyruvate and acetate [123]. The biofilm formed by the PcPCL1606 wild-type strain but not its Psl-like-derived mutant, contains a polymer that binds to banana lectin, which also binds to mannose residues [70,124].
The ∆pslAB mutant of PAO1 is severely attenuated in biofilm initiation and biofilm development in flow-cell chamber experiments [110,111]. Interestingly, similar results were observed in the biocontrol strain PcPCL1606, in which the ∆pslE mutant was severely affected in early surface attachment and development of a mature biofilm architecture compared to the wild-type strain [70]. The biofilms developed by PAO1 and PcPCL1606 wild-type strains showed an intricate architecture in flow-cell chambers, which consisted of a multilayer of cells that covered the chamber surfaces [70,110,111]. However, the biofilm phenotype of the PAO1 ∆pslAB and PcPCL1606 ∆pslE mutant strains consisted of a monolayer of loosely aggregated cells, which suggests that this exopolysaccharide could also be important for cell-to-cell interactions [70,110,111]. Similarly, the Psl-like polysaccharide of the phytopathogen PssUMAF0158 is also involved in biofilm architecture [59]. Compared to the more developed biofilm of wild-type PssUMAF0158, the PssUMAF0158 ∆pslE biofilms consisted of scattered cell aggregates across the flow-cell chamber surface [59]. These cell aggregates were disrupted in the double mutant ∆wssE,pslE strain, which did not produce both cellulose and Psl-like polysaccharides [59]. Curiously, this phenotype was also observed in some P. aeruginosa strains, where the cell aggregates formed by their derived ∆psl mutants were disrupted in the ∆psl∆pel double mutants, affected in both Psl and Pel polysaccharide production [116]. The fact that cellulose and Pel polysaccharides are both involved in the formation of pellicle biofilms [101], and that the cell aggregates formed by these Pseudomonas ∆psl strains are disrupted when either cellulose or Pel is not produced, indicates that both polysaccharides could play redundant structural roles within biofilms, as has been previously suggested [59,101]. Indeed, it is not common to find both genomic regions encoding cellulose and Pel in the same Pseudomonas strain. Thus, just 12 out of 600 Pseudomonas genomes (2%) that have been analyzed in a recent study [72]—which belong to four different groups: P. asplenii, P. fluorescens, P. fragi and P. oryzihabitans—possess both clusters (Table 2), although whether they are functional remains unknown. With these recent data, the identity and coverage of both clusters have been analyzed in these 12 Pseudomonas spp. strains using the wss operon of SBW25 and pel operon of PAO1 as references.

Table 2.
Pseudomonas spp. strains obtained from Blanco–Romero et al., (2020) that have been reported to contain the wss and pel clusters.
Swarming motility and biosurfactant synthesis are coordinated with Psl production in P. aeruginosa, as the PAO1 Psl-deficient strain exhibited a hyperswarming phenotype due to an increase in rhamnolipid production, and vice versa [118]. Curiously, the link found between biofilm formation, rhamnolipid production, and motility in the bacterial phytopathogen PssUMAF0158 seems opposite to that described in P. aeruginosa. Therefore, the PssUMAF0158 ∆pslE mutant is impaired in swarming motility compared to the wild-type strain, and this impairment could be due to a reduction in surfactant production, as the rhlA gene involved in rhamnolipid precursor synthesis is downregulated in the mutant compared to the wild-type strain [59].
The PAO1 ∆pslAB mutant is deficient in biofilm initiation due to its reduced ability to interact with biotic and abiotic surfaces [16,110,111]. In line with these data, the recently described Psl-like exopolysaccharides of the bacterial phytopathogen PssUMAF0158 and biocontrol agent PcPCL1606 have also been reported to be involved in early surface interactions. The PssUMAF0158 ∆pslE mutant is impaired in early adhesion to mango leaves [59] and the PcPCL1606 ∆pslE mutant was impaired in early surface attachment to polystyrene micro-well plates and avocado root surfaces [70]. Moreover, biofilm formation by PssUMAF0158 and PcPCL1606 strains through Psl-like exopolysaccharide biosynthesis also contributes to the lifestyles displayed by these bacteria during interaction with their plant host [59,70]. The inability to produce some extracellular matrix components, such as cellulose and Psl-like exopolysaccharides, seems to predispose PssUMAF0158 to the pathogenic lifestyle, as the mutants impaired in the production of these exopolysaccharides are significantly more virulent than the wild-type [31,59]. Consequently, the biofilm lifestyle of PssUMAF0158 could predominate during the epiphytic phase, as has been previously suggested [50]. Furthermore, the Psl-like exopolysaccharide of PcPCL1606 contributes to the biocontrol activity of this bacterium against white rot root disease caused by Rosellinia necatrix in avocado plants [70]. Thus, PcPCL1606 ∆pslE is severely compromised in disease suppression, probably because early attachment and biofilm impairments could lead to inefficient colonization of roots, which is a prerequisite for efficient disease control [70,125,126,127]. The presence of a psl-like cluster in some environmental pseudomonads with different lifestyles suggests that this polysaccharide might constitute a general feature of biofilm formation of these bacteria, providing different functions depending on the genetic context and niche of production.
4. Brief Summary and Future Perspectives
The Pseudomonas genus includes species with high metabolic and physiologic versatility, as well as broad potential for adaptation to fluctuating environmental conditions, which accounts for their ability to colonize such a wide variety of environments [9]. Within the Pseudomonas genus, the P. aeruginosa species has been traditionally used as a model bacterium for the study of biofilm formation due to its impact in clinical settings [10]. The extracellular matrix of P. aeruginosa is predominantly composed of polysaccharides [11], and interestingly, some of them are also produced by several pathogenic and beneficial plant-associated Pseudomonas [72]. These exopolysaccharides display structural and/or protective roles in plant-associated bacteria. The ecological significance derived from their production is dependent on the lifestyles displayed by these bacteria during plant–bacteria interactions (Figure 1). To date, alginate polysaccharides seem to play minor structural roles in the biofilms of plant-associated Pseudomonas in vitro, which correlates with previous results observed in P. aeruginosa [13,59,70,80,82]. However, its functions that have been described in vivo, together with those directed towards protection against external stressors, such as desiccation, seem more prominent [12,78,89,90,128]. Cellulose is not produced by the P. aeruginosa PAO1 and PA14 model strains, but is produced by several plant-associated bacteria, including P. syringae and P. fluorescens, frequently constituting the main architectural component of their biofilms [93,94]. Instead, PAO1 and PA14 strains produce Pel, which is poorly conserved among environmental nonaeruginosa Pseudomonas and is responsible for pellicle formation, as previously described for cellulose [101,102]. Overall, cellulose is described as a major component of the biofilm architecture produced by several plant-associated Pseudomonas [31,96,105]. The Psl polysaccharide, which was first described in P. aeruginosa, is a key component of the biofilm architecture of this bacterium [102,110,111]. For many years, the role of Psl in biofilm formation by some environmental nonaeruginosa pseudomonads was unknown. However, the involvement of a Psl-like polysaccharide in the biofilm architecture and lifestyles of two plant-associated Pseudomonas species has been recently described for the first time [59,70].
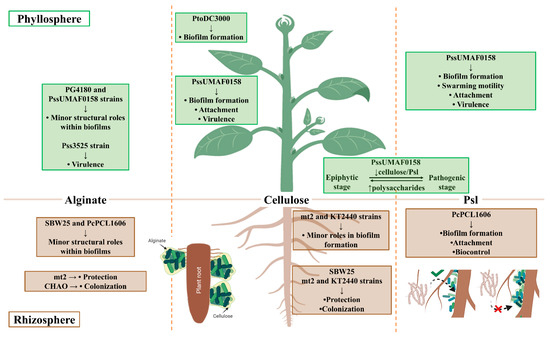
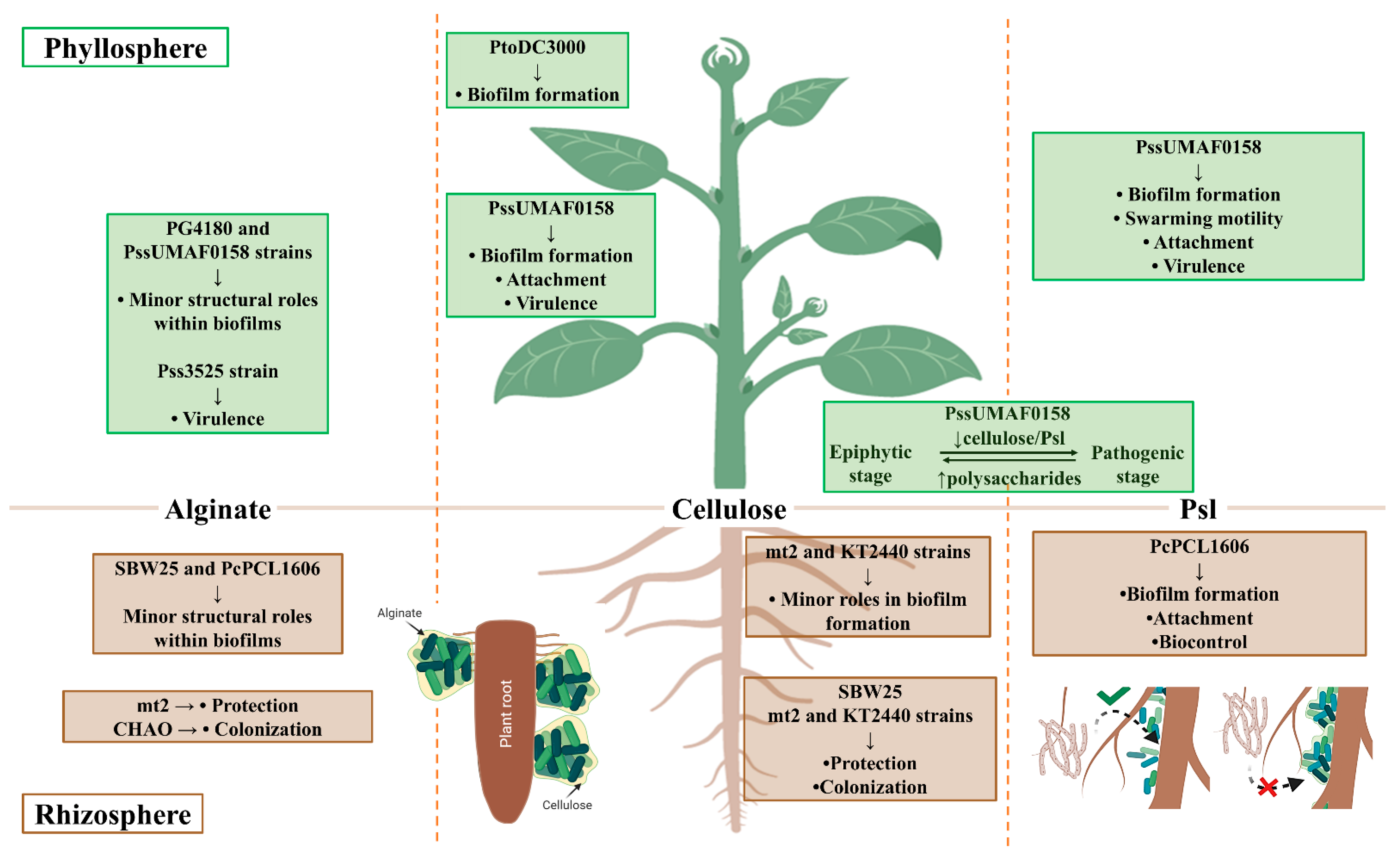
Figure 1.
Summary of functions described for exopolysaccharides produced by plant-associated Pseudomonas. P. syringae pv. syringae UMAF0158 (PssUMAF0158), P. syringae pv. syringae 3525 (Pss3525), P. syringae pv. tomato DC3000 (PtoDC3000), P. syringae pv. glycinea PG4180 (PG4180), P. fluorescens SBW25 (SBW25), P. putida mt2 (mt2), P. putida KT2440 (KT2440), P. fluorescens CHAO (CHAO), and P. chlororaphis PCL1606 (PcPCL1606). Image created with Biorender.com.
Despite all the knowledge developed from biofilm studies, numerous aspects remain underexplored, for example, how different components interact within the extracellular matrix. Lectin staining has allowed detection of some polysaccharides, such as Psl and Pel polysaccharides, within the biofilms of P. aeruginosa [100,129], as well as alginate and levan within the biofilms of P. syringae [81], but how they are located with respect to one another has not been specified. Similarly, whether Psl and cellulose can interact in the extracellular matrix of the PssUMAF0158 strain remains unknown. The involvement of Psl in biofilm formation by environmental Pseudomonas has been overlooked for a long time, and its revealed importance in the biofilm architecture and influence on the bacterial lifestyle of the phytopathogen PssUMAF0158 and biocontrol agent PcPCL1606 could lead future studies towards determining the functions and capacities of this component in other bacterial species. Furthermore, future studies should also contemplate the compositional and structural analysis of the Psl-like polysaccharide produced by environmental pseudomonads to determine its level of resemblance to the archetypal P. aeruginosa.
On another note, it is currently known that specific climate factors, such as temperature, pH, light, and humidity, influence biofilm formation [130,131,132,133]. However, little information exists regarding the direct impact that climate conditions can have on plant–bacteria interactions through biofilm formation or biofilm-related processes. For example, it has been described that white light exposure, specifically blue light, increases the attachment of PtoDC3000 to Arabidopsis thaliana leaves [134]. Recently, the impact of temperature on the biofilm architecture of P. aeruginosa, guided by exopolysaccharide synthesis, has been revised [135]. More studies should focus on the direct impact that specific and combined different climate factors have on biofilm formation, particularly the impact derived from such changes on the bacterial ecology during plant–bacteria associations.
Finally, studies on extracellular polysaccharides produced in monospecies biofilms have provided interesting information regarding their roles in biofilm architecture, as well as their influence on host–bacteria interactions, but future works should be more directed towards polymicrobial biofilms. This is because environmental habitats, such as those encountered on plant surfaces, are known to harbor complex microbial assemblages [136,137,138], in which usually different species, and even different kingdoms, interact. Currently, more details regarding multispecies biofilms are being revealed [139,140,141], but there is still much work to do regarding this issue.
Author Contributions
Z.H.-P., A.d.V., F.M.C. and J.A.G.-B. conceived the study. Z.H.-P. wrote the manuscript. A.d.V., F.M.C. and J.A.G.-B. contributed critically to the final version of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by grants from CICE-Junta de Andalucía; Proyecto de Excelencia-Junta de Andalucía, grant number P12-AGR-1473; Spanish Plan Nacional I+D+I, grant number AGL2017-83368-C2-1-R and co-financed by Proyecto FEDER UMA18-FEDERJA-046. Zaira Heredia-Ponce is financed by a FPU contract (FPU15-03644).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Costerton, J.W.; Lewandowski, Z. Microbial biofilms. Annu. Rev. Microbiol. 1995, 49, 711–745. [Google Scholar] [CrossRef]
- Costerton, J.W. Introduction to biofilm. Int. J. Antimicrob. Agents 1999, 11, 217–221. [Google Scholar] [CrossRef]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Marshall, K.C. Planktonic versus sessile life of prokaryotes. In The Prokaryotes, 3rd ed.; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; Volume 2, pp. 3–15. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wuertz, S. Bacteria and archaea on Earth and their abundance in biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Bar-On, Y.M.; Milo, R. Towards a quantitative view of the global ubiquity of biofilms. Nat. Rev. Microbiol. 2019, 17, 199–200. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, K.K. What drives bacteria to produce a biofilm? FEMS Microbiol. Lett. 2004, 136, 163–173. [Google Scholar] [CrossRef]
- Yin, W.; Wang, Y.; Liu, L.; He, J. Biofilms: The microbial "protective clothing" in extreme environments. Int. J. Mol. Sci. 2019, 20, 3423. [Google Scholar] [CrossRef]
- Silby, M.W.; Winstanley, C.; Godfrey, S.A.; Levy, S.B.; Jackson, R.W. Pseudomonas genomes: Diverse and adaptable. FEMS Microbiol. Rev. 2011, 35, 652–680. [Google Scholar] [CrossRef]
- Diggle, S.P.; Whiteley, M. Microbe profile: Pseudomonas aeruginosa: Opportunistic pathogen and lab rat. Microbiology 2020, 166, 30–33. [Google Scholar] [CrossRef] [PubMed]
- Franklin, M.J.; Nivens, D.E.; Weadge, J.T.; Howell, L. Biosynthesis of the Pseudomonas aeruginosa extracellular polysaccharides, alginate, Pel and Psl. Front. Microbiol. 2011, 2, 167. [Google Scholar] [CrossRef]
- Hentzer, M.; Teitzel, G.M.; Balzer, G.J.; Heydorn, A.; Molin, S.; Givskov, M.; Parsek, M.R. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 2001, 183, 5395–5401. [Google Scholar] [CrossRef]
- Wozniak, D.J.; Wyckoff, T.J.O.; Starkey, M.; Keyser, R.; Azadi, P.; O’Toole, G.A.; Parsek, M.R. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. USA 2003, 100, 7907–7912. [Google Scholar] [CrossRef] [PubMed]
- Stapper, A.P.; Narasimhan, G.; Ohman, D.E.; Barakat, J.; Hentzer, M.; Molin, S.; Kharazmi, A.; Høiby, N.; Mathee, K. Alginate production affects Pseudomonas aeruginosa biofilm development and architecture, but is not essential for biofilm formation. J. Med. Microbiol. 2004, 53, 679–690. [Google Scholar] [CrossRef] [PubMed]
- Leid, J.G.; Willson, C.J.; Shirtliff, M.E.; Hassett, D.J.; Parsek, M.R.; Jeffers, A.K. The exopolysaccharide alginate protects Pseudomonas aeruginosa biofilm bacteria from IFN- γ-mediated macrophage killing. J. Immunol. 2005, 175, 7512–7518. [Google Scholar] [CrossRef]
- Ma, L.; Jackson, K.D.; Landry, R.M.; Parsek, M.R.; Wozniak, D.J. Analysis of Pseudomonas aeruginosa conditional Psl variants reveals roles for the Psl polysaccharide in adhesion and maintaining biofilm structure postattachment. J. Bacteriol. 2006, 188, 8213–8221. [Google Scholar] [CrossRef]
- Colvin, K.M.; Gordon, V.D.; Murakami, K.; Borlee, B.R.; Wozniak, D.J.; Wong, G.C.; Parsek, M.R. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011, 7, e1001264. [Google Scholar] [CrossRef]
- Ma, L.; Wang, S.; Wang, D.; Parsek, M.R.; Wozniak, D.J. The roles of biofilm matrix polysaccharide Psl in mucoid Pseudomonas aeruginosa biofilms. FEMS Immunol. Med. Microbiol. 2012, 65, 377–380. [Google Scholar] [CrossRef]
- Billings, N.; Millan, M.R.; Caldara, M.; Rusconi, R.; Tarasova, Y.; Stocker, R.; Ribbeck, K. The extracellular matrix component Psl provides fast-acting antibiotic defense in Pseudomonas aeruginosa biofilms. PLoS Pathog. 2013, 9, e1003526. [Google Scholar] [CrossRef]
- Ciofu, O.; Tolker-Nielsen, T. Tolerance and resistance of Pseudomonas aeruginosa biofilms to antimicrobial agents-how P. aeruginosa can escape antibiotics. Front. Microbiol. 2019, 10, 913. [Google Scholar] [CrossRef] [PubMed]
- Elrod, R.P.; Braun, A.C. Pseudomonas aeruginosa: Its role as a plant pathogen. J. Bacteriol. 1942, 44, 633–645. [Google Scholar] [CrossRef]
- Walker, T.S.; Bais, H.P.; Déziel, E.; Schweizer, H.P.; Rahme, L.G.; Fall, R.; Vivanco, J.M. Pseudomonas aeruginosa-plant root interactions. Pathogenicity, biofilm formation, and root exudation. Plant Physiol. 2004, 134, 320–331. [Google Scholar] [CrossRef]
- Hirano, S.S.; Upper, C.D. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae-a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 2000, 64, 624–653. [Google Scholar] [CrossRef] [PubMed]
- Lugtenberg, B.J.J.; Dekkers, L.; Bloemberg, G.V. Molecular determinants of rhizosphere colonization by Pseudomonas. Annu. Rev. Phytopathol. 2001, 39, 461–490. [Google Scholar] [CrossRef] [PubMed]
- Kennelly, M.M.; Cazorla, F.M.; de Vicente, A.; Ramos, C.; Sundin, G.W. Pseudomonas syringae diseases of fruit trees: Progress toward understanding and control. Plant Dis. 2007, 91, 4–17. [Google Scholar] [CrossRef]
- Mercado-Blanco, J.; Bakker, P.A.H.M. Interactions between plants and beneficial Pseudomonas spp.: Exploiting bacterial traits for crop protection. Antonie Leeuwenhoek 2007, 92, 367–389. [Google Scholar] [CrossRef]
- Danhorn, T.; Fuqua, C. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 2007, 61, 401–422. [Google Scholar] [CrossRef] [PubMed]
- Leveau, J.H. A brief from the leaf: Latest research to inform our understanding of the phyllosphere microbiome. Curr. Opin. Microbiol. 2019, 49, 41–49. [Google Scholar] [CrossRef]
- Yu, J.; Peñaloza-Vázquez, A.; Chakrabarty, A.M.; Bender, C.L. Involvement of the exopolysaccharide alginate in the virulence and epiphytic fitness of Pseudomonas syringae pv. syringae. Mol. Microbiol. 1999, 33, 712–720. [Google Scholar] [CrossRef]
- Rigano, L.A.; Siciliano, F.; Enrique, R.; Sendín, L.; Filippone, P.; Torres, P.S.; Qüesta, J.; Dow, J.M.; Castagnaro, A.P.; Vojnov, A.A.; et al. Biofilm formation, epiphytic fitness, and canker development in Xanthomonas axonopodis pv. citri. Mol. Plant Microbe Interact. 2007, 20, 1222–1230. [Google Scholar] [CrossRef]
- Arrebola, E.; Carrión, V.J.; Gutiérrez-Barranquero, J.A.; Rodríguez-Palenzuela, P.; Cazorla, F.M.; de Vicente, A. Cellulose production in Pseudomonas syringae pv. syringae: A compromise between epiphytic and pathogenic lifestyles. FEMS Microbiol. Ecol. 2015, 91, 1–12. [Google Scholar] [CrossRef]
- Kidambi, S.P.; Sundin, G.W.; Palmer, D.A.; Chakrabarty, A.M.; Bender, C.L. Copper as a signal for alginate synthesis in Pseudomonas syringae pv. syringae. Microbiology 1995, 61, 2172–2179. [Google Scholar] [CrossRef] [PubMed]
- Ophir, T.; Gutnick, D.L. A role for exopolysaccharides in the protection of microorganisms from desiccation. Appl. Environ. Microbiol. 1994, 60, 740–745. [Google Scholar] [CrossRef]
- Dong, C.J.; Wang, L.L.; Li, Q.; Shang, Q.M. Bacterial communities in the rhizosphere, phyllosphere and endosphere of tomato plants. PLoS ONE 2019, 14, e0223847. [Google Scholar] [CrossRef]
- Fageria, N.K.; Stone, L.F. Physical, chemical, and biological changes in the rhizosphere and nutrient availability. J. Plant Nutr. 2006, 29, 1327–1356. [Google Scholar] [CrossRef]
- Thies, J.E.; Grossman, J.M. The soil habitat and soil ecology. In Biological Approaches to Sustainable Soil Systems, 1st ed.; Uphoff, N., Ball, A.S., Fernandes, E., Herren, H., Husson, O., Laing, M., Palm, C., Pretty, J., et al., Eds.; CRC Press: Boca Raton, FL, USA, 2006; Chapter 5; p. 20. [Google Scholar]
- Bogino, P.C.; Oliva, M.; Sorroche, F.G.; Giordano, W. The role of bacterial biofilms and surface components in plant-bacterial associations. Int. J. Mol. Sci. 2013, 14, 15838–15859. [Google Scholar] [CrossRef]
- Rudrappa, T.; Biedrzycki, M.L.; Bais, H.P. Causes and consequences of plant-associated biofilms. FEMS Microbiol. Ecol. 2008, 64, 153–166. [Google Scholar] [CrossRef]
- Davey, M.E.; O’Toole, G.A. Microbial biofilms: From ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 2000, 64, 847–867. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.M.; de Leij, F. Rhizosphere. In eLS; Key Concepts; John Wiley & Sons, Ltd.: Chichester, UK, 2012. [Google Scholar] [CrossRef]
- Sandhya, V.; Ali, S.Z. The production of exopolysaccharide by Pseudomonas putida GAP-P45 under various abiotic stress conditions and its role in soil aggregation. Microbiology 2015, 84, 512–519. [Google Scholar] [CrossRef]
- Costa, O.Y.A.; Raaijmakers, J.M.; Kuramae, E.E. Microbial extracellular polymeric substances: Ecological function and impact on soil aggregation. Front. Microbiol. 2018, 9, 1636. [Google Scholar] [CrossRef]
- Sjulin, T.M.; Beer, S.V. Mechanism of wilt induction by amylovorin in Cotoneaster shoots and its relation to wilting of shoots infected by Erwinia amylovora. Phytopathology 1978, 68, 89. [Google Scholar] [CrossRef]
- Koczan, J.M.; McGrath, M.J.; Zhao, Y.; Sundin, G.W. Contribution of Erwinia amylovora exopolysaccharides amylovoran and levan to biofilm formation: Implications in pathogenicity. Phytopathology 2009, 99, 1237–1244. [Google Scholar] [CrossRef]
- Koczan, J.M.; Lenneman, B.R.; McGrath, M.J.; Sundin, G.W. Cell surface attachment structures contribute to biofilm formation and xylem colonization by Erwinia amylovora. Appl. Environ. Microbiol. 2011, 77, 7031–7039. [Google Scholar] [CrossRef] [PubMed]
- Mori, Y.; Inoue, K.; Ikeda, K.; Nakayashiki, H.; Higashimoto, C.; Ohnishi, K.; Kiba, A.; Hikichi, Y. The vascular plant-pathogenic bacterium Ralstonia solanacearum produces biofilms required for its virulence on the surfaces of tomato cells adjacent to intercellular spaces. Mol. Plant Pathol. 2016, 17, 890–902. [Google Scholar] [CrossRef] [PubMed]
- Marques, L.L.R.; Ceri, H.; Manfio, G.P.; Reid, D.M.; Olson, M.E. Characterization of biofilm formation by Xylella fastidiosa in vitro. Plant Dis. 2020, 86, 633–638. [Google Scholar] [CrossRef]
- Killiny, N.; Martinez, R.H.; Dumenyo, C.K.; Cooksey, D.A.; Almeida, R.P. The exopolysaccharide of Xylella fastidiosa is essential for biofilm formation, plant virulence, and vector transmission. Mol. Plant Microbe Interact. 2013, 26, 1044–1053. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gomila, M.; Busquets, A.; Mulet, M.; García-Valdés, E.; Lalucat, J. Clarification of taxonomic status within the Pseudomonas syringae species group based on a phylogenomic analysis. Front. Microbiol. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Gutiérrez-Barranquero, J.A.; Cazorla, F.M.; de Vicente, A. Pseudomonas syringae pv. syringae associated with mango trees, a particular pathogen within the “hodgepodge” of the Pseudomonas syringae complex. Front. Plant Sci. 2019, 10, 570. [Google Scholar] [CrossRef]
- Lindow, S.E.; Brandl, M.T. Microbiology of the phyllosphere. Appl Environ. Microbiol. 2003, 69, 1875–1883. [Google Scholar] [CrossRef]
- Xin, X.F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 139–148. [Google Scholar] [CrossRef]
- Ichinose, Y.; Taguchi, F.; Mukaihara, T. Pathogenicity and virulence factors of Pseudomonas syringae. J. Gen. Plant Pathol. 2013, 79, 285–296. [Google Scholar] [CrossRef]
- Sundin, G.W.; Kidambi, S.P.; Ullrich, M.; Bender, C.L. Resistance to ultraviolet light in Pseudomonas syringae: Sequence and functional analysis of the plasmid-encoded rulAB genes. Gene 1996, 177, 77–81. [Google Scholar] [CrossRef]
- Cazorla, F.M.; Arrebola, E.; Sesma, A.; Pérez-García, A.; Codina, J.C.; Murillo, J.; de Vicente, A. Copper resistance in Pseudomonas syringae strains isolated from mango is encoded mainly by plasmids. Phytopathology 2002, 92, 909–916. [Google Scholar] [CrossRef]
- Gutiérrez-Barranquero, J.A.; de Vicente, A.; Carrión, V.J.; Sundin, G.W.; Cazorla, F.M. Recruitment and rearrangement of three different genetic determinants into a conjugative plasmid increase copper resistance in Pseudomonas syringae. Appl. Environ. Microbiol. 2013, 79, 1028–1033. [Google Scholar] [CrossRef]
- Gutiérrez-Barranquero, J.A.; Cazorla, F.M.; de Vicente, A.; Sundin, G.W. Complete sequence and comparative genomic analysis of eight native Pseudomonas syringae plasmids belonging to the pPT23A family. BMC Genom. 2017, 18, 365. [Google Scholar] [CrossRef]
- Aprile, F.; Heredia-Ponce, Z.; Cazorla, F.M.; de Vicente, A.; Gutiérrez-Barranquero, J.A. A large Tn7-like transposon confers hyper-resistance to copper in Pseudomonas syringae pv. syringae. Appl. Environ. Microbiol. 2021, 87, e02528-20. [Google Scholar] [CrossRef]
- Heredia-Ponce, Z.; Gutiérrez-Barranquero, J.A.; Purtschert-Montenegro, G.; Eberl, L.; Cazorla, F.M.; de Vicente, A. Biological role of EPS from Pseudomonas syringae pv. syringae UMAF0158 extracellular matrix, focusing on a Psl-like polysaccharide. NPJ Biofilms Microbiomes 2020, 6, 37. [Google Scholar] [CrossRef]
- Chen, Y.; Yan, F.; Chai, Y.; Liu, H.; Kolter, R.; Losick, R.; Guo, J.H. Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ. Microbiol. 2013, 15, 848–864. [Google Scholar] [CrossRef] [PubMed]
- Gal, M.; Preston, G.M.; Massey, R.C.; Spiers, A.J.; Rainey, P.B. Genes encoding a cellulosic polymer contribute toward the ecological success of Pseudomonas fluorescens SBW25 on plant surfaces. Mol. Ecol. 2003, 12, 3109–3121. [Google Scholar] [CrossRef]
- Garrido-Sanz, D.; Arrebola, E.; Martínez-Granero, F.; García-Méndez, S.; Muriel, C.; Blanco-Romero, E.; Martín, M.; Rivilla, R.; Redondo-Nieto, M. Classification of isolates from the Pseudomonas fluorescens complex into phylogenomic groups based in group-specific markers. Front. Microbiol. 2017, 8, 413. [Google Scholar] [CrossRef] [PubMed]
- Ganeshan, G.; Kumar, A.M. Pseudomonas fluorescens, a potential bacterial antagonist to control plant diseases. J. Plant Interact. 2005, 1, 123–134. [Google Scholar] [CrossRef]
- Cazorla, F.M.; Duckett, S.B.; Bergström, E.T.; Noreen, S.; Odijk, R.; Lugtenberg, B.J.J.; Thomas-Oates, J.E.; Bloemberg, G.V. Biocontrol of avocado dematophora root rot by antagonistic Pseudomonas fluorescens PCL1606 correlates with the production of 2-hexyl 5-propyl resorcinol. Mol. Plant Microbe Interact. 2006, 19, 418–428. [Google Scholar] [CrossRef]
- Anderson, A.J.; Kim, Y.C. Insights into plant-beneficial traits of probiotic Pseudomonas chlororaphis isolates. J. Med. Microbiol. 2020, 69, 361–371. [Google Scholar] [CrossRef]
- Calderón, C.E.; Pérez-García, A.; de Vicente, A.; Cazorla, F.M. The dar genes of Pseudomonas chlororaphis PCL1606 are crucial for biocontrol activity via production of the antifungal compound 2-hexyl, 5-propyl resorcinol. Mol. Plant-Microbe Interact. 2013, 26, 554–565. [Google Scholar] [CrossRef]
- Calderón, C.E.; de Vicente, A.; Cazorla, F.M. Role of 2-hexyl, 5-propyl resorcinol production by Pseudomonas chlororaphis PCL1606 in the multitrophic interactions in the avocado rhizosphere during the biocontrol process. FEMS Microbiol. Ecol. 2014, 89, 20–31. [Google Scholar] [CrossRef]
- Calderón, C.E.; Tienda, S.; Heredia-Ponce, Z.; Arrebola, E.; Cárcamo-Oyarce, G.; Eberl, L.; Cazorla, F.M. The compound 2-hexyl, 5-propyl resorcinol has a key role in biofilm formation by the biocontrol rhizobacterium Pseudomonas chlororaphis PCL1606. Front. Microbiol. 2019, 10, 369. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhuo, T.; Hu, X.; Fan, X.; Zou, H. Identification of a Pseudomonas putida as biocontrol agent for tomato bacterial wilt disease. Biol. Control 2017, 114, 45–50. [Google Scholar] [CrossRef]
- Heredia-Ponce, Z.M.; Gutiérrez-Barranquero, J.A.; Purtschert-Montenegro, G.; Eberl, L.; de Vicente, A.; Cazorla, F.M. Role of extracellular matrix components in the formation of biofilms and their contribution to the biocontrol activity of Pseudomonas chlororaphis PCL1606. Environ. Microbiol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Pandin, C.; Le Coq, D.; Canette, A.; Aymerich, S.; Briandet, R. Should the biofilm mode of life be taken into consideration for microbial biocontrol agents? Microb. Biotechnol. 2017, 10, 719–734. [Google Scholar] [CrossRef]
- Blanco-Romero, E.; Garrido-Sanz, D.; Rivilla, R.; Redondo-Nieto, M.; Martín, M. In silico characterization and phylogenetic distribution of extracellular matrix components in the model rhizobacteria Pseudomonas fluorescens F113 and other Pseudomonads. Microorganisms 2020, 8, 1740. [Google Scholar] [CrossRef]
- Nilsson, M.; Chiang, W.C.; Fazli, M.; Gjermansen, M.; Givskov, M.; Tolker-Nielsen, T. Influence of putative exopolysaccharide genes on Pseudomonas putida KT2440 biofilm stability. Environ. Microbiol. 2011, 13, 1357–1369. [Google Scholar] [CrossRef] [PubMed]
- Evans, L.R.; Linker, A. Production and characterization of the slime polysaccharide of Pseudomonas aeruginosa. J. Bacteriol. 1973, 116, 915–924. [Google Scholar] [CrossRef]
- Fegan, M.; Francis, P.; Hayward, A.C.; Davis, G.H.; Fuerst, J.A. Phenotypic conversion of Pseudomonas aeruginosa in cystic fibrosis. J. Clin. Microbiol. 1990, 28, 1143–1146. [Google Scholar] [CrossRef] [PubMed]
- Terry, J.M.; Piña, S.E.; Mattingly, S.J. Environmental conditions which influence mucoid conversion Pseudomonas aeruginosa PAO1. Infect. Immun. 1991, 59, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 2002, 15, 194–222. [Google Scholar] [CrossRef]
- Simpson, J.A.; Smith, S.E.; Dean, R.T. Alginate inhibition of the uptake of Pseudomonas aeruginosa by macrophages. J. Gen. Microbiol. 1988, 134, 29–36. [Google Scholar] [CrossRef] [PubMed]
- McIntyre-Smith, A.; Schneiderman, J.; Zhou, K. Alginate does not appear to be essential for biofilm production by PAO1 Pseudomonas aeruginosa. J. Exp. Microbiol. Immunol. 2010, 14, 63–68. [Google Scholar]
- Ghafoor, A.; Hay, I.D.; Rehm, B.H.A. Role of exopolysaccharides in Pseudomonas aeruginosa biofilm formation and architecture. Appl. Environ. Microbiol. 2011, 77, 5238–5246. [Google Scholar] [CrossRef]
- Laue, H.; Schenk, A.; Li, H.; Lambertsen, L.; Neu, T.R.; Molin, S.; Ullrich, M.S. Contribution of alginate and levan production to biofilm formation by Pseudomonas syringae. Microbiology 2006, 152, 2909–2918. [Google Scholar] [CrossRef]
- Noirot-Gros, M.F.; Forrester, S.; Malato, G.; Larsen, P.E.; Noirot, P. CRISPR interference to interrogate genes that control biofilm formation in Pseudomonas fluorescens. Sci. Rep. 2019, 9, 15954. [Google Scholar] [CrossRef]
- Goltermann, L.; Tolker-Nielsen, T. Importance of the exopolysaccharide matrix in antimicrobial tolerance of Pseudomonas aeruginosa aggregates. Antimicrob. Agents Chemother. 2017, 61, 1–7. [Google Scholar] [CrossRef]
- Rudolph, K.W.E.; Gross, M.; Ebrahim-Nesbat, F.; Nöllenburg, M.; Zomorodian, A.; Wydra, K.; Neugebauer, M.; Hettwer, U.; El-Shouny, W.; Sonnerberg, B.; et al. The role of extracellular polysaccharides as virulence factors for phytopathogenic pseudomonads and xanthomonads. In Molecular Mechanisms of Bacterial Virulence; Kado, C.I., Crosa, J.H., Eds.; Springer: Dordrech, The Netherlands, 1994; Volume 3, pp. 357–358. [Google Scholar]
- Fett, W.F.; Dunn, M.F. Exopolysaccharides produced by phytopathogenic Pseudomonas syringae pathovars in infected leaves of susceptible hosts. Plant Physiol. 1989, 89, 5–9. [Google Scholar] [CrossRef]
- Peñaloza-Vázquez, A.; Kidambi, S.P.; Chakrabarty, A.M.; Bender, C.L. Characterization of the alginate biosynthetic gene cluster in Pseudomonas syringae pv. syringae. Microbiology 1997, 179, 4464–4472. [Google Scholar] [CrossRef]
- Willis, D.K.; Holmstadt, J.J.; Kinscherf, T.G. Genetic evidence that loss of virulence associated with gacS or gacA mutations in Pseudomonas syringae B728a does not result from effects on alginate production. Appl. Environ. Microbiol. 2001, 67, 1400–1403. [Google Scholar] [CrossRef]
- Schenk, A.; Weingart, H.; Ullrich, M. The alternative sigma factor AlgT, but not alginate synthesis, promotes in planta multiplication of Pseudomonas syringae pv. glycinea. Microbiology 2008, 154, 413–421. [Google Scholar] [CrossRef]
- Nielsen, L.; Li, X.; Halverson, L.J. Cell-cell and cell-surface interactions mediated by cellulose and a novel exopolysaccharide contribute to Pseudomonas putida biofilm formation and fitness under water-limiting conditions. Environ. Microbiol. 2011, 13, 1342–1356. [Google Scholar] [CrossRef]
- Chang, W.S.; van de Mortel, M.; Nielsen, L.; Nino de Guzman, G.; Li, X.; Halverson, L.J. Alginate production by Pseudomonas putida creates a hydrated microenvironment and contributes to biofilm architecture and stress tolerance under water-limiting conditions. J. Bacteriol. 2007, 189, 8290–8299. [Google Scholar] [CrossRef] [PubMed]
- Bianciotto, V.; Andreotti, S.; Balestrini, R.; Bonfante, P.; Perotto, S. Mucoid mutants of the biocontrol strain Pseudomonas fluorescens CHA0 show increased ability in biofilm formation on mycorrhizal and nonmycorrhizal carrot roots. Mol. Plant Microbe Interact. 2001, 14, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Ramos-González, M.I.; Campos, M.J.; Ramos, J.L. Analysis of Pseudomonas putida KT2440 gene expression in the maize rhizosphere: In vivo expression technology capture and identification of root-activated promoters. J. Bacteriol. 2005, 187, 5504. [Google Scholar] [CrossRef]
- Ude, S.; Arnold, D.L.; Moon, C.D.; Timms-Wilson, T.; Spiers, A.J. Biofilm formation and cellulose expression among diverse environmental Pseudomonas isolates. Environ. Microbiol. 2006, 8, 1997–2011. [Google Scholar] [CrossRef]
- Spiers, A.J.; Deeni, Y.Y.; Folorunso, A.O.; Koza, A.; Moshynets, O.; Zawadzki, K. Cellulose expression in Pseudomonas fluorescens SBW25 and other environmental pseudomonads. In Cellulose-Medical, Pharmaceutical and Electronic Applications; van de Ven, T., Godbout, L., Eds.; IntechOpen: Rijeka, Croatia, 2013; Chapter 1. [Google Scholar] [CrossRef]
- Augimeri, R.V.; Varley, A.J.; Strap, J.L. Establishing a role for bacterial cellulose in environmental interactions: Lessons learned from diverse biofilm-producing Proteobacteria. Front. Microbiol. 2015, 6, 1282. [Google Scholar] [CrossRef] [PubMed]
- Spiers, A.J.; Bohannon, J.; Gehrig, S.M.; Rainey, P.B. Biofilm formation at the air-liquid interface by the Pseudomonas fluorescens SBW25 wrinkly spreader requires an acetylated form of cellulose. Mol. Microbiol. 2003, 50, 15–27. [Google Scholar] [CrossRef]
- Armitano, J.; Méjean, V.; Jourlin-Castelli, C. Floating biofilm in Gram-negative bacteria. Environ. Microbiol. Rep. 2014, 6, 534–544. [Google Scholar] [CrossRef]
- Ardré, M.; Dufour, D.; Rainey, P.B. Causes and biophysical consequences of cellulose production by Pseudomonas fluorescens SBW25 at the air-liquid interface. J. Bacteriol. 2019, 201, e00110-19. [Google Scholar] [CrossRef]
- Farias, G.A.; Olmedilla, A.; Gallegos, M.T. Visualization and characterization of Pseudomonas syringae pv. tomato DC3000 pellicles. Microb. Biotechnol. 2019, 12, 688–702. [Google Scholar] [CrossRef] [PubMed]
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358. [Google Scholar] [CrossRef]
- Mann, E.E.; Wozniak, D.J. Pseudomonas biofilm matrix composition and niche biology. FEMS Microbiol. 2012, 36, 893–916. [Google Scholar] [CrossRef]
- Friedman, L.; Kolter, R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 2004, 186, 4457–4465. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Mendoza, D.; Aragón, I.M.; Prada-Ramírez, H.A.; Romero-Jiménez, L.; Ramos, C.; Gallegos, M.T.; Sanjuán, J. Responses to elevated c-di-GMP levels in mutualistic and pathogenic plant-interacting bacteria. PLoS ONE 2014, 9, e91645. [Google Scholar] [CrossRef]
- Cazorla, F.M.; Torés, J.A.; Olalla, L.; Pérez-García, A.; Farré, J.M.; de Vicente, A. Bacterial apical necrosis of mango in Southern Spain: A disease caused by Pseudomonas syringae pv. syringae. Phytopathology 1998, 88, 614–620. [Google Scholar] [CrossRef]
- Prada-Ramírez, H.A.; Pérez-Mendoza, D.; Felipe, A.; Martínez-Granero, F.; Rivilla, R.; Sanjuán, J.; Gallegos, M.T. AmrZ regulates cellulose production in Pseudomonas syringae pv. tomato DC3000. Mol. Microbiol. 2016, 99, 960–977. [Google Scholar] [CrossRef] [PubMed]
- Rainey, P.B.; Travisano, M. Adaptive radiation in a heterogeneous environment. Nature 1998, 394, 69–72. [Google Scholar] [CrossRef] [PubMed]
- Spiers, A.J.; Kahn, S.G.; Bohannon, J.; Travisano, M.; Rainey, P.B. Adaptive divergence in experimental populations of Pseudomonas fluorescens. Genetics 2002, 161, 33–46. [Google Scholar]
- Nakazawa, T. Travels of a Pseudomonas, from Japan around the world. Environ. Microbiol. 2002, 4, 782–786. [Google Scholar] [CrossRef]
- Svenningsen, N.B.; Martínez-García, E.; Nicolaisen, M.H.; de Lorenzo, V.; Nybroe, O. The biofilm matrix polysaccharides cellulose and alginate both protect Pseudomonas putida mt-2 against reactive oxygen species generated under matric stress and copper exposure. Microbiology 2018, 164, 883–888. [Google Scholar] [CrossRef]
- Jackson, K.D.; Starkey, M.; Kremer, S.; Parsek, M.R.; Wozniak, D.J. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 2004, 186, 4466–4475. [Google Scholar] [CrossRef] [PubMed]
- Matsukawa, M.; Greenberg, E.P. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 2004, 186, 4449–4456. [Google Scholar] [CrossRef]
- Byrd, M.S.; Sadovskaya, I.; Vinogradov, E.; Lu, H.; Sprinkle, A.B.; Richardson, S.H.; Ma, L.; Ralston, B.; Parsek, M.R.; Anderson, E.M.; et al. Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol. Microbiol. 2009, 73, 622–638. [Google Scholar] [CrossRef] [PubMed]
- Goodman, A.L.; Kulasekara, B.; Rietsch, A.; Boyd, D.; Smith, R.S.; Lory, S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 2004, 7, 745–754. [Google Scholar] [CrossRef] [PubMed]
- Hickman, J.W.; Tifrea, D.F.; Harwood, C.S. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA 2005, 102, 14422–14427. [Google Scholar] [CrossRef] [PubMed]
- Starkey, M.; Hickman, J.H.; Ma, L.; Zhang, N.; De Long, S.; Hinz, A.; Palacios, S.; Manoil, C.; Kirisits, M.J.; Starner, T.D.; et al. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J. Bacteriol. 2009, 191, 3492–3503. [Google Scholar] [CrossRef] [PubMed]
- Colvin, K.M.; Irie, Y.; Tart, C.S.; Urbano, R.; Whitney, J.C.; Ryder, C.; Howell, P.L.; Wozniak, D.J.; Parsek, M.R. The Pel and Psl polysaccharides provide Pseudomonas aeruginosa structural redundancy within the biofilm matrix. Environ. Microbiol. 2012, 14, 1913–1918. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Parsek, M.R.; Wozniak, D.J.; Ma, L.Z. A spider web strategy of type IV pili-mediated migration to build a fibre-like Psl polysaccharide matrix in Pseudomonas aeruginosa biofilms. Environ. Micriobiol 2013, 15, 2238–2253. [Google Scholar] [CrossRef]
- Wang, S.; Yu, S.; Zhang, Z.; Wei, Q.; Yan, L.; Ai, G.; Liu, H.; Ma, L.Z. Coordination of swarming motility, biosurfactant synthesis, and biofilm matrix exopolysaccharide production in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2014, 80, 6724–6732. [Google Scholar] [CrossRef] [PubMed]
- Periasamy, S.; Nair, H.A.S.; Lee, K.W.K.; Ong, J.; Goh, J.Q.J.; Kjelleberg, S.; Rice, S.A. Pseudomonas aeruginosa PAO1 exopolysaccharides are important for mixed species biofilm community development and stress tolerance. Front. Microbiol. 2015, 6, 851. [Google Scholar] [CrossRef]
- Ma, L.; Lu, H.; Sprinkle, A.; Parsek, M.R.; Wozniak, D.J. Pseudomonas aeruginosa Psl is a galactose- and mannose-rich exopolysaccharide. J. Bacteriol. 2007, 189, 8353–8356. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Jiang, K.; Li, W.; Zhang, Q.; Yan, G.; Guo, K.; Zhang, S.; Liu, Y. GP73 N-glycosylation at Asn144 reduces hepatocellular carcinoma cell motility and invasiveness. Oncotarget 2016, 7, 23530–23541. [Google Scholar] [CrossRef] [PubMed]
- Fett, W.F.; Osman, S.F.; Dunn, M.F. Characterization of exopolysaccharides produced by plant-associated fluorescent Pseudomonads. Appl. Environ. Microbiol. 1989, 55, 579–583. [Google Scholar] [CrossRef]
- Singh, S.S.; Devi, S.K.; Ng, T.B. Banana lectin: A brief review. Molecules 2014, 19, 18817–18827. [Google Scholar] [CrossRef] [PubMed]
- Bloemberg, G.V.; Lugtenberg, B.J.J. Molecular basis of plant growth promotion and biocontrol by rhizobacteria. Curr. Opin. Plant Biol. 2001, 4, 343–350. [Google Scholar] [CrossRef]
- Chin-A-Woeng, T.F.C.; Bloemberg, G.V.; Mulders, I.H.M.; Dekkers, L.C.; Lugtenberg, B.J.J. Root colonization by phenazine-1-carboxamide-producing bacterium Pseudomonas chlororaphis PCL1391 is essential for biocontrol of tomato foot and root rot. Mol. Plant Microbe Interact. 2007, 13, 1340–1345. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Schnider-Keel, U.; Lejbølle, K.B.; Baehler, E.; Haas, D.; Keel, C. The sigma factor AlgU (AlgT) controls exopolysaccharide production and tolerance towards desiccation and osmotic stress in the biocontrol agent Pseudomonas fluorescens CHA0. Appl. Environ. Microbiol. 2001, 67, 5683–5693. [Google Scholar] [CrossRef] [PubMed]
- Reichhardt, C.; Wong, C.; da Silva, D.P.; Wozniak, D.J.; Parsek, M.R. CdrA interactions within the Pseudomonas aeruginosa biofilm matrix safeguard it from proteolysis and promote cellular packing. mBio 2018, 9, e01376-18. [Google Scholar] [CrossRef]
- Hoštacká, A.; Čižnár, I.; Štefkovičová, M. Temperature and pH affect the production of bacterial biofilm. Folia Microbiol. 2010, 55, 75–78. [Google Scholar] [CrossRef]
- Townsley, L.; Yildiz, F.H. Temperature affects c-di-GMP signalling and biofilm formation in Vibrio cholerae. Environ. Microbiol. 2015, 17, 4290–4305. [Google Scholar] [CrossRef]
- Hakimzadeh, A.; Okshevsky, M.; Maisuria, V.; Déziel, E.; Tufenkji, N. Exposure to freeze-thaw conditions increases virulence of Pseudomonas aeruginosa to Drosophila melanogaster. Environ. Sci. Technol. 2018, 52, 14180–14186. [Google Scholar] [CrossRef] [PubMed]
- Khelissa, S.O.; Abdallah, M.; Jama, C.; Barras, A.; Chihib, N.E. Comparative study on the impact of growth conditions on the physiology and the virulence of Pseudomonas aeruginosa biofilm and planktonic cells. J. Food Prot. 2019, 82, 1357–1363. [Google Scholar] [CrossRef]
- Río-Álvarez, I.; Rodríguez-Herva, J.J.; Martínez, P.M.; González-Melendi, P.; García-Casado, G.; Rodríguez-Palenzuela, P.; López-Solanilla, E. Light regulates motility, attachment and virulence in the plant pathogen Pseudomonas syringae pv. tomato DC3000. Environ. Microbiol. 2014, 16, 2072–2085. [Google Scholar] [CrossRef]
- Kim, S.; Li, X.H.; Hwang, H.J.; Lee, J.H. Thermoregulation of Pseudomonas aeruginosa biofilm formation. Appl. Environ. Microbiol. 2020, 86, 1–11. [Google Scholar] [CrossRef]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef] [PubMed]
- Reinhold-Hurek, B.; Bünger, W.; Burbano, C.S.; Sabale, M.; Hurek, T. Root shaping their microbiome: Global hotspots for microbial activity. Annu. Rev. Phytopathol. 2015, 53, 403–424. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Company, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden word within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Burmølle, M.; Webb, J.S.; Rao, D.; Hansen, L.H.; Sørensen, S.J.; Kjelleberg, S. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl. Environ. Microbiol. 2006, 72, 3916–3923. [Google Scholar] [CrossRef]
- Lopes, S.P.; Ceri, H.; Azevedo, N.F.; Pereira, M.O. Antibiotic resistance of mixed biofilms in cystic fibrosis: Impact of emerging microorganisms on treatment of infection. Int. J. Antimicrob. Agents 2012, 40, 260–263. [Google Scholar] [CrossRef] [PubMed]
- Molina-Santiago, C.; Pearson, J.R.; Navarro, Y.; Berlanga-Clavero, M.V.; Caraballo-Rodriguez, A.M.; Petras, D.; García-Martín, M.; Lamon, G.; Haberstein, B.; Cazorla, F.M.; et al. The extracellular matrix protects Bacillus subtilis colonies from Pseudomonas invasion and modulates plant co-colonization. Nat. Commun. 2019, 10, 1919. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).