Benchmarking DNA Extraction Methods for Phylogenomic Analysis of Sub-Antarctic Rhodococcus and Williamsia Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Benchmarking Extraction and Purification of gDNA
2.3. Quantity, Fragment Size and Purity of Extracted DNA
2.4. gDNA Sequencing and Quality Assessment
2.5. 16S rRNA Gene Sequencing and Preliminary Phylogenetic Analysis
2.6. Genome Sequence Analyses to Identify Rhodococcus Species
2.7. Biochemical Characterization of Strains 1139 and 1159
2.7.1. Enzymatic Tests to Characterize Bacterial Strains
2.7.2. Growth on Different Carbon and Nitrogen Sources and Impact of Phosphate Concentration on Growth
2.7.3. Salt Tolerance Testing
2.7.4. Determining Optimum Temperature for Growth in MSM-F Broth
2.7.5. Modelling Minimum and Maximum Growth Temperatures
2.7.6. Triacylglycerol (TAG) Analysis
2.7.7. Fatty Acid Methyl Ester (FAME) Analysis by GC–MS
2.8. Analysis of Mycolic Acids (MA)
3. Results
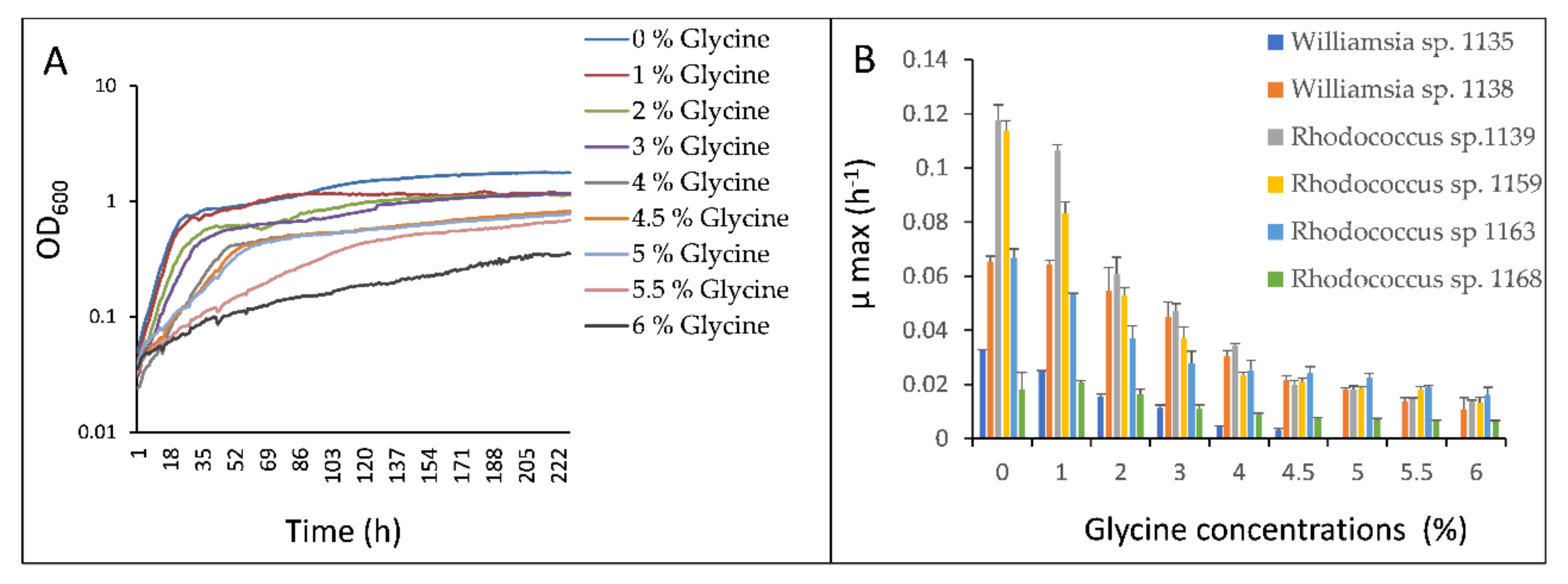
3.1. Impacts of Glycine on Bacterial Growth
3.2. Quantity and Purity of Extracted DNA
3.3. DNA Sequencing and Quality Score of Sequenced DNA
3.4. Preliminary Strain Identification Based on Sequencing 16S rRNA Gene PCR Amplicons
3.5. Taxonomy of Rhodococcus Isolates Based on Analysis of Sequenced Genomes
3.6. Taxonomy of Williamsia Strains Based on Mycolic Acid and Genome Sequence Analyses
3.7. Biochemical Characterization of the Rhodococcus Strains 1139 and 1159
3.7.1. Enzymatic Characterization of Bacterial Strains and Carbon Source Utilization
3.7.2. Identifying Preferred Nitrogen Sources and Impact of Phosphate Ion Concentration
3.7.3. Salt Tolerance
3.7.4. Strains 1139 and 1159 Are Psychrotolerant and Predicted to Survive at Sub-Zero Temperatures
3.7.5. MA Analysis by UPLC–MS/MS
3.7.6. TAG Analysis by UPLC–MS/MS
3.7.7. FAME Analysis by GC–MS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Mayjonade, B.; Gouzy, J.; Donnadieu, C.; Pouilly, N.; Marande, W.; Callot, C.; Langlade, N.; Muños, S. Extraction of high-molecular-weight genomic DNA for long-read sequencing of single molecules. Biotechniques 2016, 61, 203–205. [Google Scholar] [CrossRef] [PubMed]
- Buermans, H.; Den Dunnen, J. Next generation sequencing technology: Advances and applications. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 1932–1941. [Google Scholar] [CrossRef] [PubMed]
- Healey, A.; Furtado, A.; Cooper, T.; Henry, R.J. Protocol: A simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 2014, 10, 21. [Google Scholar] [CrossRef]
- Kitagawa, W.; Kimura, N.; Kamagata, Y. A novel p-nitrophenol degradation gene cluster from a gram-positive bacterium, Rhodococcus opacus SAO101. J. Bacteriol. 2004, 186, 4894–4902. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.-Q.; Xu, L.; Tang, Y.-Q.; Chen, F.-M.; Liu, W.-Q.; Wu, X.-L. Degradation of pyridine by one Rhodococcus strain in the presence of chromium (VI) or phenol. J. Hazard. Mater. 2011, 191, 62–68. [Google Scholar] [CrossRef]
- Inoue, D.; Tsunoda, T.; Yamamoto, N.; Ike, M.; Sei, K. 1, 4-Dioxane degradation characteristics of Rhodococcus aetherivorans JCM 14343. Biodegradation 2018, 29, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, H.; Steinbüchel, A. Triacylglycerols in prokaryotic microorganisms. Appl. Microbiol. Biotechnol. 2002, 60, 367–376. [Google Scholar] [CrossRef] [PubMed]
- Holder, J.W.; Ulrich, J.C.; DeBono, A.C.; Godfrey, P.A.; Desjardins, C.A.; Zucker, J.; Zeng, Q.; Leach, A.L.; Ghiviriga, I.; Dancel, C. Comparative and functional genomics of Rhodococcus opacus PD630 for biofuels development. PLoS Genet. 2011, 7, e1002219. [Google Scholar] [CrossRef]
- Shavandi, M.; Mohebali, G.; Haddadi, A.; Shakarami, H.; Nuhi, A. Emulsification potential of a newly isolated biosurfactant-producing bacterium, Rhodococcus sp. strain TA6. Colloids Surf. B. Biointerfaces 2011, 82, 477–482. [Google Scholar] [CrossRef]
- Dhakal, D.; Shrestha, A.; Thuan, N.H.; Rayamajhi, V.; Mishra, R.; Magar, R.T.; Sohng, J.K. Bioactive Compounds from Nocardia: Biosynthesis and Production. In Pharmaceuticals from Microbes; Arora, D., Sharma, C., Jaglan, S., Lichtfouse, E., Eds.; Springer: Cham, Switzerland, 2019; Volume 28, pp. 49–74. [Google Scholar] [CrossRef]
- Mischitz, M.; Faber, K.; Willetts, A. Isolation of a highly enantioselective epoxide hydrolase from Rhodococcus sp. NCIMB 11216. Biotechnol. Lett. 1995, 17, 893–898. [Google Scholar] [CrossRef]
- Hébert, L.; Bidaud, P.; Goux, D.; Benachour, A.; Laugier, C.; Petry, S. Study of lysozyme resistance in Rhodococcus equi. Curr. Microbiol. 2014, 68, 352–357. [Google Scholar] [CrossRef]
- Amaro, A.; Duarte, E.; Amado, A.; Ferronha, H.; Botelho, A. Comparison of three DNA extraction methods for Mycobacterium bovis, Mycobacterium tuberculosis and Mycobacterium avium subsp. avium. Lett. Appl. Microbiol. 2008, 47, 8–11. [Google Scholar] [CrossRef]
- Marmur, J.A. Procedure for the isolation of deoxyribonucleic acid from microorganisms. J. Mol. Biol. 1961, 3, 201–218. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, NY, USA, 1989; ISBN 0879693096. [Google Scholar]
- Wu, G.D.; Lewis, J.D.; Hoffmann, C.; Chen, Y.-Y.; Knight, R.; Bittinger, K.; Hwang, J.; Chen, J.; Berkowsky, R.; Nessel, L. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010, 10, 206. [Google Scholar] [CrossRef] [PubMed]
- Gontang, E.A.; Fenical, W.; Jensen, P.R. Phylogenetic diversity of gram-positive bacteria cultured from marine sediments. Appl. Environ. Microbiol. 2007, 73, 3272–3282. [Google Scholar] [CrossRef] [PubMed]
- Schorn, M.A.; Alanjary, M.M.; Aguinaldo, K.; Korobeynikov, A.; Podell, S.; Patin, N.; Lincecum, T.; Jensen, P.R.; Ziemert, N.; Moore, B.S. Sequencing rare marine actinomycete genomes reveals high density of unique natural product biosynthetic gene clusters. Microbiology 2016, 162, 2075–2086. [Google Scholar] [CrossRef]
- Hammes, W.; Schleifer, K.; Kandler, O. Mode of action of glycine on the biosynthesis of peptidoglycan. J. Bacteriol. 1973, 116, 1029–1053. [Google Scholar] [CrossRef]
- Sato, H.; Diena, B.; Greenberg, L. The production of spheroplasts by rapid-growing non-virulent mycobacteria. Can. J. Microbiol. 1965, 11, 807–810. [Google Scholar] [CrossRef]
- Haynes, J.A.; Britz, M.L. The effect of growth conditions of Corynebacterium glutamicum on the transformation frequency obtained by electroporation. Microbiology 1990, 136, 255–263. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.-W.; De Meyer, S. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Švec, P.; Černohlávková, J.; Busse, H.-J.; Vojtková, H.; Pantu, R.; Cnockaert, M.; Mašlaňová, I.; Králová, S.; Vandamme, P.; Sedláček, I. Classification of strain CCM 4446T as Rhodococcus degradans sp. nov. Int. J. Syst. Evol. Microbiol. 2015, 65, 4381–4387. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.D.; Kim, I.S.; Kim, Y.J.; Young, Y. Rhodococcus cavernicola sp. nov., isolated from a cave, and Rhodococcus degradans is a later heterosynonym of Rhodococcus qingshengii. Int. J. Syst. Evol. Microbiol. 2020, 70, 4409–4415. [Google Scholar] [CrossRef] [PubMed]
- Nouioui, I.; Carro, L.; García-López, M.; Meier-Kolthoff, J.; Woyke, T.; Kyrpides, N.; Klenk, H.-P.; Goodfellow, M.; Göker, M. Genome-based taxonomic classification of the phylum Actinobacteria. Front. Microbiol. 2018, 9, 2007. [Google Scholar] [CrossRef] [PubMed]
- Táncsics, A.; Benedek, T.; Farkas, M.; Máthé, I.; Márialigeti, K.; Szoboszlay, S.; Kukolya, J.; Kriszt, B. Sequence analysis of 16S rRNA, gyrB and catA genes and DNA–DNA hybridization reveal that Rhodococcus jialingiae is a later synonym of Rhodococcus qingshengii. Int. J. Syst. Evol. Microbiol. 2014, 64, 298–301. [Google Scholar] [CrossRef]
- Nahar, A.; Baker, A.L.; Charleston, M.A.; Bowman, J.P.; Britz, M.L. Draft genome sequences of three sub-Antarctic Rhodococcus spp., including two movel psychrophilic genomospecies. Microbiol. Resour. Announc. 2017, 5, e00898-17. [Google Scholar] [CrossRef]
- Nahar, A.; Baker, A.L.; Charleston, M.A.; Bowman, J.P.; Britz, M.L. Draft genome sequences of two novel sub-Antarctic Williamsia species. Microbiol. Resour. Announc. 2017, 5, e01047-17. [Google Scholar] [CrossRef] [PubMed]
- Nahar, A.; Baker, A.L.; Charleston, M.A.; Britz, M.L. Draft genome sequence of subantarctic Rhodococcus sp. strain 1139. Microbiol. Resour. Announc. 2017, 5, e00090-17. [Google Scholar] [CrossRef]
- Monod, J. The growth of bacterial cultures. Ann. Rev. Microbiol. 1949, 3, 371–394. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Chen, I.-M.A.; Chu, K.; Palaniappan, K.; Pillay, M.; Ratner, A.; Huang, J.; Huntemann, M.; Varghese, N.; White, J.R.; Seshadri, R. IMG/M v. 5.0: An integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2018, 47, D666–D677. [Google Scholar] [CrossRef] [PubMed]
- Lincoln, S.A.; Hamilton, T.L.; Juárez, A.G.V.; Schedler, M.; Macalady, J.L.; Müller, R.; Freeman, K.H. Draft genome sequence of the piezotolerant and crude oil-degrading bacterium Rhodococcus qingshengii strain TUHH-12. Microbiol. Resour. Announc. 2015, 3, e00268-15. [Google Scholar] [CrossRef]
- Parte, A.C. LPSN-List of prokaryotic names with standing in nomenclature (bacterio. net), 20 years on. Int. J. Syst. Evol. Microbiol. 2018, 68, 1825–1829. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, msw054. [Google Scholar] [CrossRef]
- Táncsics, A.; Benedek, T.; Szoboszlay, S.; Veres, P.G.; Farkas, M.; Máthé, I.; Márialigeti, K.; Kukolya, J.; Lányi, S.; Kriszt, B. The detection and phylogenetic analysis of the alkane 1-monooxygenase gene of members of the genus Rhodococcus. Syst. Appl. Microbiol. 2015, 38, 1–7. [Google Scholar] [CrossRef]
- Ratkowsky, D.; Olley, J.; McMeekin, T.; Ball, A. Relationship between temperature and growth rate of bacterial cultures. J. Bacteriol. 1982, 149, 1–5. [Google Scholar] [CrossRef]
- Elzhov, T.V.; Mullen, K.M.; Spiess, A.-N.; Bolker, B.; Mullen, M.K.M.; Suggests, M. Package ‘minpack.lm’. 2016. Available online: https://rdocumentation.org/packages/minpack.lm/versions/1.2-1 (accessed on 1 December 2018).
- McLean, S.; Davies, N.W.; Nichols, D.S.; Mcleod, B.J. Triacylglycerol estolides, a new class of mammalian lipids, in the paracloacal gland of the brushtail possum (Trichosurus vulpecula). Lipids 2015, 50, 591–604. [Google Scholar] [CrossRef]
- Wright, E.S.; Baum, D.A. Exclusivity offers a sound yet practical species criterion for bacteria despite abundant gene flow. BMC Genom. 2018, 19, 724. [Google Scholar] [CrossRef]
- Nahar, A.; Baker, A.L.; Nichols, D.S.; Bowman, J.P.; Britz, M.L. Application of thin-layer chromatography-flame ionization detection (TLC-FID) to total lipid quantitation in mycolic-acid synthesizing Rhodococcus and Williamsia species. Int. J. Mol. Sci. 2020, 21, 1670. [Google Scholar] [CrossRef] [PubMed]
- Australian Government, Department of Agriculture, Water and the Environment, World Heritage Places, Macquarie Island. Available online: Htpp://environment.gov.au/heritage/places/world/Macquarie-island (accessed on 20 May 2021).
- Sangal, V.; Goodfellow, M.; Jones, A.L.; Schwalbe, E.C.; Blom, J.; Hoskisson, P.A.; Sutcliffe, I.C. Next-generation systematics: An innovative approach to resolve the structure of complex prokaryotic taxa. Sci. Rep. 2016, 6, 38392. [Google Scholar] [CrossRef] [PubMed]
- Baek, I.; Kim, M.; Lee, I.; Na, S.-I.; Goodfellow, M.; Chun, J. Phylogeny trumps chemotaxonomy: A case study involving Turicella otitidis. Front. Microbiol. 2018, 9, 834. [Google Scholar] [CrossRef]
- Tyler, A.D.; Christianson, S.; Knox, N.C.; Mabon, P.; Wolfe, J.; Van Domselaar, G.; Graham, M.R.; Sharma, M.K. Comparison of sample preparation methods used for the next-generation sequencing of Mycobacterium tuberculosis. PLoS ONE 2016, 11, e0148676. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Chuvochina, M.; Chaumeil, P.A.; Rike, C.; Mussig, A.J.; Hugenholtz, P. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 2020, 38, 1079–1086. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Hishinuma, F.; Izaki, K.; Takahashi, H. Effects of glycine and D-amino acids on growth of various microorganisms. Agric. Biol. Chem. 1969, 33, 1577–1586. [Google Scholar] [CrossRef]
- Lechevalier, M.; Prauser, H.; Labeda, D.; Ruan, J.-S. 2 new genera of nocardioform actinomycetes: Amycolata gen. nov. and Amycolatopsis gen. nov. Int. J. Syst. Evol. Microbiol. 1986, 36, 29–37. [Google Scholar] [CrossRef]
- Kampfer, P.; Anderson, M.A.; Raine, F.A.; Kroppenstedt, R.M.; Salkinoja-Salonen, M. Williamsia muralis gen. nov., sp. nov., isolated from the indoor environment of a children’s day care centre. Int. J. Syst. Bacteriol. 1999, 49, 681–687. [Google Scholar] [CrossRef][Green Version]
- Sazak, A.; Sahin, N. Williamsia limnetica sp. nov., isolated from a limnetic lake sediment. Int. J. Syst. Evol. Microbiol. 2012, 62, 1414–1418. [Google Scholar] [CrossRef] [PubMed]
- Sutcliffe, I.C. Cell envelope composition and organisation in the genus Rhodococcus. Antonie Van Leeuwenhoek 1998, 74, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Sutcliffe, I.C.; Brown, A.K.; Dover, L.G. The rhodococcal cell envelope: Composition, organisation and biosynthesis. In Biology of Rhodococcus; Microbiology Monographs 16; Alvarez, H.M., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 30–71. [Google Scholar] [CrossRef]
- Goodfellow, M.; Alderson, G. The Actinomycete-genus Rhodococcus: Home for the ‘rhodochrous’ complex. J. Gen. Microbiol. 1977, 100, 99–122. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.-L.; He, J.; Wang, Z.-C.; Wang, K.; Li, W.-J.; Tang, S.K.; Li, S.-P. Rhodococcus qingshengii sp. nov., a carbendazim-degrading bacterium. Int. J. Syst. Evol. Microbiol. 2007, 57, 2754–2757. [Google Scholar] [CrossRef]


| Parameters | Method 1 Isolate II Genomic DNA Kit (Bioline) | Method 2 Ultraclean Microbial DNA Isolation Kit (MO BIO) | Method 3 QIAAMP Mini Kit (Qiagen) | Method 4 Classical Extraction | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0% a | 2% a | 4.5% a | E. coli | 0% a | 2% a | 4.5% a | E. coli | 0% a | 2% a | 4.5% a | E. coli | 0% a | 2% a | 4.5% a | |
| Mean concentration of DNA (ng/µL) ± s.d. b | 10 ± 1 | 39 ± 3.2 | 69 ± 2 | 295 ± 2 | 25.7 ± 1.1 | 45.7 ± 4.9 | 60.2 ± 6.7 | 74 ± 3.5 | 17 ± 2.6 | 42.2 ± 3.2 | 71.7 ± 3.5 | 79.4 ± 2.5 | 1.5 ± 0.35 | 6 ± 1.64 | 30.4 ± 4.1 |
| Mean GQN ± s.d. b | 0.9 ± 0.07 | 1.0 ± 0.3 | 0.2 ± 0.4 | 6.5 ± 0.3 | 5.9 ± 0.06 | 8.1 ± 0.04 | 8.4 ± 0.12 | 9.2 ± 0.03 | 6.5 ± 0.56 | 7.8 ± 0.35 | 7.6 ± 0.35 | 8.6 ± 0.1 | 1.8 ± 0.04 | 6.5 ± 0.28 | 9.0 ± 0.07 |
| Fragment size range (bp) b | 823–9763 | 557–1321 | 27–388 | 991–39,079 | 2019–40,109 | 2849–54,726 | 2755–54,903 | 1913–48,525 | 8549–49,782 | 7605–55,301 | 2891–50,707 | 6787–52,428 | 0–14 | 22,636–44,933 | 16,591–42,748 |
| Average fragment size (bp) b | 9554 | 490 | 128 | 13,975 | 21,246 | 18,923 | 17,284 | 26,027 | 24,249 | 26,555 | 23,410 | 32,406 | 2.0 | 33,581 | 28,430 |
| Mean A260/A280 ± s.d. c | 2.0 ± 0.09 | 2.0 ± 0.04 | 2.0 ± 0.06 | 2.1 ± 0.03 | 2.0 ± 0.02 | 1.99 ± 0.04 | 1.88 ± 0 | 1.98 ± 0.03 | 2.1 ± 10.04 | 2.1 ± 0.01 | 2.1 ± 0.01 | 2.1 ± 0.09 | 1.96 ± 0.09 | 1.69 ± 0.04 | 1.70 ± 0.06 |
| Mean A260/A230 ± s.d. c | 2.1 ± 0.03 | 1.9 ± 0.12 | 1.8 ± 0.03 | 1.9 ± 0.06 | 1.3 ± 0.12 | 1.4 ± 0.16 | 1.6 ± 0.06 | 1.6 ± 0.09 | 1.1 ± 0.03 | 2.1 ± 0.03 | 2.1 ± 0.03 | 2.1 ± 0.06 | 0.91 ± 0.3 | 1.65 ± 0.12 | 1.71 ± 0.03 |
| Final volume (µL) | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 100 | 100 | 100 | 200 | 50 | 50 | 50 |
| Strain Name | ng/µL a | GQN a | Fragment Size Range (Kbp) a | Average Size (Kbp) a | A 260/280 b | A 260/230 b | Q30 (%) c | N50 d (Kbp) | L50 d | Contigs d | GC Content (%) d |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Williamsia sp. 1135 | 66.1 | 8.1 | 13.28–44.50 | 28.721 | 1.92 | 1.84 | 64.36 | 142.77 | 14 | 109 | 64.7 |
| Williamsia sp. 1138 | 319 | 9.8 | 57.24–62.78 | 59.424 | 1.93 | 1.93 | 63.92 | 191.09 | 12 | 54 | 64.8 |
| R. qingshengii 1139 | 169.7 | 9.3 | 41.57–56.44 | 48.133 | 1.94 | 1.75 | 62.70 | 94.97 | 24 | 192 | 62.3 |
| R. erythropolis 1159 | 106.2 | 8.7 | 11.63–48.44 | 27.142 | 1.78 | 1.78 | 65.11 | 196.55 | 13 | 114 | 62.3 |
| Rhodococcus sp. 1163 | 40.5 | 8.7 | 15.73–47.69 | 32.799 | 1.99 | 1.87 | 64.67 | 329.55 | 5 | 43 | 62.3 |
| Rhodococcus sp. 1168 | 66.3 | 9.5 | 9.72–51.78 | 27.334 | 2.01 | 1.88 | 66.25 | 154.59 | 10 | 97 | 62.1 |
| Panel A | |||||||||
| Strain No. | Species | Provenance | Pairwise ANI and AF (%) a | ||||||
| CCM 2595 | NBRC 15567 | TUHH-12 | JCM 15477 | Djl-6-2 | 1139 | 1159 | |||
| CCM 2595 | R. erythropolis | NCBI reference genome for the species | 98.83 AF = 88.64 | 95.55 AF = 64.79 | 95.60 AF = 79.47 | 95.57 AF = 85.27 | 95.67 AF = 81.41 | 98.86 AF = 84.01 | |
| NBRC 15567 = JCM 3201 b | R. erythropolis | Type strains, https://lpsn.dsmz.de | 98.83 AF = 99.99 | 95.49 AF = 65.56 | 95.49 AF = 81.12 | 95.54 AF = 85.42 | 95.53 AF = 82.11 | 98.78 AF = 84.47 | |
| TUHH-12 | R. qingshengii | NCBI-cited genome publication for this species (2015) | 95.547 AF = 72.99 | 95.49 AF = 71.14 | 98.58 AF = 68.04 | 98.77 AF = 72.62 | 98.73 AF = 69.10 | 95.43 AF = 68.31 | |
| JCM 15477 c | R. qingshengii | Type strain, https://lpsn.dsmz.de | 95.60 AF = 89.60 | 95.49 AF = 88.16 | 98.57 AF = 68.11 | 98.83 AF = 89.45 | 98.67 AF = 85.23 | 95.47 AF = 83.86 | |
| Djl-6-2 | R. qingshengii | Related to original isolate djl-6 type strain, https://lpsn.dsmz.de | 95.57 AF = 85.47 | 95.54 AF = 85.32 | 98.79 AF = 66.68 | 98.82 AF = 82.23 | 98.91 AF = 82.90 | 95.53 AF = 80.41 | |
| 1139 | Rhodococcus sp. | This study | 95.66 AF = 89.01 | 95.52 AF = 86.46 | 98.73 AF = 67.09 | 98.67 AF = 82.59 | 98.91 AF = 87.35 | 95.56 AF = 83.34 | |
| 1159 | Rhodococcus sp. | This study | 98.86 AF = 92.59 | 98.78 AF = 89.77 | 95.44 AF = 66.72 | 95.48 AF = 81.83 | 95.54 AF = 85.57 | 95.55 AF = 83.95 | |
| Panel B | |||||||||
| Strain | Species | Provenance | Pairwise ANI and AF (%) | ||||||
| PAMC 28707 | 1163 | 1168 | |||||||
| PAMC 28705 PAMC 28707 | Rhodococcus sp. (identical strains) | Published genome Korean Polar Research Institute (IMG/M) | 94.50 AF = 84.60 | 93.58 AF = 74.36 | |||||
| 1163 | Rhodococcus sp. | This study | 94.50 AF = 80.13 | 93.75 AF = 74.20 | |||||
| 1168 | Rhodococcus sp. | This study | 93.55 AF = 79.27 | 93.74 AF = 83.80 | |||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nahar, A.; Baker, A.L.; Nichols, D.S.; Bowman, J.P.; Britz, M.L. Benchmarking DNA Extraction Methods for Phylogenomic Analysis of Sub-Antarctic Rhodococcus and Williamsia Species. Microorganisms 2021, 9, 1253. https://doi.org/10.3390/microorganisms9061253
Nahar A, Baker AL, Nichols DS, Bowman JP, Britz ML. Benchmarking DNA Extraction Methods for Phylogenomic Analysis of Sub-Antarctic Rhodococcus and Williamsia Species. Microorganisms. 2021; 9(6):1253. https://doi.org/10.3390/microorganisms9061253
Chicago/Turabian StyleNahar, Akhikun, Anthony L. Baker, David S. Nichols, John P. Bowman, and Margaret L. Britz. 2021. "Benchmarking DNA Extraction Methods for Phylogenomic Analysis of Sub-Antarctic Rhodococcus and Williamsia Species" Microorganisms 9, no. 6: 1253. https://doi.org/10.3390/microorganisms9061253
APA StyleNahar, A., Baker, A. L., Nichols, D. S., Bowman, J. P., & Britz, M. L. (2021). Benchmarking DNA Extraction Methods for Phylogenomic Analysis of Sub-Antarctic Rhodococcus and Williamsia Species. Microorganisms, 9(6), 1253. https://doi.org/10.3390/microorganisms9061253








