Utilization of Monosaccharides by Hungateiclostridium thermocellum ATCC 27405 through Adaptive Evolution
Abstract
:1. Introduction
2. Materials and Methods
2.1. H. thermocellum ATCC 27,405 Growth Conditions and Medium Preparation
2.2. Phosphoric Acid Swollen Cellulose and Cellulosome Isolation
2.3. Avicelase, CMCase, and Xylanase Assays
2.4. Measurement of Residual Reducing Sugar, Carbon Dioxide, and Ethanol
2.5. RNA Isolation and Library Preparation and Sequencing
2.6. Differential Gene Expression Analysis
2.7. Sample Preparation for Genomic Analysis
2.8. Library Construction and Sequencing
2.9. Read Mapping, Variant Calling and Annotation
3. Results and Discussion
3.1. Growth Patterns, Sugar Utilization, Cellulosomal Enzyme Activities and Ethanol Productivity of CGs1, FAs1, and GAs1
3.2. Adaptive Laboratory Evolution
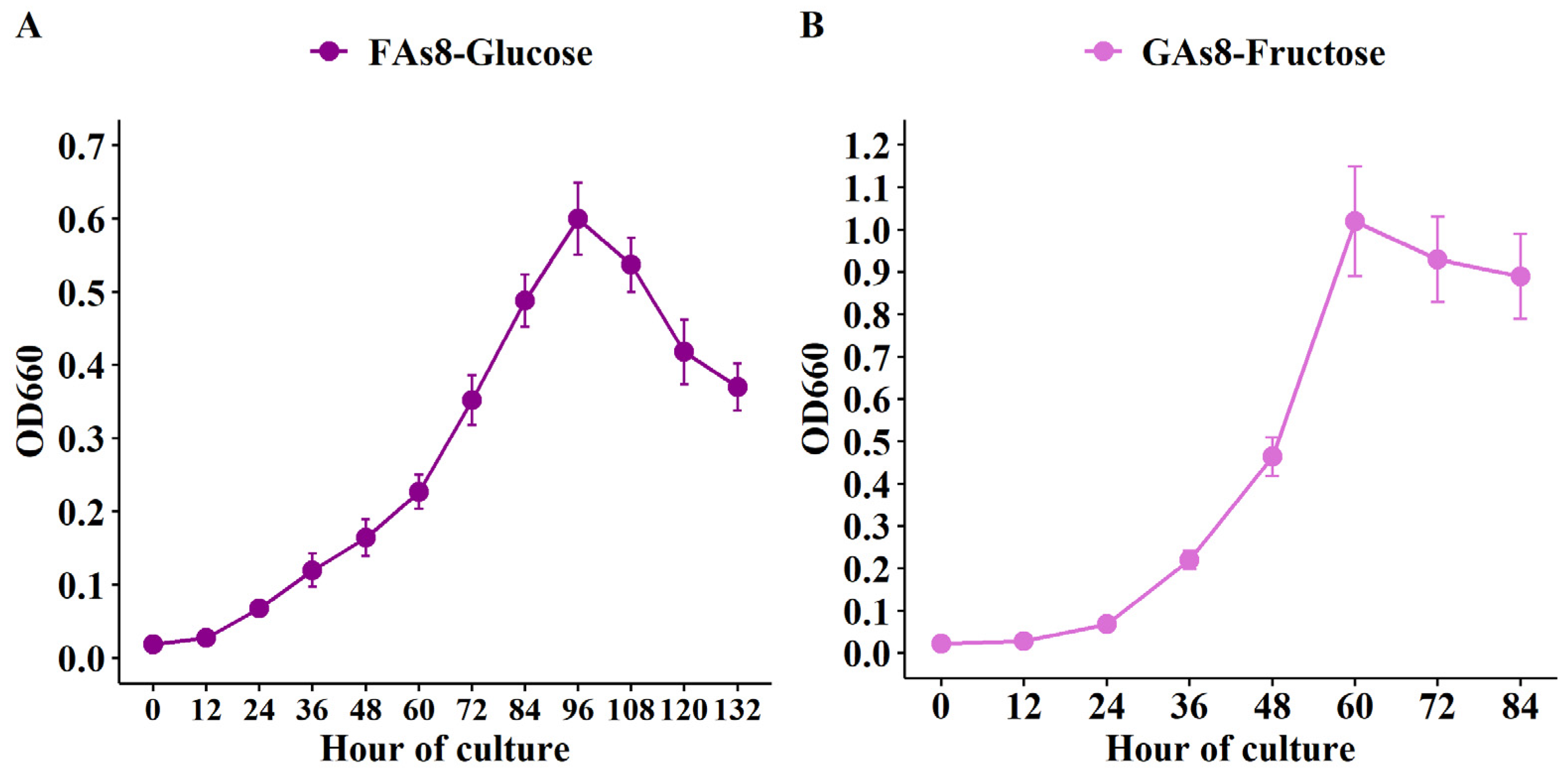
3.3. Growth of FAs8 on Glucose-Based Medium and Growth of GAs8 on Fructose-Based Medium
3.4. Transcriptomic Profiling of CGs1, FAs1, GAs1 and Their Evolved Strains FAs8 and GAs8
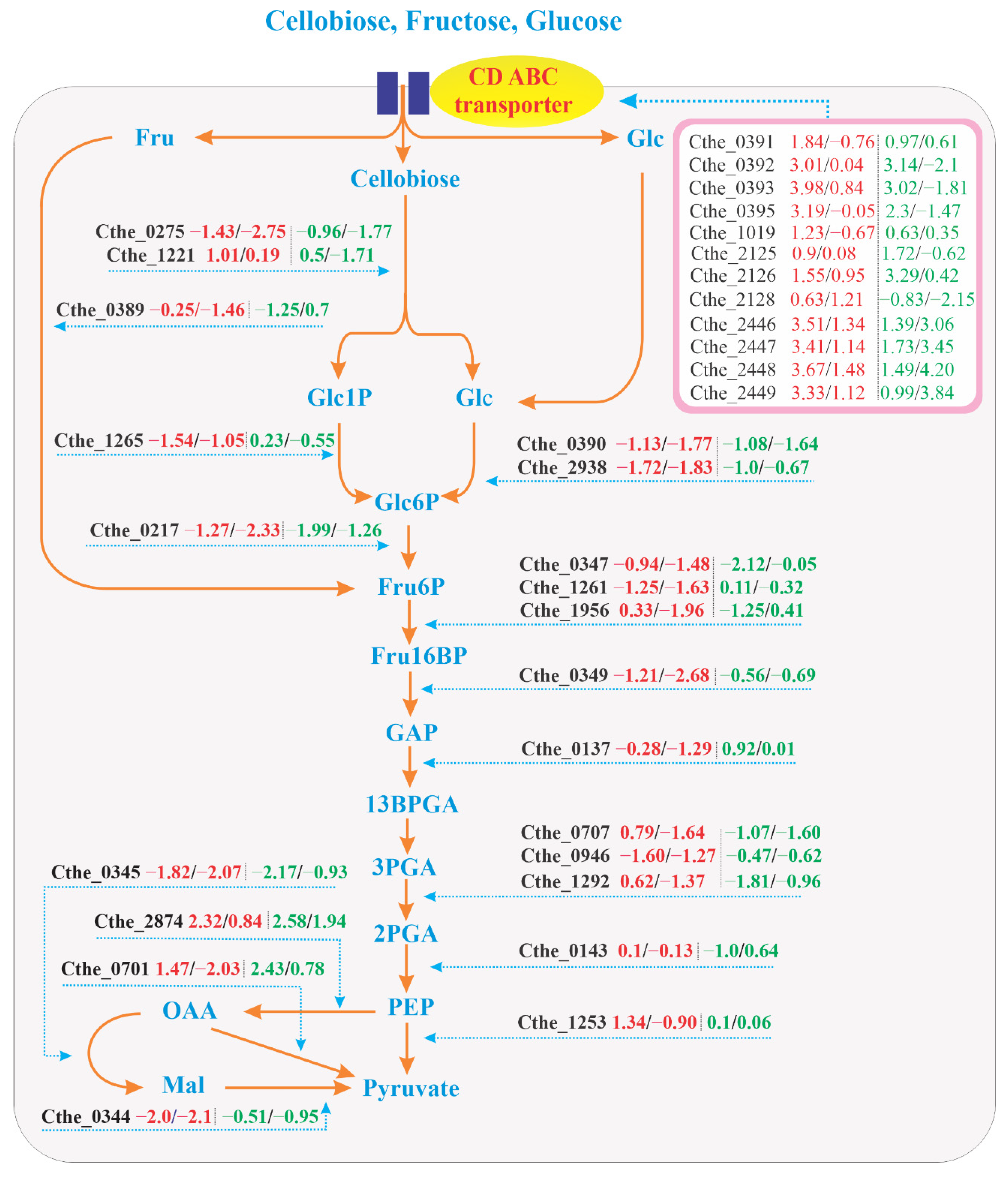
3.5. EMP Pathway
3.5.1. Genes Upstream of Phosphoenolpyruvate (PEP)
3.5.2. Genes Downstream of PEP
3.6. EMP Pathway
3.7. Energy Generation and Redox Balance
3.8. Other DEGs in Different Categories
3.9. Genomic Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 2PGA | 2-phosphoglycerate |
| 3PGA | 3-phosphoglycerate |
| 13BPGA | 1,3-bisphosphoglycerate |
| A | Adenine |
| ALE | Adaptive laboratory evolution |
| BSA | Bovine serum albumin |
| CAZymes | Carbohydrate-active enzymes |
| CBM | Carbohydrate-binding module |
| CBP | Cellobiose phosphorylase |
| CDS | Coding sequence |
| CGs | Cellobiose-grown cells |
| CMC | Carboxymethylcellulose |
| DE | Differential expression |
| DEGs | Differentially expressed genes |
| FAs | Fructose-adapted cells |
| Fd | Ferredoxin |
| Fru | Fructose |
| Fru6P | Fructose 6-phosphate |
| Fru16BP | Fructose 1,6-biphosphate |
| FBA | Fructose-bisphosphate aldolase |
| G | Guanine |
| Glc | Glucose |
| GAP | Glyceraldehyde 3-phosphate |
| GAs | Glucose-adapted cells |
| Glc1P | Glucose 1-phosphate |
| Glc6P | Glucose 6-phosphate |
| GCK | Glucokinase |
| GH | Glycoside hydrolase |
| GHnc | Glycoside hydrolase family “Non classified” |
| GA3P | Glyceraldehyde 3-phosphate |
| GTP | Guanosine-5′-triphosphate |
| indel | Insertion/Deletion |
| ITP | Inosine triphosphate |
| Mal | Malate |
| MDH | Malate dehydrogenase |
| ME | Malic enzyme |
| nbdA | ATP-binding protein |
| NGS | Next-generation sequencing |
| non-syn | Nonsynonymous |
| OAA | Oxaloacetate |
| ODC | Oxaloacetate decarboxylase |
| PGI | Glucose-6-phosphate isomerase |
| PEP | Phosphoenolpyruvate |
| PFK | Phosphofructokinase |
| PGAM | Phosphoglycerate mutase |
| PPDK | Pyruvate phosphate dikinase |
| PYK | Pyruvate kinase |
| SNVs | Single nucleotide variations |
| syn | Synonymous |
References
- Dror, T.W.; Morag, E.; Rolider, A.; Bayer, E.A.; Lamed, R.; Shoham, Y. Regulation of the Cellulosomal celS (cel48A) Gene of Clostridium thermocellum Is Growth Rate Dependent. J. Bacteriol. 2003, 185, 3042–3048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dror, T.W.; Rolider, A.; Bayer, E.A.; Lamed, R.; Shoham, Y. Regulation of Expression of Scaffoldin-Related Genes in Clostridium thermocellum. J. Bacteriol. 2003, 185, 5109–5116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahel-Raifer, H.; Jindou, S.; Bahari, L.; Nataf, Y.; Shoham, Y.; Bayer, E.A.; Borovok, I.; Lamed, R. The unique set of putative membrane-associated anti-σ factors in Clostridium thermocellum suggests a novel extracellular carbohydrate-sensing mechanism involved in gene regulation. FEMS Microbiol. Lett. 2010, 308, 84–93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bahari, L.; Gilad, Y.; Borovok, I.; Kahel-Raifer, H.; Dassa, B.; Nataf, Y.; Shoham, Y.; Lamed, R.; Bayer, E.A. Glycoside hydrolases as components of putative carbohydrate biosensor proteins in Clostridium thermocellum. J. Ind. Microbiol. Biotechnol. 2011, 38, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Sander, K.; Wilson, C.M.; Rodriguez, M.; Klingeman, D.M.; Rydzak, T.; Davison, B.H.; Brown, S.D. Clostridium thermocellum DSM 1313 transcriptional responses to redox perturbation. Biotechnol. Biofuels 2015, 8, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.; Klingeman, D.M.; Brown, S.D.; Cox, C.D. The LacI family protein GlyR3 co-regulates the celC operon and manB in Clostridium thermocellum. Biotechnol. Biofuels 2017, 10, 163. [Google Scholar] [CrossRef] [Green Version]
- De Ora, L.O.; Lamed, R.; Liu, Y.-J.; Xu, J.; Cui, Q.; Feng, Y.; Shoham, Y.; Bayer, E.A.; Muñoz-Gutiérrez, I. Regulation of biomass degradation by alternative σ factors in cellulolytic clostridia. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Linville, J.; Rodriguez, M., Jr.; Land, M.; Syed, M.; Engle, N.; Tschaplinski, T.; Mielenz, J.; Cox, C. Industrial Robustness: Understanding the Mechanism of Tolerance for the Populus Hydrolysate-Tolerant Mutant Strain of Clostridium thermocellum. PLoS ONE 2013, 8, e78829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holwerda, E.K.; Olson, D.G.; Ruppertsberger, N.M.; Stevenson, D.M.; Murphy, S.; Maloney, M.I.; Lanahan, A.A.; Amador-Noguez, D.; Lynd, L.R. Metabolic and evolutionary responses of Clostridium thermocellum to genetic interventions aimed at improving ethanol production. Biotechnol. Biofuels 2020, 13, 1–20. [Google Scholar] [CrossRef]
- Raman, B.; McKeown, C.; Rodriguez, M., Jr.; Brown, S.; Mielenz, J. Transcriptomic Analysis of Clostridium thermocellum ATCC 27405 Cellulose Fermentation. BMC Microbiol. 2011, 11. Available online: www.biomedcentral.com/1471-2180/11/134 (accessed on 15 May 2021). [CrossRef] [Green Version]
- Wilson, C.; Yang, S.; Rodriguez, M., Jr.; Ma, Q.; Johnson, C.; Dice, L.; Xu, Y.; Brown, S. Clostridium thermocellum Transcriptomic Profiles after Exposure to Furfural or Heat Stress. Biotechnol. Biofuels 2013, 6. Available online: www.biotechnologyforbiofuels.com/content/6/1/131 (accessed on 20 May 2020). [CrossRef]
- Wilson, C.; Rodriquez, M., Jr.; Johnson, C.; Martin, S.; Chu, T.; Wolfinger, R.; Hauser, L.; Land, M.; Klingeman, D.; Syed, M.; et al. Global Transcriptome Analysis of Clostridium thermocellum ATCC 27405 during Growth on Dilute Acid Pretreated Populus and Switchgrass. Biotechnol. Biofuels 2013, 6, 1–18. Available online: www.biotechnologyforbiofuels.com/content/6/1/179 (accessed on 10 June 2021). [CrossRef] [PubMed] [Green Version]
- Linville, J.; Rodriguez, M., Jr.; Brown, S.; Mielenz, J.; Cox, C. Transcriptomic Analysis of Clostridium thermocellum Populus Hydrolysate-Tolerant Mutant Strain Shows Increased Cellular Efficiency in Response to Populus Hydrolysate Compared to the Wild Type Strain. BMC Microbiol. 2014, 14, 1–17. Available online: www.biomedcentral.com/1471-2180/14/215 (accessed on 20 May 2020). [CrossRef] [Green Version]
- Wei, H.; Fu, Y.; Magnusson, L.; Baker, J.O.; Maness, P.-C.; Xu, Q.; Yang, S.; Bowersox, A.; Bogorad, I.; Wang, W.; et al. Comparison of transcriptional profiles of Clostridium thermocellum grown on cellobiose and pretreated yellow poplar using RNA-Seq. Front. Microbiol. 2014, 5, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, L.; Perot, S.J.; Stevenson, D.; Jacobson, T.; Lanahan, A.A.; Amador-Noguez, D.; Olson, D.G.; Lynd, L.R. Metabolome analysis reveals a role for glyceraldehyde 3-phosphate dehydrogenase in the inhibition of C. thermocellum by ethanol. Biotechnol. Biofuels 2017, 10, 1–11. [Google Scholar] [CrossRef]
- Xiong, W.; Lo, J.; Chou, K.J.; Wu, C.; Magnusson, L.; Dong, T.; Maness, P. Isotope-Assisted Metabolite Analysis Sheds Light on Central Carbon Metabolism of a Model Cellulolytic Bacterium Clostridium thermocellum. Front. Microbiol. 2018, 9, 1947. [Google Scholar] [CrossRef]
- Gold, N.D.; Martin, V.J.J. Global View of the Clostridium thermocellum Cellulosome Revealed by Quantitative Proteomic Analysis. J. Bacteriol. 2007, 189, 6787–6795. [Google Scholar] [CrossRef] [Green Version]
- Raman, B.; Pan, C.; Hurst, G.; Rodriguez, M., Jr.; McKeown, C.; Lankford, P.; Samatova, N.; Mielenz, J. Impact of Pretreated Switchgrass and Biomass Carbohydrates on Clostridium thermocellum ATCC 27405 Cellulosome Composition: A Quantitative Proteomic Analysis. PLoS ONE 2009, 4, e5271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burton, E.; Martin, V.J. Proteomic analysis of Clostridium thermocellum ATCC 27405 reveals the upregulation of an alternative transhydrogenase–malate pathway and nitrogen assimilation in cells grown on cellulose. Can. J. Microbiol. 2012, 58, 1378–1388. [Google Scholar] [CrossRef]
- Rydzak, T.; McQueen, P.D.; Krokhin, O.V.; Spicer, V.; Ezzati, P.; Dwivedi, R.C.; Shamshurin, D.; Levin, D.B.; Wilkins, J.A.; Sparling, R. Proteomic analysis of Clostridium thermocellum core metabolism: Relative protein expression profiles and growth phase-dependent changes in protein expression. BMC Microbiol. 2012, 12, 214. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.; Giannone, R.J.; Dice, L.; Yang, Z.K.; Engle, N.L.; Tschaplinski, T.J.; Hettich, R.L.; Brown, S.D. Clostridium thermocellum ATCC27405 transcriptomic, metabolomic and proteomic profiles after ethanol stress. BMC Genom. 2012, 13, 336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dumitrache, A.; Klingeman, D.M.; Natzke, J.; Rodriguez, M., Jr.; Giannone, R.J.; Hettich, R.L.; Davison, B.H.; Brown, S.D. Specialized activities and expression differences for Clostridium thermocellum biofilm and planktonic cells. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poudel, S.; Giannone, R.J.; Rodriguez, M.; Raman, B.; Martin, M.Z.; Engle, N.L.; Mielenz, J.R.; Nookaew, I.; Brown, S.D.; Tschaplinski, T.J.; et al. Integrated omics analyses reveal the details of metabolic adaptation of Clostridium thermocellum to lignocellulose-derived growth inhibitors released during the deconstruction of switchgrass. Biotechnol. Biofuels 2017, 10, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitham, J.; Moon, J.-W.; Rodriguez, M.; Engle, N.L.; Klingeman, D.M.; Rydzak, T.; Abel, M.M.; Tschaplinski, T.J.; Guss, A.M.; Brown, S.D. Clostridium thermocellum LL1210 pH homeostasis mechanisms informed by transcriptomics and metabolomics. Biotechnol. Biofuels 2018, 11, 98. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, D.M.; Weimer, P.J. Expression of 17 Genes in Clostridium thermocellum ATCC 27405 during Fermentation of Cellulose or Cellobiose in Continuous Culture. Appl. Environ. Microbiol. 2005, 71, 4672–4678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Lynd, L. Regulation of Cellulase Synthesis in Batch and Continuous Cultures of Clostridium thermocellum. J. Bacteriol. 2005, 187, 99–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prawitwong, P.; Waeonukul, R.; Tachaapaikoon, C.; Pason, P.; Ratanakhanokchai, K.; Deng, L.; Sermsathanaswadi, J.; Septiningrum, K.; Mori, Y.; Kosugi, A. Direct glucose production from lignocellulose using Clostridium thermocellum cultures supplemented with a thermostable β-glucosidase. Biotechnol. Biofuels 2013, 6, 184. [Google Scholar] [CrossRef] [Green Version]
- Zhang, P.; Wang, B.; Xiao, Q.; Wu, S. A kinetics modeling study on the inhibition of glucose on cellulosome of Clostridium thermocellum. Bioresour. Technol. 2015, 190, 36–43. [Google Scholar] [CrossRef]
- Johnson, E.A.; Bouchot, F.; Demain, A.L. Regulation of Cellulase Formation in Clostridium thermocellum. Microbiology 1985, 131, 2303–2308. [Google Scholar] [CrossRef] [Green Version]
- Nochur, S.; Roberts, M.; Demain, A. Mutation of Clostridium thermocellum in the Presence of Certain Carbon Sources. FEMS Microbiol. Lett. 1990, 71, 199–204. [Google Scholar] [CrossRef]
- Nochur, S.V.; Demain, A.L.; Roberts, M.F. Carbohydrate utilization by Clostridium thermocellum: Importance of internal pH in regulating growth. Enzym. Microb. Technol. 1992, 14, 338–349. [Google Scholar] [CrossRef]
- Patni, N.J.; Alexander, J.K. Utilization of Glucose by Clostridium thermocellum: Presence of Glucokinase and Other Glycolytic Enzymes in Cell Extracts. J. Bacteriol. 1971, 105, 220–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hernández, P.E. Transport of D-glucose in Clostridium thermocellum ATCC-27405. J. Gen. Appl. Microbiol. 1982, 28, 469–477. [Google Scholar] [CrossRef]
- Strobel, H.J.; Caldwell, F.C.; Dawson, K.A. Carbohydrate Transport by the Anaerobic Thermophile Clostridium thermocellum LQRI. Appl. Environ. Microbiol. 1995, 61, 4012–4015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dragosits, M.; Mattanovich, D. Adaptive laboratory evolution—Principles and applications for biotechnology. Microb. Cell Factories 2013, 12, 64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Conrad, T.M.; Lewis, N.; Palsson, B. Ø Microbial laboratory evolution in the era of genome-scale science. Mol. Syst. Biol. 2011, 7, 509. [Google Scholar] [CrossRef]
- Tian, L.; Papanek, B.; Olson, D.G.; Rydzak, T.; Holwerda, E.K.; Zheng, T.; Zhou, J.; Maloney, M.; Jiang, N.; Giannone, R.J.; et al. Simultaneous achievement of high ethanol yield and titer in Clostridium thermocellum. Biotechnol. Biofuels 2016, 9, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yayo, J.; Kuil, T.; Olson, D.G.; Lynd, L.R.; Holwerda, E.K.; van Maris, A.J.A. Laboratory Evolution and Reverse Engineering of Clostridium thermocellum for Growth on Glucose and Fructose. Appl. Environ. Microbiol. 2021, 87. [Google Scholar] [CrossRef]
- Johnson, E.A.; Madia, A.; Demain, A.L. Chemically Defined Minimal Medium for Growth of the Anaerobic Cellulolytic Thermophile Clostridium thermocellum. Appl. Environ. Microbiol. 1981, 41, 1060–1062. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Lynd, L.R. Quantification of cell and cellulase mass concentrations during anaerobic cellulose fermentation: Development of an enzyme-linked immunosorbent assay-based method with application to Clostridium thermocellum batch cultures. Anal. Chem. 2003, 75, 219–227. [Google Scholar] [CrossRef]
- Xiong, W.; Lin, P.P.; Magnusson, L.; Warner, L.; Liao, J.; Maness, P.-C.; Chou, K.J. CO2-fixing one-carbon metabolism in a cellulose-degrading bacterium Clostridium thermocellum. Proc. Natl. Acad. Sci. USA 2016, 113, 13180–13185. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.-H.P.; Cui, J.; Lynd, L.; Kuang, L.R. A Transition from Cellulose Swelling to Cellulose Dissolution byo-Phosphoric Acid: Evidence from Enzymatic Hydrolysis and Supramolecular Structure. Biomacromolecules 2006, 7, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Brice, L.A.S.; Shao, X.; Izquierdo, J.A.; Lynd, L.R. Optimization of Affinity Digestion for the Isolation of Cellulosomes from Clostridium thermocellum. Prep. Biochem. Biotechnol. 2013, 44, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.P.; Hong, J.; Ye, X. Cellulase Assays. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2009; Volume 581, pp. 213–231. [Google Scholar]
- Ribeiro, L.F.; De Lucas, R.C.; Vitcosque, G.L.; Ribeiro, L.F.; Ward, R.J.; Rubio, M.V.; Damásio, A.R.; Squina, F.M.; Gregory, R.C.; Walton, P.H.; et al. A novel thermostable xylanase GH10 from Malbranchea pulchella expressed in Aspergillus nidulans with potential applications in biotechnology. Biotechnol. Biofuels 2014, 7, 115. [Google Scholar] [CrossRef] [Green Version]
- Miller, G.L. Use of Dinitrosalicylic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Saqib, A.A.N.; Whitney, P.J. Differential behaviour of the dinitrosalicylic acid (DNS) reagent towards mono- and di-saccharide sugars. Biomass Bioenergy 2011, 35, 4748–4750. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Johnson, E.; Sakajoh, M.; Halliwell, G.; Madia, A.; Demain, A. Saccharification of Complex Cellulosic Substrates by the Cellulase System from Clostridium thermocellum. Appl. Environ. Microbiol. 1982, 43, 1125–1132. [Google Scholar] [CrossRef] [Green Version]
- Bayer, E.; Shoham, Y.; Lamed, R. The cellulosome: An exocellular organelle for degrading plant cell wall polysaccharides. In Glycomicrobiology; Doyle, R., Ed.; Kluwer Academic/Plenum Publishers: Amsterdam, The Netherlands, 2000; pp. 387–439. [Google Scholar]
- Nataf, Y.; Bahari, L.; Kahel-Raifer, H.; Borovok, I.; Lamed, R.; Bayer, E.A.; Sonenshein, A.L.; Shoham, Y. Clostridium thermocellum cellulosomal genes are regulated by extracytoplasmic polysaccharides via alternative sigma factors. Proc. Natl. Acad. Sci. USA 2010, 107, 18646–18651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riederer, A.; Takasuka, T.E.; Makino, S.-I.; Stevenson, D.M.; Bukhman, Y.; Elsen, N.L.; Fox, B.G. Global Gene Expression Patterns in Clostridium thermocellum as Determined by Microarray Analysis of Chemostat Cultures on Cellulose or Cellobiose. Appl. Environ. Microbiol. 2011, 77, 1243–1253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waeonukul, R.; Kosugi, A.; Tachaapaikoon, C.; Pason, P.; Ratanakhanokchai, K.; Prawitwong, P.; Deng, L.; Saito, M.; Mori, Y. Efficient saccharification of ammonia soaked rice straw by combination of Clostridium thermocellum cellulosome and Thermoanaerobacter brockii β-glucosidase. Bioresour. Technol. 2012, 107, 352–357. [Google Scholar] [CrossRef]
- Koeck, D.; Koellmeier, T.; Zverlov, V.; Liebl, W.; Schwarz, W. Differences in biomass degradation between newly isolated environmental strains of Clostridium thermocellum and heterogeneity in the size of the cellulosomal scaffoldin. Syst. Appl. Microbiol. 2015, 38, 424–432. [Google Scholar] [CrossRef]
- Sand, A.; Holwerda, E.K.; Ruppertsberger, N.M.; Maloney, M.; Olson, D.; Nataf, Y.; Borovok, I.; Sonenshein, A.L.; Bayer, E.A.; Lamed, R.; et al. Three cellulosomal xylanase genes in Clostridium thermocellumare regulated by both vegetative SigA (σA) and alternative SigI6 (σI6) factors. FEBS Lett. 2015, 589, 3133–3140. [Google Scholar] [CrossRef]
- Xu, Q.; Resch, M.; Podkaminer, K.; Yang, S.; Baker, J.; Donohoe, B.; Wilson, C.; Klingeman, D.; Olson, D.; Decker, S.; et al. Dramatic Performance of Clostridium thermocellum Explained by Its Wide Range of Cellulase Modalities. Sci. Adv. 2016, 2, e1501254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verbeke, T.J.; Garcia, G.M.; Elkins, J.G. The effect of switchgrass loadings on feedstock solubilization and biofuel production by Clostridium thermocellum. Biotechnol. Biofuels 2017, 10, 233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.-K.; Himmel, M.E.; Bomble, Y.J.; Westpheling, J. Expression of a Cellobiose Phosphorylase from Thermotoga maritima in Caldicellulosiruptor bescii Improves the Phosphorolytic Pathway and Results in a Dramatic Increase in Cellulolytic Activity. Appl. Environ. Microbiol. 2018, 84, e02348-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamed, R.; Kenig, R.; Morag, E.; Calzada, J.; De Micheo, F.; Bayer, E. Efficient Cellulose Solubilization by a Combined Cellulosome Beta-Glucosidase System. Appl. Biochem. Biotechnol. 1991, 27, 173–183. [Google Scholar] [CrossRef]
- Zhou, J.; Olson, D.G.; Argyros, D.A.; Deng, Y.; van Gulik, W.M.; van Dijken, J.P.; Lynd, L.R. Atypical Glycolysis in Clostridium thermocellum. Appl. Environ. Microbiol. 2013, 79, 3000–3008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, R.A.; Layton, D.S.; Guss, A.; Olson, D.; Lynd, L.R.; Trinh, C.T. Elucidating central metabolic redox obstacles hindering ethanol production in Clostridium thermocellum. Metab. Eng. 2015, 32, 207–219. [Google Scholar] [CrossRef] [Green Version]
- Olson, D.; Hörl, M.; Fuhrer, T.; Cui, J.; Zhou, J.; Maloney, M.I.; Amador-Noguez, D.; Tian, L.; Sauer, U.; Lynd, L.R. Glycolysis without pyruvate kinase in Clostridium thermocellum. Metab. Eng. 2017, 39, 169–180. [Google Scholar] [CrossRef] [Green Version]
- Nataf, Y.; Yaron, S.; Stahl, F.; Lamed, R.; Bayer, E.A.; Scheper, T.-H.; Sonenshein, A.L.; Shoham, Y. Cellodextrin and Laminaribiose ABC Transporters in Clostridium thermocellum. J. Bacteriol. 2009, 191, 203–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rangel, A.E.T.; Croft, T.; Barrios, A.F.G.; Reyes, L.H.; Maness, P.-C.; Chou, K.J. Transcriptomic analysis of a Clostridium thermocellum strain engineered to utilize xylose: Responses to xylose versus cellobiose feeding. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Biswas, R.; Wilson, C.M.; Giannone, R.J.; Klingeman, D.M.; Rydzak, T.; Shah, M.B.; Hettich, R.L.; Brown, S.D.; Guss, A.M. Improved growth rate in Clostridium thermocellum hydrogenase mutant via perturbed sulfur metabolism. Biotechnol. Biofuels 2017, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Retchless, A.C.; Lawrence, J.G. Ecological Adaptation in Bacteria: Speciation Driven by Codon Selection. Mol. Biol. Evol. 2012, 29, 3669–3683. [Google Scholar] [CrossRef] [Green Version]
- Plotkin, J.B.; Kudla, G. Synonymous but not the same: The causes and consequences of codon bias. Nat. Rev. Genet. 2010, 12, 32–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thakur, M.; Badugu, S.; Muniyappa, K. UvrA and UvrC Subunits of the Mycobacterium Tuberculosis UvrABC Excinuclease Interact Independently of UvrB and DNA. FEBS Lett. 2020, 594, 851–863. [Google Scholar] [CrossRef]
- Elgrably-Weiss, M.; Schlosser-Silverman, E.; Rosenshine, I.; Altuvia, S. DeoT, a DeoR-type transcriptional regulator of multiple target genes. FEMS Microbiol. Lett. 2006, 254, 141–148. [Google Scholar] [CrossRef] [Green Version]
- Soisson, S.; MacDougall-Shackleton, B.; Schleif, R.; Wolberger, C. The 1.6 Å Crystal Structure of the AraC Sugar-Binding and Dimerization Domain Complexed with D-Fucose. J. Mol. Biol. 1997, 273, 226–237. [Google Scholar] [CrossRef] [Green Version]
- Gallegos, M.-T.; Michán, C.; Ramos, J.L. The XylS/AraC family of regulators. Nucleic Acids Res. 1993, 21, 807–810. [Google Scholar] [CrossRef] [Green Version]

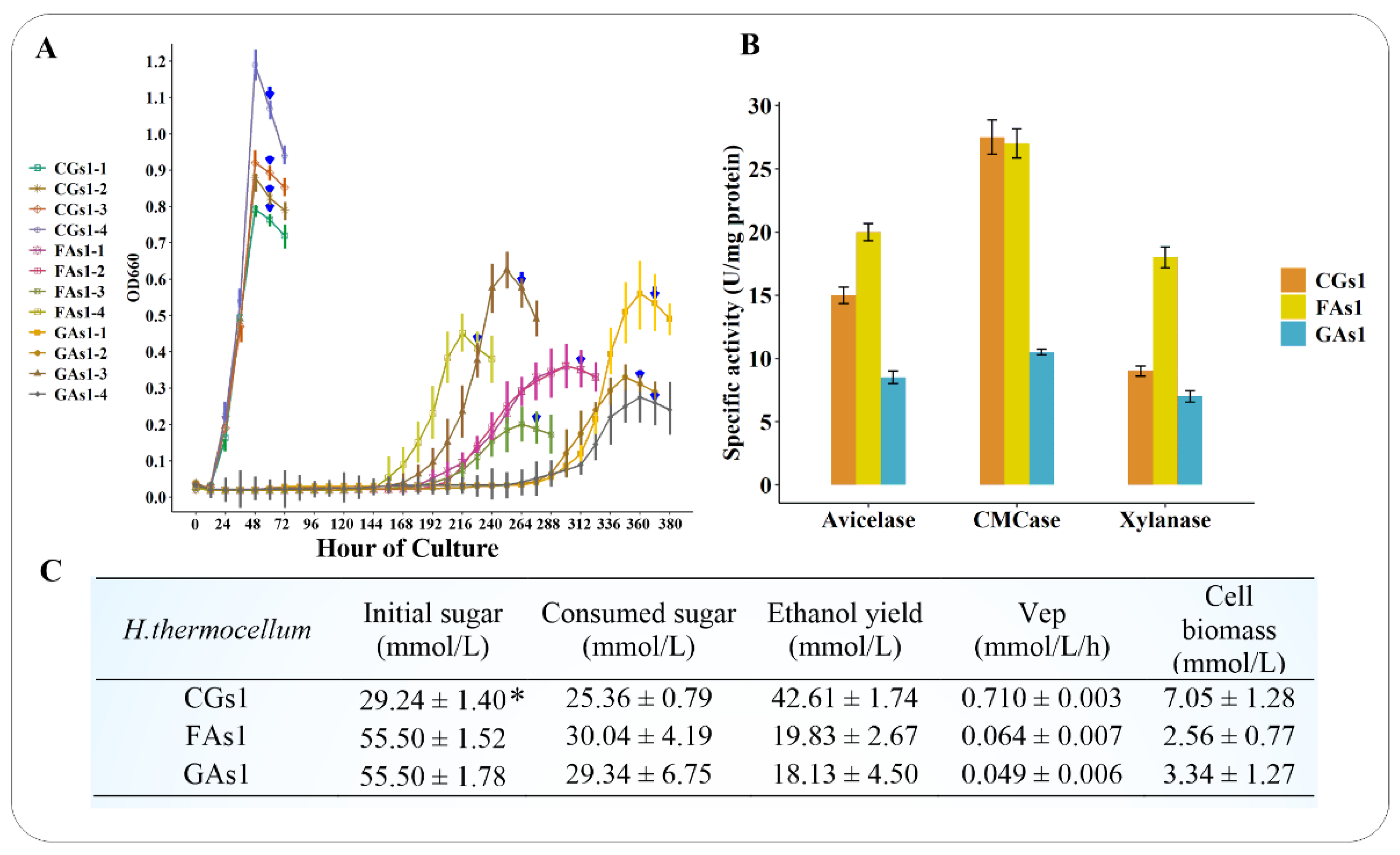
| Fructose-Adapted Cells | Consumed Sugar (mmol/L) | Ethanol (mmol/L) | CO2 (mmol/L) | Cell Biomass (mmol/L) | Ethanol Yield (mmol Eth/mmol Sugar) |
|---|---|---|---|---|---|
| FAs1 | 30.04 ± 4.19 | 19.83 ± 2.67 | 3.24 ± 0.46 | 2.56 ± 0.77 | 0.66 ± 0.03 |
| FAs2 | 15.24 ± 1.67 | 7.610 ± 1.13 | 1.79 ± 0.24 | 3.68 ± 1.24 | 0.50 ± 0.02 |
| FAs3 | 17.94 ± 2.63 | 8.110 ± 1.68 | 2.14 ± 0.28 | 5.22 ± 2.4 | 0.45 ± 0.02 |
| FAs4 | 18.22 ± 2.80 | 8.260 ± 1.36 | 2.09 ± 0.31 | 5.54 ± 2.43 | 0.45 ± 0.03 |
| FAs5 | 22.58 ± 1.59 | 11.30 ± 1.21 | 2.74 ± 0.22 | 6.04 ± 1.12 | 0.50 ± 0.02 |
| FAs6 | 26.21 ± 2.50 | 11.97 ± 0.98 | 2.83 ± 0.14 | 8.33 ± 1.14 | 0.46 ± 0.04 |
| FAs7 | 30.37 ± 1.84 | 14.45 ± 1.24 | 2.98 ± 0.28 | 9.32 ± 0.83 | 0.48 ± 0.05 |
| FAs8 | 34.23 ± 2.74 | 17.54 ± 1.06 | 3.68 ± 0.29 | 9.77 ± 1.37 | 0.51 ± 0.06 |
| Glucose-Adapted Cells | |||||
| GAs1 | 29.34 ± 6.75 | 18.13 ± 4.50 | 4.42 ± 1.10 | 3.34 ± 1.27 | 0.62 ± 0.04 |
| GAs2 | 17.19 ± 2.23 | 8.800 ± 3.20 | 2.85 ± 1.25 | 3.75 ± 1.83 | 0.51 ± 0.02 |
| GAs3 | 18.51 ± 2.92 | 9.130 ± 1.20 | 3.03 ± 0.70 | 4.48 ± 0.66 | 0.49 ± 0.03 |
| GAs4 | 21.05 ± 2.12 | 9.910 ± 1.72 | 3.15 ± 0.80 | 5.87 ± 2.36 | 0.47 ± 0.04 |
| GAs5 | 21.52 ± 3.47 | 10.00 ± 1.30 | 3.24 ± 0.49 | 6.30 ± 1.73 | 0.46 ± 0.04 |
| GAs6 | 23.41 ± 2.50 | 11.83 ± 1.23 | 3.29 ± 0.42 | 6.18 ± 2.25 | 0.51 ± 0.02 |
| GAs7 | 23.76 ± 3.50 | 12.28 ± 3.85 | 3.35 ± 0.82 | 6.13 ± 2.86 | 0.52 ± 0.06 |
| GAs8 | 24.88 ± 2.74 | 12.35 ± 1.18 | 3.41 ± 0.41 | 7.27 ± 4.49 | 0.50 ± 0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ha-Tran, D.M.; Nguyen, T.T.M.; Lo, S.-C.; Huang, C.-C. Utilization of Monosaccharides by Hungateiclostridium thermocellum ATCC 27405 through Adaptive Evolution. Microorganisms 2021, 9, 1445. https://doi.org/10.3390/microorganisms9071445
Ha-Tran DM, Nguyen TTM, Lo S-C, Huang C-C. Utilization of Monosaccharides by Hungateiclostridium thermocellum ATCC 27405 through Adaptive Evolution. Microorganisms. 2021; 9(7):1445. https://doi.org/10.3390/microorganisms9071445
Chicago/Turabian StyleHa-Tran, Dung Minh, Trinh Thi My Nguyen, Shou-Chen Lo, and Chieh-Chen Huang. 2021. "Utilization of Monosaccharides by Hungateiclostridium thermocellum ATCC 27405 through Adaptive Evolution" Microorganisms 9, no. 7: 1445. https://doi.org/10.3390/microorganisms9071445
APA StyleHa-Tran, D. M., Nguyen, T. T. M., Lo, S.-C., & Huang, C.-C. (2021). Utilization of Monosaccharides by Hungateiclostridium thermocellum ATCC 27405 through Adaptive Evolution. Microorganisms, 9(7), 1445. https://doi.org/10.3390/microorganisms9071445







