Analysis of clasp2 Transcription Pattern in Male Germ Cells during Spermatogenesis: A Comparative Study in Zebrafish (Danio rerio) and Guppy (Poecilia reticulata)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
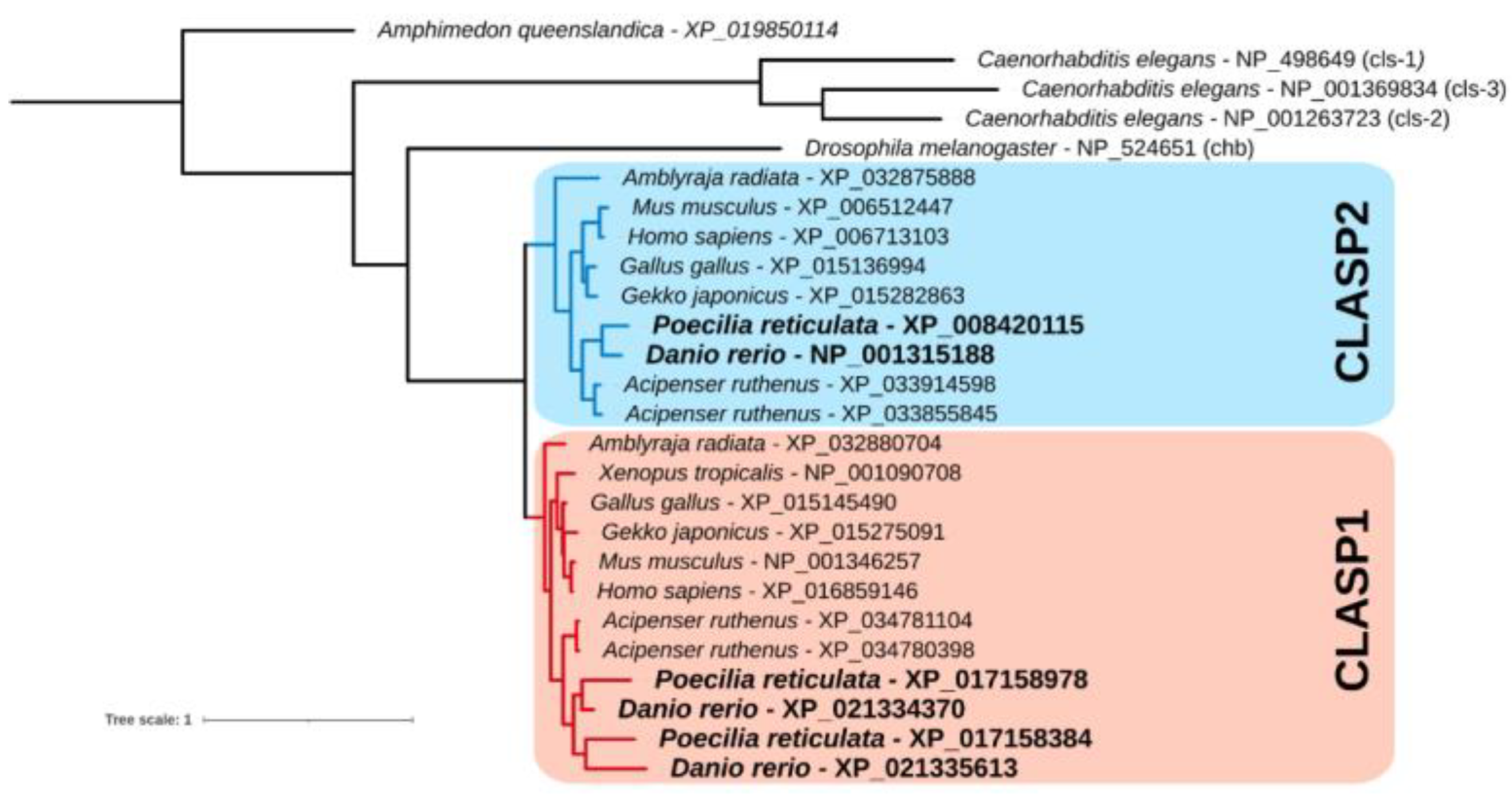
2.1. Phylogenetic Analysis
2.2. Dissection of Animals and Organs
2.3. RNA Extraction
2.4. Reverse Transcriptase PCR
2.5. Quantitative Real-Time PCR
2.6. Synthesis of Riboprobes for clasp2 in Zebrafish and Guppy
2.7. In Situ Hybridization
2.8. Histology (Haematoxylin and Eosin Staining)
2.9. Statistical Analysis
| RNAeasy minikit (Qiagen) | cat # 74104 |
| Superscript III First-Strand Synthesis kit | cat # 12574026 |
| TOPO™ TA Cloning™ | cat # K4575J10 |
| Dig-RNA label mix (Roche) | cat # 11277073910 |
| HNPP/Fast Red Fluorescent Detection set | cat # 11758888001 |
| Anti-Dig antibody (Roche) | cat # 11093274910 |
| Proteinase K | cat # 25530049 |
| Ethanol | cat # 100983 |
3. Results
3.1. Phylogenetic Tree of the Clasp Gene Family
3.2. Quantitative Analysis of clasp2 Transcription Level in Different Organs of Zebrafish and Guppy
3.3. Clasp2 Transcript Is Transcribed in Spermatozoa of Adult Zebrafish Testis
3.4. Clasp2 Transcript Is Transcribed in Spermatozeugmata of Adult Guppy Testis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Drabek, K.; van Ham, M.; Stepanova, T.; Draegestein, K.; van Horssen, R.; Sayas, C.L.; Akhmanova, A.; Ten Hagen, T.; Smits, R.; Fodde, R.; et al. Role of CLASP2 in microtubule stabilization and the regulation of persistent motility. Curr. Biol. 2006, 16, 2259–2264. [Google Scholar] [CrossRef]
- Al-Bassam, J.; Chang, F. Regulation of microtubule dynamics by TOG-domain proteins XMAP215/Dis1 and CLASP. Trends Cell Biol. 2011, 21, 604–614. [Google Scholar] [CrossRef]
- Lawrence, E.J.; Arpag, G.; Norris, S.R.; Zanic, M. Human CLASP2 specifically regulates microtubule catastrophe and rescue. Mol. Biol. Cell 2018, 29, 1168–1177. [Google Scholar] [CrossRef]
- Pereira, A.L.; Pereira, A.J.; Maia, A.R.; Drabek, K.; Sayas, C.L.; Hergert, P.J.; Lince-Faria, M.; Matos, I.; Duque, C.; Stepanova, T.; et al. Mammalian CLASP1 and CLASP2 cooperate to ensure mitotic fidelity by regulating spindle and kinetochore function. Mol. Biol. Cell 2006, 17, 4526–4542. [Google Scholar] [CrossRef]
- Matsui, T.; Watanabe, T.; Matsuzawa, K.; Kakeno, M.; Okumura, N.; Sugiyama, I.; Itoh, N.; Kaibuchi, K. PAR3 and aPKC regulate Golgi organization through CLASP2 phosphorylation to generate cell polarity. Mol. Biol. Cell 2015, 26, 751–761. [Google Scholar] [CrossRef]
- Adachi, A.; Kano, F.; Tsuboi, T.; Fujita, M.; Maeda, Y.; Murata, M. Golgi-associated GSK3beta regulates the sorting process of post-Golgi membrane trafficking. J. Cell Sci. 2010, 123, 3215–3225. [Google Scholar] [CrossRef]
- Dillon, G.M.; Tyler, W.A.; Omuro, K.C.; Kambouris, J.; Tyminski, C.; Henry, S.; Haydar, T.F.; Beffert, U.; Ho, A. CLASP2 Links Reelin to the Cytoskeleton during Neocortical Development. Neuron 2017, 93, 1344–1358.e1345. [Google Scholar] [CrossRef]
- Sayas, C.L.; Basu, S.; van der Reijden, M.; Bustos-Moran, E.; Liz, M.; Sousa, M.; Van Ijcken, W.F.; Avila, J.; Galjart, N. Distinct Functions for Mammalian CLASP1 and -2 During Neurite and Axon Elongation. Front. Cell Neurosci. 2019, 13, 5. [Google Scholar] [CrossRef]
- Zhu, B.; Qi, L.; Liu, S.; Liu, W.; Ou, Z.; Chen, M.; Liu, L.; Zu, X.; Wang, J.; Li, Y. CLASP2 is involved in the EMT and early progression after transurethral resection of the bladder tumor. BMC Cancer 2017, 17, 105. [Google Scholar] [CrossRef]
- Chen, L.; Xiong, W.; Guo, W.; Su, S.; Qi, L.; Zhu, B.; Mo, M.; Jiang, H.; Li, Y. Significance of CLASP2 expression in prognosis for muscle-invasive bladder cancer patients: A propensity score-based analysis. Urol. Oncol. 2019, 37, 800–807. [Google Scholar] [CrossRef] [PubMed]
- Valter, A.; Luhari, L.; Pisarev, H.; Truumees, B.; Planken, A.; Smolander, O.P.; Oselin, K. Genomic alterations as independent prognostic factors to predict the type of lung cancer recurrence. Gene 2023, 885, 147690. [Google Scholar] [CrossRef]
- Beffert, U.; Dillon, G.M.; Sullivan, J.M.; Stuart, C.E.; Gilbert, J.P.; Kambouris, J.A.; Ho, A. Microtubule plus-end tracking protein CLASP2 regulates neuronal polarity and synaptic function. J. Neurosci. 2012, 32, 13906–13916. [Google Scholar] [CrossRef]
- Siino, V.; Amato, A.; Di Salvo, F.; Caldara, G.F.; Filogamo, M.; James, P.; Vasto, S. Impact of diet-induced obesity on the mouse brain phosphoproteome. J. Nutr. Biochem. 2018, 58, 102–109. [Google Scholar] [CrossRef]
- Langlais, P.; Dillon, J.L.; Mengos, A.; Baluch, D.P.; Ardebili, R.; Miranda, D.N.; Xie, X.; Heckmann, B.L.; Liu, J.; Mandarino, L.J. Identification of a role for CLASP2 in insulin action. J. Biol. Chem. 2012, 287, 39245–39253. [Google Scholar] [CrossRef]
- Kruse, R.; Krantz, J.; Barker, N.; Coletta, R.L.; Rafikov, R.; Luo, M.; Hojlund, K.; Mandarino, L.J.; Langlais, P.R. Characterization of the CLASP2 Protein Interaction Network Identifies SOGA1 as a Microtubule-Associated Protein. Mol. Cell Proteomics 2017, 16, 1718–1735. [Google Scholar] [CrossRef] [PubMed]
- Karki, P.; Ke, Y.; Zhang, C.O.; Li, Y.; Tian, Y.; Son, S.; Yoshimura, A.; Kaibuchi, K.; Birukov, K.G.; Birukova, A.A. SOCS3-microtubule interaction via CLIP-170 and CLASP2 is critical for modulation of endothelial inflammation and lung injury. J. Biol. Chem. 2021, 296, 100239. [Google Scholar] [CrossRef]
- Karki, P.; Ke, Y.; Tian, Y.; Ohmura, T.; Sitikov, A.; Sarich, N.; Montgomery, C.P.; Birukova, A.A. Staphylococcus aureus-induced endothelial permeability and inflammation are mediated by microtubule destabilization. J. Biol. Chem. 2019, 294, 3369–3384. [Google Scholar] [CrossRef]
- Maton, G.; Edwards, F.; Lacroix, B.; Stefanutti, M.; Laband, K.; Lieury, T.; Kim, T.; Espeut, J.; Canman, J.C.; Dumont, J. Kinetochore components are required for central spindle assembly. Nat. Cell Biol. 2015, 17, 953. [Google Scholar] [CrossRef] [PubMed]
- Nahaboo, W.; Zouak, M.; Askjaer, P.; Delattre, M. Chromatids segregate without centrosomes during Caenorhabditis elegans mitosis in a Ran- and CLASP-dependent manner. Mol. Biol. Cell 2015, 26, 2020–2029. [Google Scholar] [CrossRef]
- Park, E.C.; Lee, H.; Hong, Y.; Kim, M.J.; Lee, Z.W.; Kim, S.I.; Kim, S.; Kim, G.H.; Han, J.K. Analysis of the expression of microtubule plus-end tracking proteins (+TIPs) during Xenopus laevis embryogenesis. Gene Expr. Patterns 2012, 12, 204–212. [Google Scholar] [CrossRef]
- Grimaldi, A.D.; Zanic, M.; Kaverina, I. Encoding the microtubule structure: Allosteric interactions between the microtubule +TIP complex master regulators and TOG-domain proteins. Cell Cycle 2015, 14, 1375–1378. [Google Scholar] [CrossRef]
- Akhmanova, A.; Hoogenraad, C.C.; Drabek, K.; Stepanova, T.; Dortland, B.; Verkerk, T.; Vermeulen, W.; Burgering, B.M.; De Zeeuw, C.I.; Grosveld, F.; et al. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell 2001, 104, 923–935. [Google Scholar] [CrossRef]
- Aonuma, M.; Miyamoto, M.; Inoue, Y.H.; Tamai, K.; Sakai, H.; Kamasawa, N.; Matsukage, A. Microtubule bundle formation and cell death induced by the human CLASP/Orbit N-terminal fragment. Cell Struct. Funct. 2005, 30, 7–13. [Google Scholar] [CrossRef]
- Moriwaki, T.; Goshima, G. Five factors can reconstitute all three phases of microtubule polymerization dynamics. J. Cell Biol. 2016, 215, 357–368. [Google Scholar] [CrossRef]
- Mathe, E.; Inoue, Y.H.; Palframan, W.; Brown, G.; Glover, D.M. Orbit/Mast, the CLASP orthologue of Drosophila, is required for asymmetric stem cell and cystocyte divisions and development of the polarised microtubule network that interconnects oocyte and nurse cells during oogenesis. Development 2003, 130, 901–915. [Google Scholar] [CrossRef]
- Miyauchi, C.; Kitazawa, D.; Ando, I.; Hayashi, D.; Inoue, Y.H. Orbit/CLASP is required for germline cyst formation through its developmental control of fusomes and ring canals in Drosophila males. PLoS ONE 2013, 8, e58220. [Google Scholar] [CrossRef]
- Inoue, Y.H.; Savoian, M.S.; Suzuki, T.; Mathe, E.; Yamamoto, M.T.; Glover, D.M. Mutations in orbit/mast reveal that the central spindle is comprised of two microtubule populations, those that initiate cleavage and those that propagate furrow ingression. J. Cell Biol. 2004, 166, 49–60. [Google Scholar] [CrossRef]
- Kitazawa, D.; Matsuo, T.; Kaizuka, K.; Miyauchi, C.; Hayashi, D.; Inoue, Y.H. Orbit/CLASP is required for myosin accumulation at the cleavage furrow in Drosophila male meiosis. PLoS ONE 2014, 9, e93669. [Google Scholar] [CrossRef]
- Shoda, T.; Yamazoe, K.; Tanaka, Y.; Asano, Y.; Inoue, Y.H. Orbit/CLASP determines centriole length by antagonising Klp10A in Drosophila spermatocytes. J. Cell Sci. 2021, 134, jcs251231. [Google Scholar] [CrossRef]
- Marchal, G.A.; Jouni, M.; Chiang, D.Y.; Perez-Hernandez, M.; Podliesna, S.; Yu, N.; Casini, S.; Potet, F.; Veerman, C.C.; Klerk, M.; et al. Targeting the Microtubule EB1-CLASP2 Complex Modulates Na(V)1.5 at Intercalated Discs. Circ. Res. 2021, 129, 349–365. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.Y.; Fu, X.H.; Cai, J.; Li, W.C.; Fan, B.Y.; Pang, Y.L.; Zhao, C.X.; Abula, M.; Kong, X.H.; Yao, X.; et al. Identification of key genes involved in recovery from spinal cord injury in adult zebrafish. Neural Regen. Res. 2022, 17, 1334–1342. [Google Scholar] [CrossRef]
- Klaus, A.; Clapes, T.; Yvernogeau, L.; Basu, S.; Weijts, B.; Maas, J.; Smal, I.; Galjart, N.; Robin, C. CLASP2 safeguards hematopoietic stem cell properties during mouse and fish development. Cell Rep. 2022, 39, 110957. [Google Scholar] [CrossRef]
- Leal, M.C.; Cardoso, E.R.; Nobrega, R.H.; Batlouni, S.R.; Bogerd, J.; Franca, L.R.; Schulz, R.W. Histological and stereological evaluation of zebrafish (Danio rerio) spermatogenesis with an emphasis on spermatogonial generations. Biol. Reprod. 2009, 81, 177–187. [Google Scholar] [CrossRef]
- Cacialli, P. Expression of Nerve Growth Factor and Its Receptor TrkA in the Reproductive System of Adult Zebrafish. Vet. Sci. 2022, 9, 225. [Google Scholar] [CrossRef]
- Cacialli, P.; D’Angelo, L.; de Girolamo, P.; Avallone, L.; Lucini, C.; Pellegrini, E.; Castaldo, L. Morpho-Functional Features of the Gonads of Danio rerio: The Role of Brain-Derived Neurotrophic Factor. Anat. Rec. 2018, 301, 140–147. [Google Scholar] [CrossRef]
- Torres-Martinez, A.; Ruiz de Dios, L.; Hernandez-Franyutti, A.; Uribe, M.C.; Sanchez, W.C. Structure of the testis and spermatogenesis of the viviparous teleost Poecilia mexicana (Poeciliidae) from an active sulfur spring cave in Southern Mexico. J. Morphol. 2019, 280, 1537–1547. [Google Scholar] [CrossRef] [PubMed]
- Piccinini, G.; Milani, L. Germline-related molecular phenotype in Metazoa: Conservation and innovation highlighted by comparative transcriptomics. Evodevo 2023, 14, 2. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Criscuolo, A.; Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): A new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010, 10, 210. [Google Scholar] [CrossRef]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef]
- Milani, L.; Cinelli, F.; Iannello, M.; Lazzari, M.; Franceschini, V.; Maurizii, M.G. Immunolocalization of Vasa, PIWI, and TDRKH proteins in male germ cells during spermatogenesis of the teleost fish Poecilia reticulata. Acta Histochem. 2022, 124, 151870. [Google Scholar] [CrossRef]
- Mahony, C.B.; Cacialli, P.; Pasche, C.; Monteiro, R.; Savvides, S.N.; Bertrand, J.Y. Hapln1b, a central organizer of the ECM, modulates kit signaling to control developmental hematopoiesis in zebrafish. Blood Adv. 2021, 5, 4935–4948. [Google Scholar] [CrossRef]
- Menke, A.L.; Spitsbergen, J.M.; Wolterbeek, A.P.; Woutersen, R.A. Normal anatomy and histology of the adult zebrafish. Toxicol. Pathol. 2011, 39, 759–775. [Google Scholar] [CrossRef]
- Girao, H.; Okada, N.; Rodrigues, T.A.; Silva, A.O.; Figueiredo, A.C.; Garcia, Z.; Moutinho-Santos, T.; Hayashi, I.; Azevedo, J.E.; Macedo-Ribeiro, S.; et al. CLASP2 binding to curved microtubule tips promotes flux and stabilizes kinetochore attachments. J. Cell Biol. 2020, 219, e201905080. [Google Scholar] [CrossRef]
- Rodgers, N.C.; Lawrence, E.J.; Sawant, A.V.; Efimova, N.; Gonzalez-Vasquez, G.; Hickman, T.T.; Kaverina, I.; Zanic, M. CLASP2 facilitates dynamic actin filament organization along the microtubule lattice. Mol. Biol. Cell 2023, 34, br3. [Google Scholar] [CrossRef]
- Mimori-Kiyosue, Y.; Grigoriev, I.; Lansbergen, G.; Sasaki, H.; Matsui, C.; Severin, F.; Galjart, N.; Grosveld, F.; Vorobjev, I.; Tsukita, S.; et al. CLASP1 and CLASP2 bind to EB1 and regulate microtubule plus-end dynamics at the cell cortex. J. Cell Biol. 2005, 168, 141–153. [Google Scholar] [CrossRef]
- Sousa, A.; Reis, R.; Sampaio, P.; Sunkel, C.E. The Drosophila CLASP homologue, Mast/Orbit regulates the dynamic behaviour of interphase microtubules by promoting the pause state. Cell Motil. Cytoskeleton 2007, 64, 605–620. [Google Scholar] [CrossRef]
- Drabek, K.; Gutierrez, L.; Vermeij, M.; Clapes, T.; Patel, S.R.; Boisset, J.C.; van Haren, J.; Pereira, A.L.; Liu, Z.; Akinci, U.; et al. The microtubule plus-end tracking protein CLASP2 is required for hematopoiesis and hematopoietic stem cell maintenance. Cell Rep. 2012, 2, 781–788. [Google Scholar] [CrossRef]
- O’Donnell, L.; O’Bryan, M.K. Microtubules and spermatogenesis. Semin. Cell Dev. Biol. 2014, 30, 45–54. [Google Scholar] [CrossRef]
- Gunes, S.; Sengupta, P.; Henkel, R.; Alguraigari, A.; Sinigaglia, M.M.; Kayal, M.; Joumah, A.; Agarwal, A. Microtubular Dysfunction and Male Infertility. World J. Mens. Health 2020, 38, 9–23. [Google Scholar] [CrossRef]
- Lehti, M.S.; Sironen, A. Formation and function of the manchette and flagellum during spermatogenesis. Reproduction 2016, 151, R43–R54. [Google Scholar] [CrossRef]
- Paduch, D.A. Testicular cancer and male infertility. Curr. Opin. Urol. 2006, 16, 419–427. [Google Scholar] [CrossRef]
- Ostrowski, K.A.; Walsh, T.J. Infertility with Testicular Cancer. Urol. Clin. N. Am. 2015, 42, 409–420. [Google Scholar] [CrossRef]





| Zebrafish | (Amplicon Size) | |
|---|---|---|
| clasp2 | F: TGGAGGCACATAAAGACC | (122 bp) |
| R: TGACTGGATGATGGGACA | ||
| Gapdh | F: GTGTAGGCGTGGACTGTGG | (151 bp) |
| R: TGGGAGTCAACCAGGACAAA |
| Guppy | (Amplicon Size) | |
|---|---|---|
| clasp2 | F: GAAGGACGTTACACGTAGAC | (181 bp) |
| R: CCCACAGATGTCTATCCCT | ||
| Gapdh | F: CTCCACTCATGGTGTCTG | (140 bp) |
| R: CAACATAGTCTACGGCAGC |
| Zebrafish | Length of Riboprobe | |
|---|---|---|
| clasp2 | F: GCATTGCTGGGGATCGATA | (532 bp) |
| R: CGTCGAAACTACGGTCTTG |
| Guppy | Length of Riboprobe | |
|---|---|---|
| clasp2 | F: CTCAGCTCAGGCTGCTTT | (494 bp) |
| R: CGCAGTTGGGAATGAGGT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ricci, S.; Lazzari, M.; Maurizii, M.G.; Franceschini, V.; Milani, L.; Cacialli, P. Analysis of clasp2 Transcription Pattern in Male Germ Cells during Spermatogenesis: A Comparative Study in Zebrafish (Danio rerio) and Guppy (Poecilia reticulata). Animals 2023, 13, 3617. https://doi.org/10.3390/ani13233617
Ricci S, Lazzari M, Maurizii MG, Franceschini V, Milani L, Cacialli P. Analysis of clasp2 Transcription Pattern in Male Germ Cells during Spermatogenesis: A Comparative Study in Zebrafish (Danio rerio) and Guppy (Poecilia reticulata). Animals. 2023; 13(23):3617. https://doi.org/10.3390/ani13233617
Chicago/Turabian StyleRicci, Serena, Maurizio Lazzari, Maria Gabriella Maurizii, Valeria Franceschini, Liliana Milani, and Pietro Cacialli. 2023. "Analysis of clasp2 Transcription Pattern in Male Germ Cells during Spermatogenesis: A Comparative Study in Zebrafish (Danio rerio) and Guppy (Poecilia reticulata)" Animals 13, no. 23: 3617. https://doi.org/10.3390/ani13233617





