MiR-206 Suppresses Triacylglycerol Accumulation via Fatty Acid Elongase 6 in Dairy Cow Mammary Epithelial Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Transfection
2.2. RNA Extraction and Real-Time Fluorescence Quantitative PCR
2.3. Prediction and Plasmid Construction of miR-206 Target Gene
2.4. Dual-Luciferase Assays
2.5. ELOVL6 Interference
2.6. TAG and Fatty Acid Assays
2.7. Statistical Analysis
3. Results
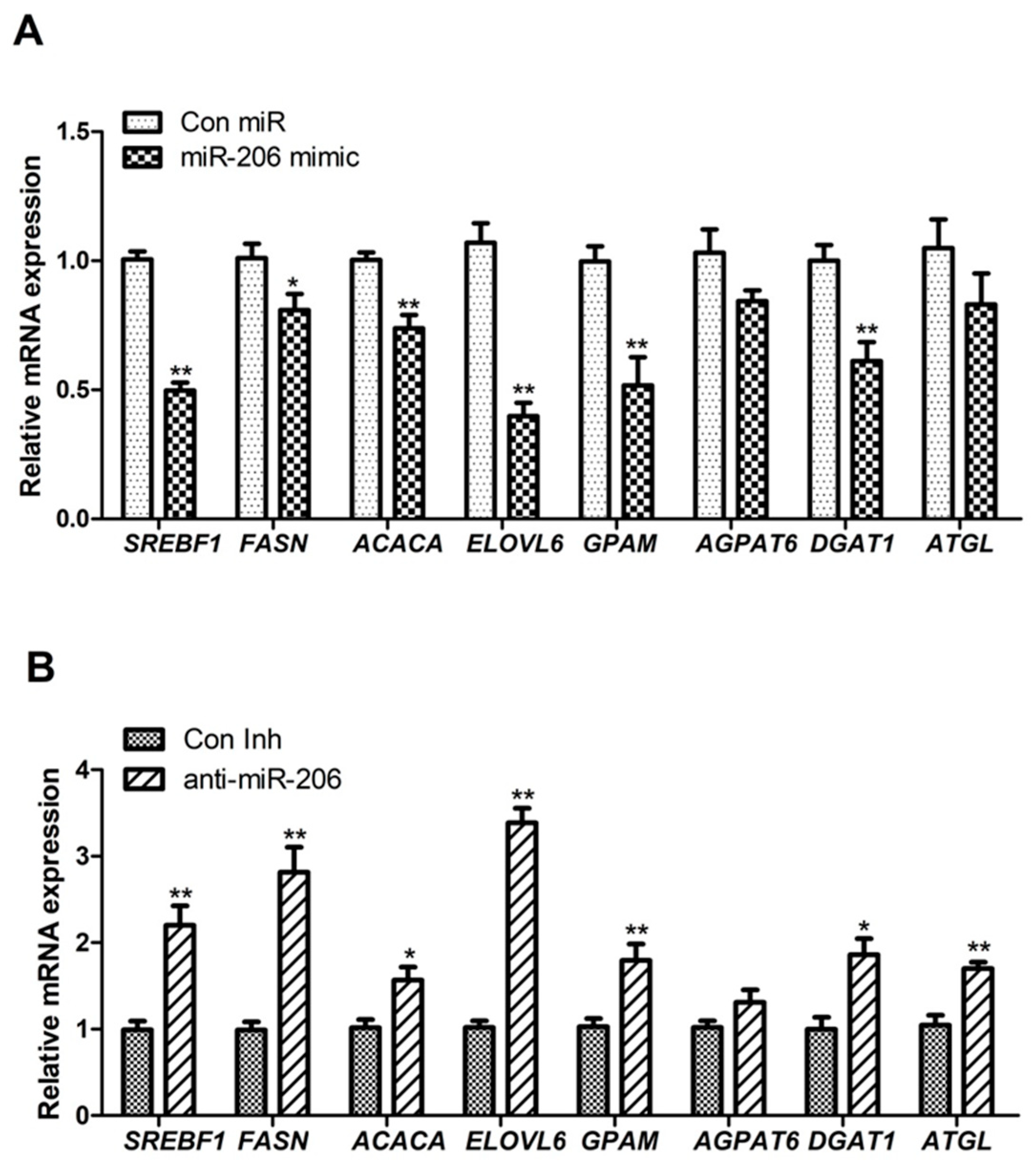
3.1. miR-206 Affects Triacylglycerol Levels and Expression of Genes Related to Lipid Metabolism in BMECs
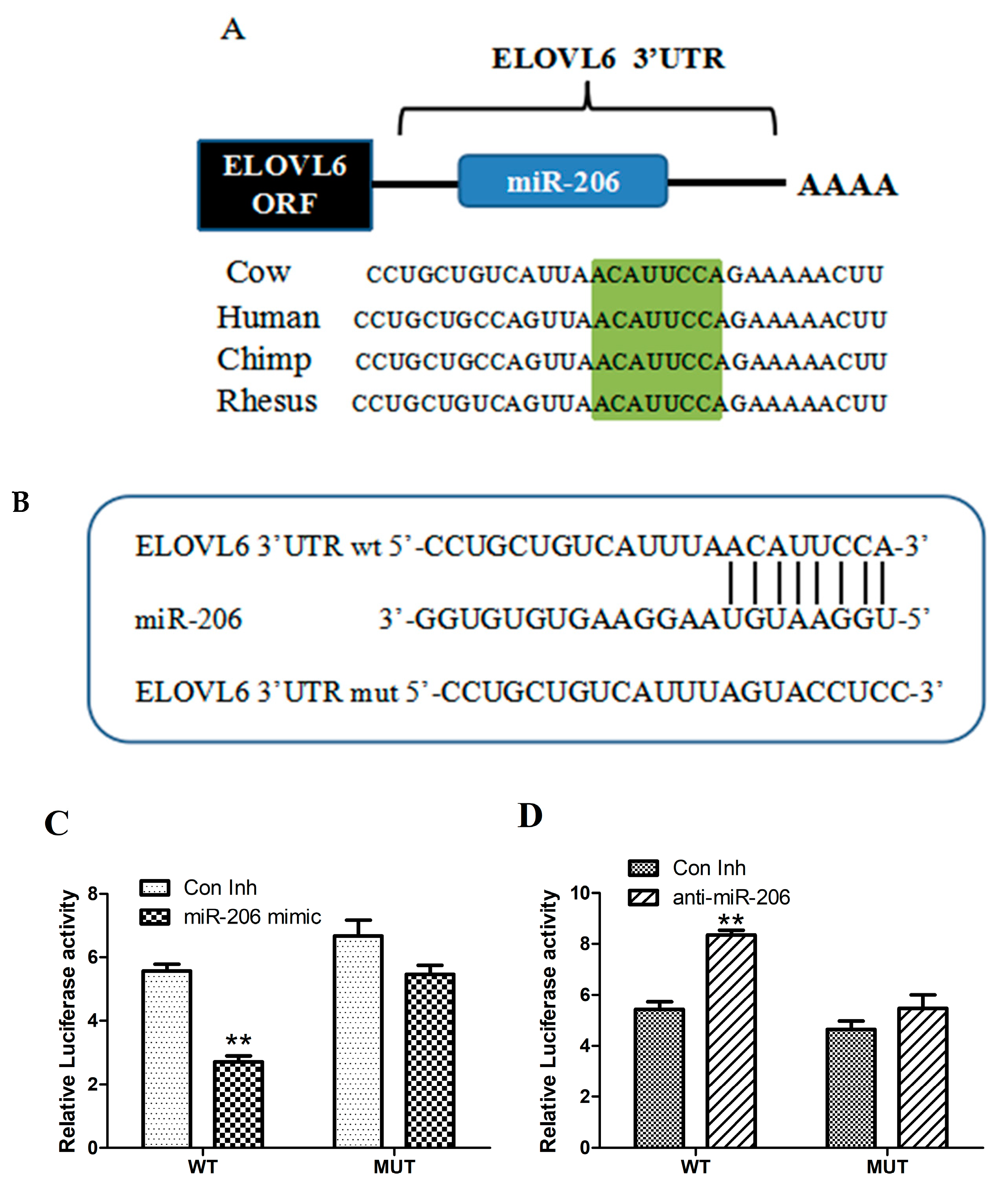
3.2. miR-206 Directly Targets the 3′-UTR of ELOVL6 mRNA
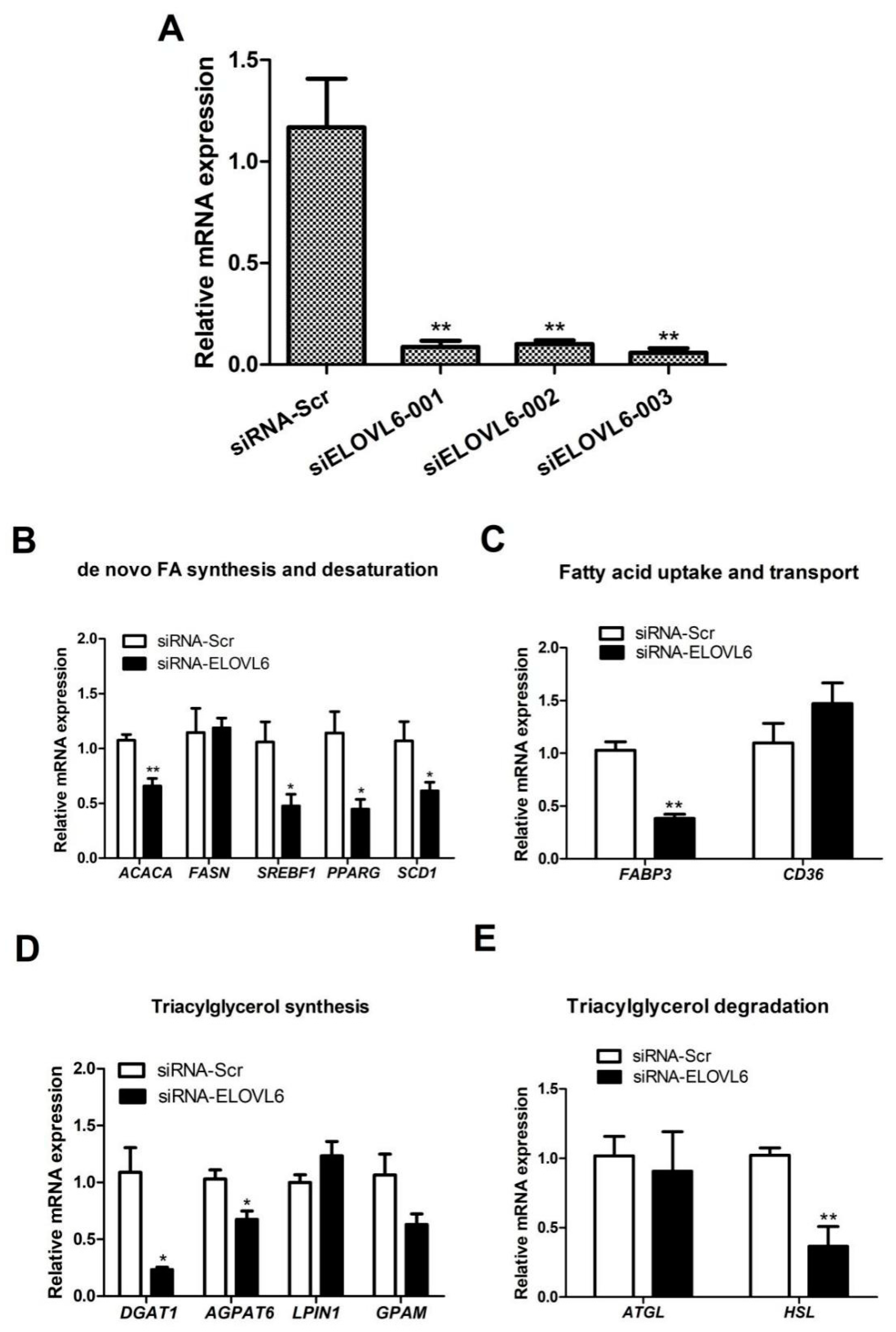
3.3. Interference with ELOVL6 Expression Decreases the Expression of Lipid Metabolism-Related Genes in BMECs
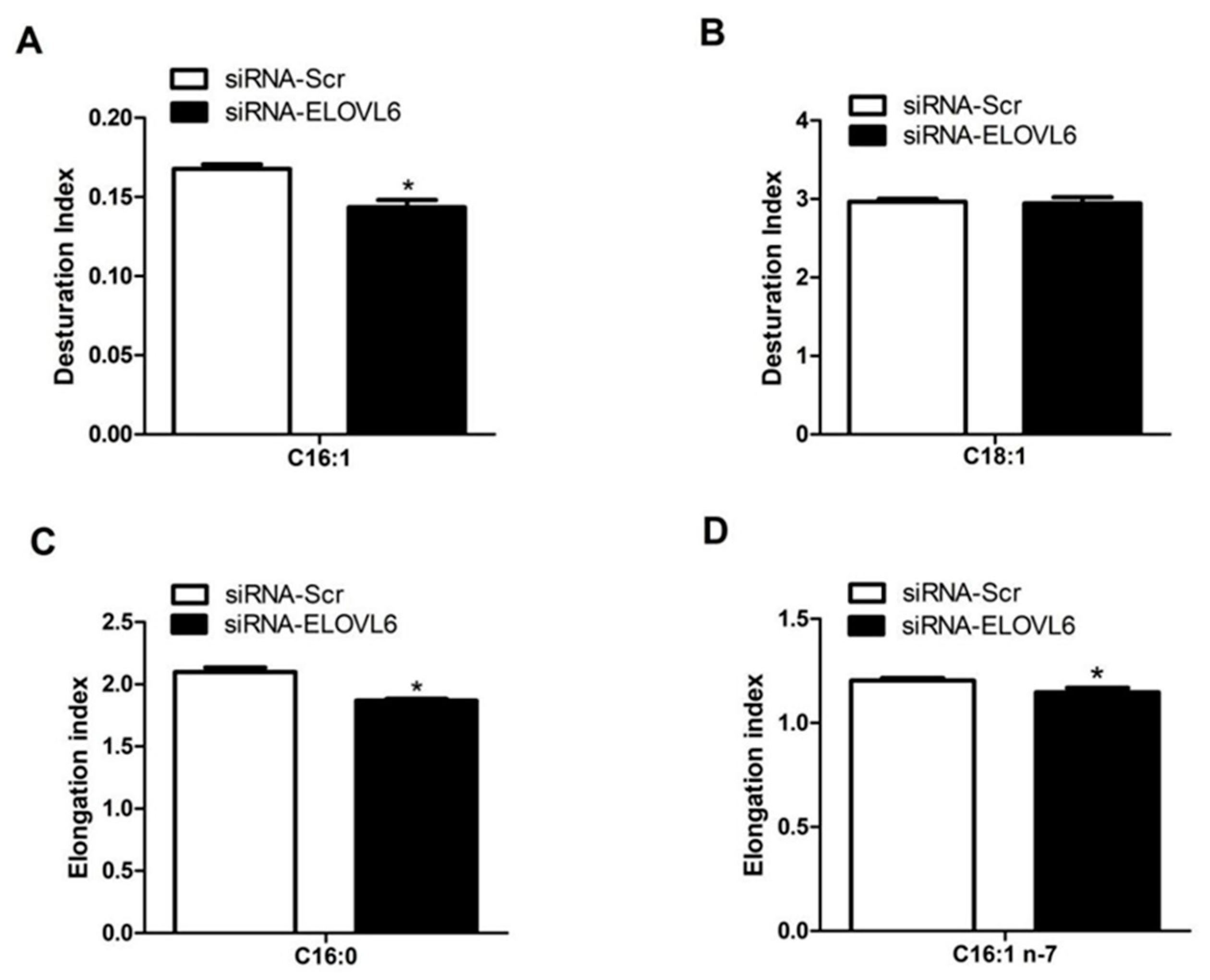
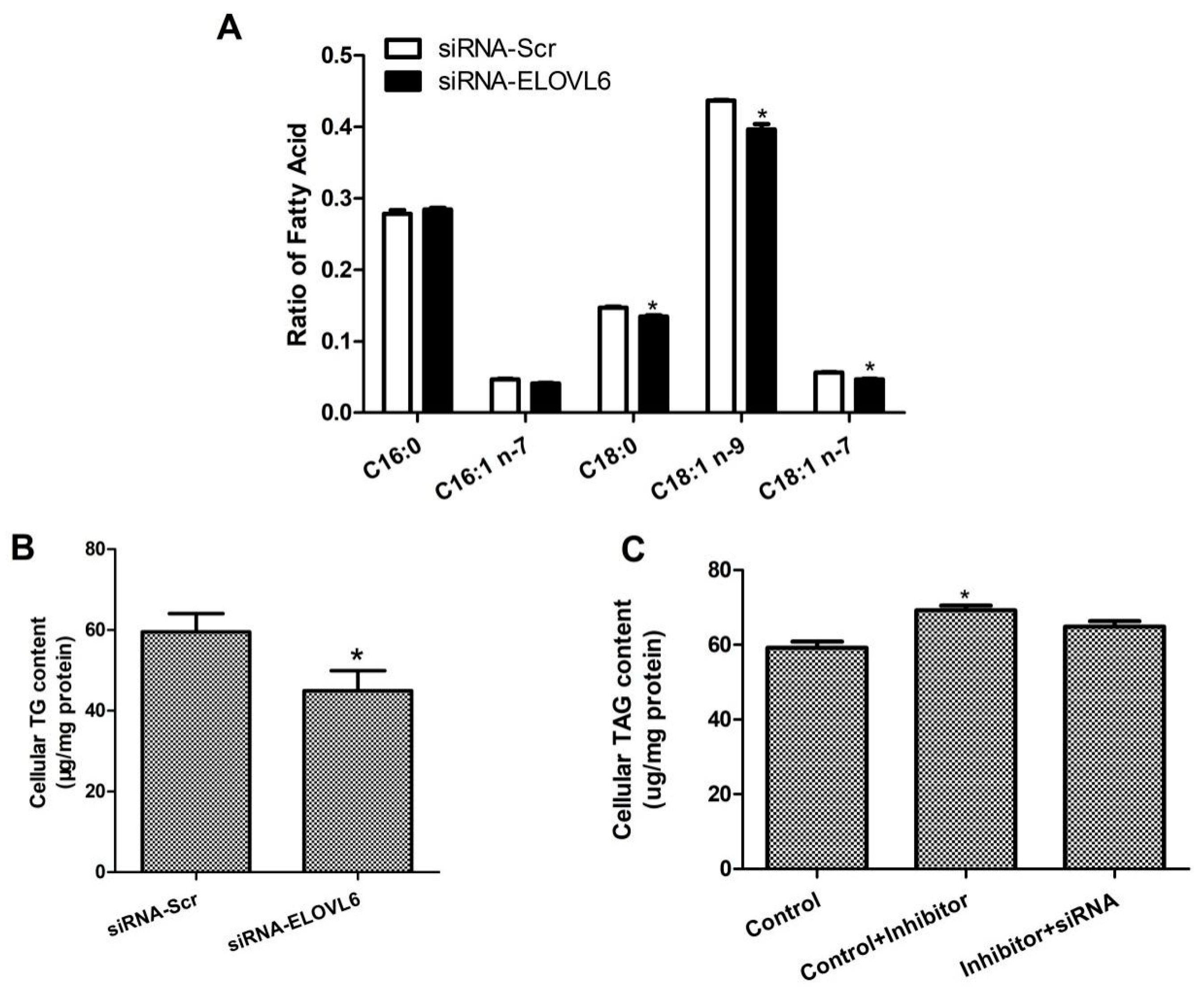
3.4. Interference with ELOVL6 Expression Affects Intracellular Fatty Acid Composition
3.5. ELOVL6 siRNA Partially Reverses the Decrease in TAG Levels Caused by Inhibition of miR-206
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Samsø Mathiasen, S.; Kanta, J.M.; Frydenberg, R.P.; Lundsgaard, A.; Guo, Z.; Fritzen, A.M.; Kiens, B.; Wiking, L.; Kleinert, M. Novel Methodology to Enrich Medium- and Short-Chain Fatty Acids in Milk Fat to Improve Metabolic Health. Food Funct. 2024, 15, 7951–7960. [Google Scholar] [CrossRef]
- Fan, X.; Zhu, W.; Qiu, L.; Zhang, G.; Zhang, Y.; Miao, Y. Elongase of Very Long Chain Fatty Acids 6 (ELOVL6) Promotes Lipid Synthesis in Buffalo Mammary Epithelial Cells. J. Anim. Physiol. Anim. Nutr. 2022, 106, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Diener, C.; Keller, A.; Meese, E. Emerging Concepts of miRNA Therapeutics: From Cells to Clinic. Trends Genet. 2022, 38, 613–626. [Google Scholar] [CrossRef]
- Zanoaga, O.; Braicu, C.; Jurj, A.; Berindan-Neagoe, I. Chapter 8—MicroRNA-Mediated Transcriptional and Posttranscriptional Regulation. In MicroRNA; Xiao, J., Ed.; Academic Press: Cambridge, MA, USA, 2022; pp. 141–152. ISBN 978-0-323-89774-7. [Google Scholar]
- Chu, M.; Zhao, Y.; Yu, S.; Hao, Y.; Zhang, P.; Feng, Y.; Zhang, H.; Ma, D.; Liu, J.; Cheng, M.; et al. miR-15b Negatively Correlates with Lipid Metabolism in Mammary Epithelial Cells. Am. J. Physiol. Cell Physiol. 2018, 314, C43–C52. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Yan, D.; Song, Z.; Zhu, X.; Liu, X.; Li, X.; Zhao, S. miR-126-3p Sensitizes Glioblastoma Cells to Temozolomide by Inactivating Wnt/β-Catenin Signaling via Targeting SOX2. Life Sci. 2019, 226, 98–106. [Google Scholar] [CrossRef]
- Tan, J.; Yang, B.; Qiu, L.; He, R.; Wu, Z.; Ye, M.; Zan, L.; Yang, W. Bta-miR-200a Regulates Milk Fat Biosynthesis by Targeting IRS2 to Inhibit the PI3K/Akt Signal Pathway in Bovine Mammary Epithelial Cells. J. Agric. Food Chem. 2024, 72, 16449–16460. [Google Scholar] [CrossRef]
- Khalilian, S.; Hosseini Imani, S.Z.; Ghafouri-Fard, S. Emerging Roles and Mechanisms of miR-206 in Human Disorders: A Comprehensive Review. Cancer Cell Int. 2022, 22, 412. [Google Scholar] [CrossRef]
- Dai, Y.; Wang, Y.M.; Zhang, W.R.; Liu, X.F.; Li, X.; Ding, X.B.; Guo, H. The Role of microRNA-1 and microRNA-206 in the Proliferation and Differentiation of Bovine Skeletal Muscle Satellite Cells. In Vitro Cell. Dev. Biol. Anim. 2016, 52, 27–34. [Google Scholar] [CrossRef]
- Chen, Z.; Chu, S.; Wang, X.; Fan, Y.; Zhan, T.; Arbab, A.A.I.; Li, M.; Zhang, H.; Mao, Y.; Loor, J.J.; et al. MicroRNA-106b Regulates Milk Fat Metabolism via ATP Binding Cassette Subfamily A Member 1 (ABCA1) in Bovine Mammary Epithelial Cells. J. Agric. Food Chem. 2019, 67, 3981–3990. [Google Scholar] [CrossRef]
- Li, C.; Tian, J.; Liu, N.; Song, D.; Steer, C.J.; Han, Q.; Song, G. MicroRNA-206 as a Potential Cholesterol-Lowering Drug Is Superior to Statins in Mice. J. Lipid Res. 2024, 65, 100576. [Google Scholar] [CrossRef]
- Wang, A.; Chen, B.; Jian, S.; Cai, W.; Xiao, M.; Du, G. miR-206-G6PD Axis Regulates Lipogenesis and Cell Growth in Hepatocellular Carcinoma Cell. Anticancer Drugs 2021, 32, 508–516. [Google Scholar] [CrossRef]
- Liu, N.; Tian, J.; Steer, C.J.; Han, Q.; Song, G. MicroRNA-206-3p Suppresses Hepatic Lipogenesis and Cholesterol Synthesis While Driving Cholesterol Efflux. Hepatology, 2023; ahead of print. [Google Scholar] [CrossRef]
- Chen, Z.; Cao, X.; Lu, Q.; Zhou, J.; Wang, Y.; Wu, Y.; Mao, Y.; Xu, H.; Yang, Z. Circ01592 Regulates Unsaturated Fatty Acid Metabolism through Adsorbing miR-218 in Bovine Mammary Epithelial Cells. Food Funct. 2021, 12, 12047–12058. [Google Scholar] [CrossRef]
- Bionaz, M.; Loor, J.J. Gene Networks Driving Bovine Milk Fat Synthesis during the Lactation Cycle. BMC Genom. 2008, 9, 366. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Jiang, N.; Wei, L.; Cai, J.; Zhang, W.; Jiang, Q.; Loor, J.J.; Liu, J. AMPK-ChREBP Axis Mediates de Novo Milk Fatty Acid Synthesis Promoted by Glucose in the Mammary Gland of Lactating Goats. Anim. Nutr. 2022, 10, 234–242. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, C.; Luo, J.; Xu, H.; Liu, J.; Loor, J.J.; Shi, H. The LXRB-SREBP1 Network Regulates Lipogenic Homeostasis by Controlling the Synthesis of Polyunsaturated Fatty Acids in Goat Mammary Epithelial Cells. J. Anim. Sci. Biotechnol. 2022, 13, 120. [Google Scholar] [CrossRef]
- Yao, D.W.; Luo, J.; He, Q.Y.; Wu, M.; Shi, H.B.; Wang, H.; Wang, M.; Xu, H.F.; Loor, J.J. Thyroid Hormone Responsive (THRSP) Promotes the Synthesis of Medium-Chain Fatty Acids in Goat Mammary Epithelial Cells. J. Dairy Sci. 2016, 99, 3124–3133. [Google Scholar] [CrossRef]
- Walden, T.B.; Timmons, J.A.; Keller, P.; Nedergaard, J.; Cannon, B. Distinct Expression of Muscle-Specific microRNAs (Myomirs) in Brown Adipocytes. J. Cell Physiol. 2009, 218, 444–449. [Google Scholar] [CrossRef] [PubMed]
- Mikkelsen, T.S.; Xu, Z.; Zhang, X.; Wang, L.; Gimble, J.M.; Lander, E.S.; Rosen, E.D. Comparative Epigenomic Analysis of Murine and Human Adipogenesis. Cell 2010, 143, 156–169. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Ma, F.; Li, W.; Ouyang, S.; Liu, Z.; Wu, J. miR-206-3p Inhibits 3T3-L1 Cell Adipogenesis via the c-Met/PI3K/Akt Pathway. Int. J. Mol. Sci. 2017, 18, 1510. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhao, C.; Zhang, N.; Kang, W.; Lu, R.; Wu, H.; Geng, Y.; Zhao, Y.; Xu, X. Serum microRNA miR-206 Is Decreased in Hyperthyroidism and Mediates Thyroid Hormone Regulation of Lipid Metabolism in HepG2 Human Hepatoblastoma Cells. Mol. Med. Rep. 2018, 17, 5635–5641. [Google Scholar] [CrossRef]
- Li, N.; Zhao, F.; Wei, C.; Liang, M.; Zhang, N.; Wang, C.; Li, Q.-Z.; Gao, X.-J. Function of SREBP1 in the Milk Fat Synthesis of Dairy Cow Mammary Epithelial Cells. Int. J. Mol. Sci. 2014, 15, 16998–17013. [Google Scholar] [CrossRef]
- Vinod, M.; Chennamsetty, I.; Colin, S.; Belloy, L.; De Paoli, F.; Schaider, H.; Graier, W.F.; Frank, S.; Kratky, D.; Staels, B.; et al. miR-206 Controls LXRα Expression and Promotes LXR-Mediated Cholesterol Efflux in Macrophages. Biochim. Biophys. Acta 2014, 1841, 827–835. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Zhu, Y.; Lin, X.; Tan, X.; Lu, B.; Li, Y. Stabilization of FASN by ACAT1-Mediated GNPAT Acetylation Promotes Lipid Metabolism and Hepatocarcinogenesis. Oncogene 2020, 39, 2437–2449. [Google Scholar] [CrossRef]
- Lin, P.; Yang, X.; Wang, L.; Zou, X.; Mu, L.; Xu, C.; Yang, X. Novel Artesunate-Metformin Conjugate Inhibits Bladder Cancer Cell Growth Associated with Clusterin/SREBP1/FASN Signaling Pathway. Korean J. Physiol. Pharmacol. 2024, 28, 219–227. [Google Scholar] [CrossRef]
- Yu, H.; Zhao, Z.; Yu, X.; Li, J.; Lu, C.; Yang, R. Bovine Lipid Metabolism Related Gene GPAM: Molecular Characterization, Function Identification, and Association Analysis with Fat Deposition Traits. Gene 2017, 609, 9–18. [Google Scholar] [CrossRef]
- Li, F.; Li, D.; Zhang, M.; Sun, J.; Li, W.; Jiang, R.; Han, R.; Wang, Y.; Tian, Y.; Kang, X.; et al. miRNA-223 Targets the GPAM Gene and Regulates the Differentiation of Intramuscular Adipocytes. Gene 2019, 685, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaka, T. Role of Fatty Acid Elongase Elovl6 in the Regulation of Energy Metabolism and Pathophysiological Significance in Diabetes. Diabetol. Int. 2021, 12, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Junjvlieke, Z.; Khan, R.; Mei, C.; Cheng, G.; Wang, S.; Raza, S.H.A.; Hong, J.; Wang, X.; Yang, W.; Zan, L. Effect of ELOVL6 on the Lipid Metabolism of Bovine Adipocytes. Genomics 2020, 112, 2282–2290. [Google Scholar] [CrossRef]
- Su, Y.-C.; Feng, Y.-H.; Wu, H.-T.; Huang, Y.-S.; Tung, C.-L.; Wu, P.; Chang, C.-J.; Shiau, A.-L.; Wu, C.-L. Elovl6 Is a Negative Clinical Predictor for Liver Cancer and Knockdown of Elovl6 Reduces Murine Liver Cancer Progression. Sci. Rep. 2018, 8, 6586. [Google Scholar] [CrossRef]
- Michler, S.; Schöffmann, F.A.; Robaa, D.; Volmer, J.; Hinderberger, D. Fatty Acid Binding to the Human Transport Proteins FABP3, FABP4, and FABP5 from a Ligand’s Perspective. J. Biol. Chem. 2024, 300, 107396. [Google Scholar] [CrossRef]
- Tăbăran, A.; Balteanu, V.A.; Gal, E.; Pusta, D.; Mihaiu, R.; Dan, S.D.; Tăbăran, A.F.; Mihaiu, M. Influence of DGAT1 K232A Polymorphism on Milk Fat Percentage and Fatty Acid Profiles in Romanian Holstein Cattle. Anim. Biotechnol. 2015, 26, 105–111. [Google Scholar] [CrossRef]
- Tzompa-Sosa, D.A.; van Valenberg, H.J.F.; van Aken, G.A.; Bovenhuis, H. Milk Fat Triacylglycerols and Their Relations with Milk Fatty Acid Composition, DGAT1 K232A Polymorphism, and Milk Production Traits. J. Dairy Sci. 2016, 99, 3624–3631. [Google Scholar] [CrossRef] [PubMed]
- Pacheco-Pappenheim, S.; Yener, S.; van Valenberg, H.J.F.; Tzompa-Sosa, D.A.; Bovenhuis, H. The DGAT1 K232A Polymorphism and Feeding Modify Milk Fat Triacylglycerol Composition. J. Dairy Sci. 2019, 102, 6842–6852. [Google Scholar] [CrossRef]
- Løvsletten, N.G.; Vu, H.; Skagen, C.; Lund, J.; Kase, E.T.; Thoresen, G.H.; Zammit, V.A.; Rustan, A.C. Treatment of Human Skeletal Muscle Cells with Inhibitors of Diacylglycerol Acyltransferases 1 and 2 to Explore Isozyme-Specific Roles on Lipid Metabolism. Sci. Rep. 2020, 10, 238. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Geng, F.; Pan, M.; Wu, X.; Zhong, Y.; Wang, C.; Tian, Z.; Cheng, C.; Zhang, R.; Puduvalli, V.; et al. Targeting DGAT1 Ameliorates Glioblastoma by Increasing Fat Catabolism and Oxidative Stress. Cell Metab. 2020, 32, 229–242.e8. [Google Scholar] [CrossRef]
- Shi, H.B.; Wu, M.; Zhu, J.J.; Zhang, C.H.; Yao, D.W.; Luo, J.; Loor, J.J. Fatty Acid Elongase 6 Plays a Role in the Synthesis of Long-Chain Fatty Acids in Goat Mammary Epithelial Cells. J. Dairy Sci. 2017, 100, 4987–4995. [Google Scholar] [CrossRef]
- Green, C.D.; Ozguden-Akkoc, C.G.; Wang, Y.; Jump, D.B.; Olson, L.K. Role of Fatty Acid Elongases in Determination of de Novo Synthesized Monounsaturated Fatty Acid Species. J. Lipid Res. 2010, 51, 1871–1877. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Deng, Y.-Y.; Liu, H.-X.; Pu, Y. LncRNA MALAT1 Promotes PPARα/CD36-Mediated Hepatic Lipogenesis in Nonalcoholic Fatty Liver Disease by Modulating miR-206/ARNT Axis. Front. Bioeng. Biotechnol. 2022, 10, 858558. [Google Scholar] [CrossRef]
- Yin, J.; Chen, X.; Zhang, F.; Zhao, M. RMRP Inhibition Prevents NAFLD Progression in Rats via Regulating miR-206/PTPN1 Axis. Mamm. Genome 2022, 33, 480–489. [Google Scholar] [CrossRef]

| Gene1 | Primer Sequences (5′-3′) | Source |
|---|---|---|
| ACACA | F: CATCTTGTCCGAAACGTCGAT | [15] |
| R: CCCTTCGAACATACACCTCCA | ||
| AGPAT6 | F: AAGCAAGTTGCCCATCCTCA | [15] |
| R: AAACTGTGGCTCCAATTTCGA | ||
| ATGL | F: CACCAGCATCCAGTTCAACCT | [15] |
| R: CTGTAGCCCTGTTTGCACATCT | ||
| CD36 | F: GTACAGATGCAGCCTCATTTCC | [15] |
| R: TGGACCTGCAAATATCAGAGGA | ||
| DGAT1 | F: CCACTGGGACCTGAGGTGTC | [15] |
| R: GCATCACCACACACCAATTCA | ||
| ELOVL6 | F: TTACAATGGACCTGTCAGCAAAT | [15] |
| R: TCATAGTCATAAACCAACCACCC | ||
| FABP3 | F: GAACTCGACTCCCAGCTTGAA | [15] |
| R: AAGCCTACCACAATCATCGAAG | ||
| FASN | F: ACCTCGTGAAGGCTGTGACTCA | [15] |
| R: TGAGTCGAGGCCAAGGTCTGAA | ||
| GPAM | F: GCAGGTTTATCCAGTATGGCATT | [15] |
| R: GGACTGATATCTTCCTGATCATCTTG | ||
| HSL | F: TCAGTGTCCAAGACAGAGCCAAT | [15] |
| R: CATGCAGCTTCAGGCTTTTG | ||
| LPIN1 | F: TGGCCACCAGAATAAAGCATG | [15] |
| R: GCTGACGCTGGACAACAGG | ||
| MRPL39 | F: AGGTTCTCTTTTGTTGGCATCC | [15] |
| R: TTGGTCAGAGCCCCAGAAGT | ||
| PPARG | F: CCAAATATCGGTGGGAGTCG | [15] |
| R: ACAGCGAAGGGCTCACTCTC | ||
| SREBF1 | F: CCAGCTGACAGCTCCATTGA | [15] |
| R: TGCGCGCCACAAGGA | ||
| UXT | F: TGTGGCCCTTGGATATGGTT | [15] |
| R: GGTTGTCGCTGAGCTCTGTG |
| Primer Name | Primer Sequences (5′-3′) |
|---|---|
| ELOVL6-3′UTR-F | GATGTTTATCATGGGAGGG |
| ELOVL6-3′UTR-R | GAATTGAAGTGCAGTGGAA |
| MiR-206 binding site | ACAUUCCA |
| Mutation site | GUACCUCC |
| ELOVL6-mut-F | CCTGCTGTCATTTAGUACCUCC |
| ELOVL6-mut-R | GCCCAAGTTTTTCGGAGGTAC |
| Name | Sequence (5′-3′) |
|---|---|
| siELOVL6-001 | GATGTTTATCATGGGAGGG |
| siELOVL6-002 | GAATTGAAGTGCAGTGGAA |
| siELOVL6-003 | ACAUUCCA |
| siRNA-Scr | GUACCUCC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Liu, Y.; Li, Y.; Zhang, Y.; Yang, C.; Yao, D. MiR-206 Suppresses Triacylglycerol Accumulation via Fatty Acid Elongase 6 in Dairy Cow Mammary Epithelial Cells. Animals 2024, 14, 2590. https://doi.org/10.3390/ani14172590
Zhao X, Liu Y, Li Y, Zhang Y, Yang C, Yao D. MiR-206 Suppresses Triacylglycerol Accumulation via Fatty Acid Elongase 6 in Dairy Cow Mammary Epithelial Cells. Animals. 2024; 14(17):2590. https://doi.org/10.3390/ani14172590
Chicago/Turabian StyleZhao, Xin, Yu Liu, Yupeng Li, Yuxin Zhang, Chunlei Yang, and Dawei Yao. 2024. "MiR-206 Suppresses Triacylglycerol Accumulation via Fatty Acid Elongase 6 in Dairy Cow Mammary Epithelial Cells" Animals 14, no. 17: 2590. https://doi.org/10.3390/ani14172590




