Functional Analysis of Genes Involved in Glycerolipids Biosynthesis (GPAT1 and GPAT2) in Pigs
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Tissue Samples and Carcass Characteristics
2.3. RNA Isolation and Reverse Transcription
2.4. Real-Time PCR Analysis
2.5. Statistical Analyses
3. Results
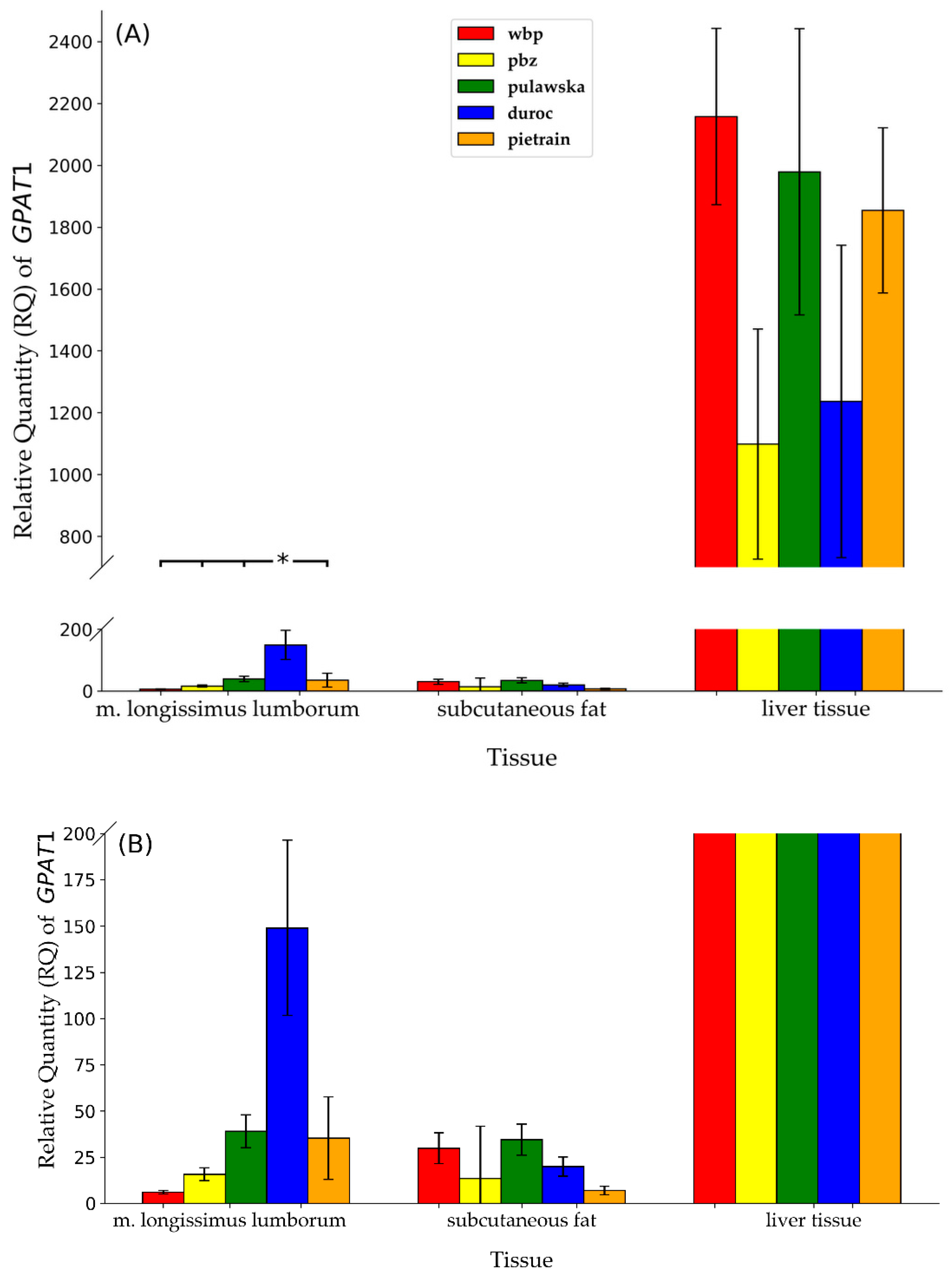
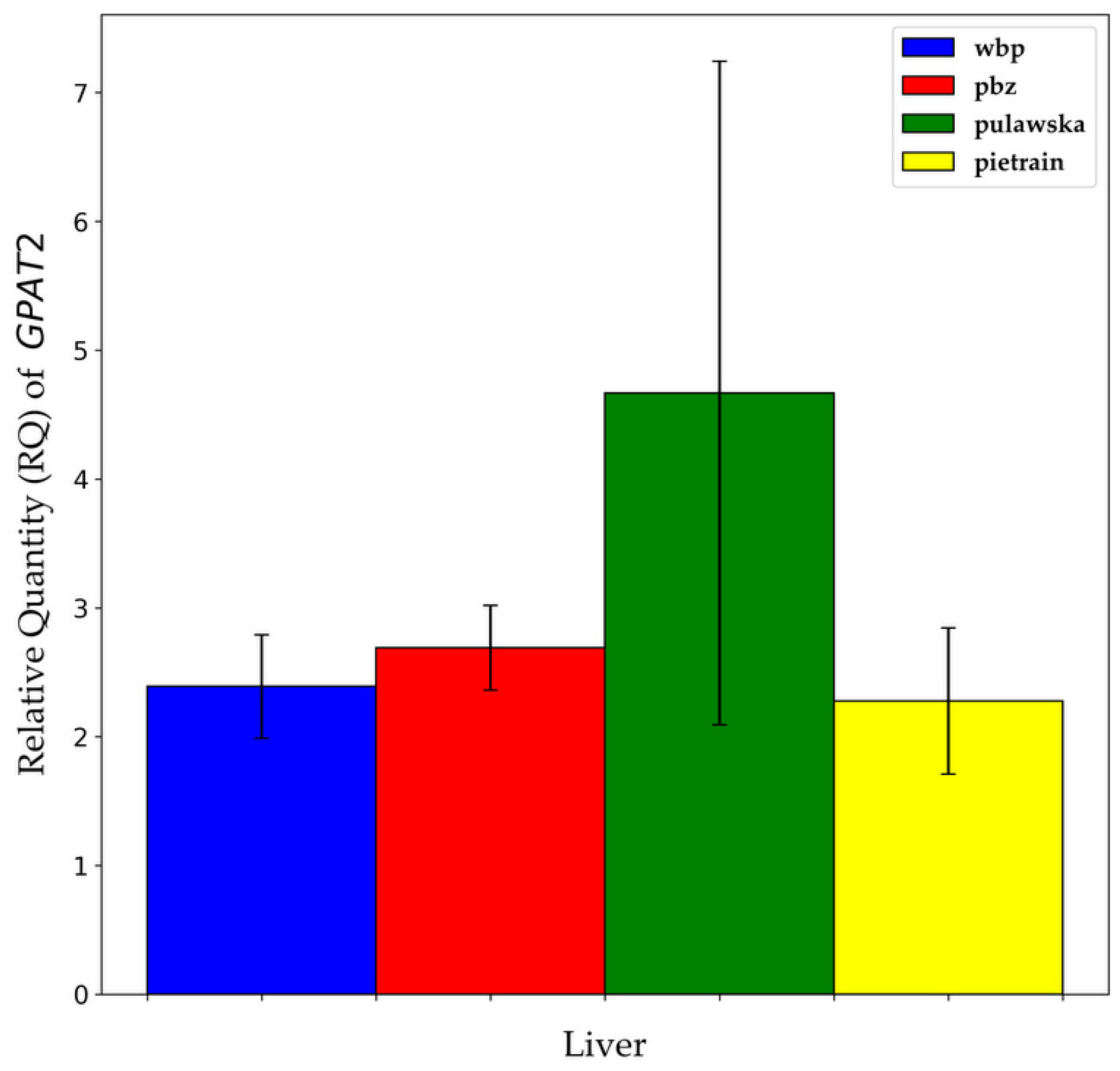
3.1. Expression Levels of GPAT1 and GPAT2 Genes in Analyzed Tissues
3.2. The mRNA Expression of GPAT1 between Tissues within Studied Breeds
3.3. GPAT1 and GPAT2 Gene Expression between Breeds within Analyzed Tissues
3.4. Correlation between the Expression Abundance of GPAT1 and GPAT2 Genes and Utility Traits in Pigs
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations:
| TAG | triacylglycerol |
| IMF | intramuscular fat |
| GPAT1 | glicerol-3-phosphate acyltransferase 1 |
| GPAT2 | glicerol-3-phosphate acyltransferase 2 |
| PL | polish landrace |
| PLW | polish large white |
| TPA | texture profile analysis |
| WBP | Warner–Bratzler shear force |
| WHC | water holding capacity |
| RFI | residual feed intake |
References
- Coleman, R.A.; Mashek, D.G. Mammalian triacylglycerol metabolism: Synthesis, lipolysis, and signaling. Chem. Rev. 2011, 111, 6359–6386. [Google Scholar] [CrossRef] [PubMed]
- Gimeno, R.E.; Cao, J. Mammalian glycerol-3-phosphate acyltransferases: New genes for an old activity. J. Lipid Res. 2008, 49, 2079–2088. [Google Scholar] [CrossRef] [PubMed]
- Hammond, L.E.; Gallagher, P.A.; Wang, S.; Hiller, S.; Kluckman, K.D.; Posey-Marcos, E.L.; Maeda, N.; Coleman, R.A. Mitochondrial glycerol-3-phosphate acyltransferase-deficient mice have reduced weight and liver triacylglycerol content and altered glycerolipid fatty acid composition. Mol. Cell. Biol. 2002, 22, 8204–8214. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Lee, D.P.; Gong, N.; Schwerbrock, N.M.; Mashek, D.G.; Gonzalez-Baró, M.R.; Stapleton, C.; Li, L.O.; Lewin, T.M.; Coleman, R.A. Cloning and functional characterization of a novel mitochondrial N-ethylmaleimide-sensitive glycerol-3-phosphate acyltransferase (GPAT2). Arch. Biochem. Biophys. 2007, 465, 347–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, J.; Kwon, E.G.; Im, S.K.; Seo, K.S.; Baik, M. Expression of fat deposition and fat removal genes is associated with intramuscular fat content in longissimus dorsi muscle of Korean cattle steers. J. Anim. Sci. 2012, 90, 2044–2053. [Google Scholar] [CrossRef] [PubMed]
- Tyra, M.; Żak, G. Analysis of relationships between fattening and slaughter performance of pigs and the level of intramuscular fat (IMF) in longissimus lumborum muscle. Ann. Anim. Sci. 2012, 12, 169–178. [Google Scholar] [CrossRef]
- Grau, R.; Hamm, R. Eine einfache Methode zur Bestimmung der Wasserbindung in Fleisch. Fleischwirtschaft 1952, 4, 295–297. [Google Scholar]
- Chomczynski, P. A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 1993, 15, 532–537. [Google Scholar]
- Piórkowska, K.; Ropka-Molik, K.; Eckert, R.; Różycki, M. The expression pattern of proteolytic enzymes of cathepsin family in two important porcine skeletal muscles. Livest. Sci. 2013, 157, 427–434. [Google Scholar] [CrossRef]
- Piórkowska, K.; Oczkowicz, M.; Różycki, M.; Ropka-Molik, K.; Piestrzyńska-Kajtoch, A. Novel porcine housekeeping genes for real-time RT-PCR experiments normalization in adipose tissue: Assessment of leptin mRNA quantity in different pig breeds. Meat Sci. 2011, 87, 191–195. [Google Scholar] [CrossRef]
- Lewin, T.M.; Schwerbrock, N.M.J.; Lee, D.P.; Coleman, R.A. Identification of a new glycerol-3-phosphate acyltransferase isoenzyme, mtGPAT2, in mitochondria. J. Biol. Chem. 2004, 279, 13488–13495. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, K.; Reue, K. Biochemistry, physiology, and genetics of GPAT, AGPAT, and lipid enzymes in triglyceride synthesis. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E1195–E1209. [Google Scholar] [CrossRef] [PubMed]
- Shi-Zheng, G.; Su-Mei, Z. Physiology, affecting factors and strategies for control of pig meat intramuscular fat. Recent Pat. Food Nutr. Agric. 2009, 1, 59–74. [Google Scholar] [CrossRef]
- Tyra, M.; Żak, G. Characteristics of the Polish breeding population of pigs in terms of intramuscular fat (IMF) content of m. longissimus dorsi. Ann. Anim. Sci. 2010, 10, 241–248. [Google Scholar]
- Freeman, T.C.; Ivens, A.; Baillie, J.K.; Beraldi, D.; Barnett, M.W.; Dorward, D.; Downing, A.; Fairbairn, L.; Kapetanovic, R.; Raza, S.; et al. A gene expression atlas of the domestic pig. BMC Biol. 2012, 10, 90. [Google Scholar] [CrossRef] [PubMed]
- BioGPS. Available online: http://biogps.org/dataset/BDS_00012/pig-atlas/ (accessed on 27 May 2019).
- Bertolesi, G.E.; Iannattone, S.; Johnston, J.; Zaremberg, V.; McFarlane, S. Identification and expression analysis of GPAT family genes during early development of Xenopus laevis. Gene Expr. Patterns 2012, 12, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Ehara, T.; Kamei, Y.; Takahashi, M.; Yuan, X.; Kanai, S.; Tamura, E.; Tanaka, M.; Yamazaki, T.; Miura, S.; Ezaki, O.; et al. Role of DNA methylation in the regulation of lipogenic glycerol-3-phosphate acyltransferase 1 gene expression in the mouse neonatal liver. Diabetes 2012, 61, 2442–2450. [Google Scholar] [CrossRef]
- Lewin, T.M.; Granger, D.A.; Kim, J.H.; Coleman, R.A. Regulation of mitochondrial sn-glycerol-3-phosphate acyltransferase activity: Response to feeding status is unique in various rat tissues and is discordant with protein expression. Arch. Biochem. Biophys. 2001, 396, 119–127. [Google Scholar] [CrossRef]
- Cao, J.; Li, J.L.; Li, D.; Tobin, J.F.; Gimeno, R.E. Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 19695–19700. [Google Scholar] [CrossRef]
- Weber, K.L.; Welly, B.T.; Van Eenennaam, A.L.; Young, A.E.; Porto-Neto, L.R.; Reverter, A.; Rincon, G. Identification of Gene Networks for Residual Feed Intake in Angus Cattle Using Genomic Prediction and RNA-seq. PLoS ONE 2016, 11, e0152274. [Google Scholar] [CrossRef]
- Claire D’Andre, H.; Paul, W.; Shen, X.; Jia, X.; Zhang, R.; Sun, L.; Zhang, X. Identification and characterization of genes that control fat deposition in chickens. J. Anim. Sci. Biotechnol. 2013, 4, 43. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Loor, J.J. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genom. 2008, 9, 366. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.N.; Wang, Y.; Xu, Y.O.; Li, Y.C.; Tian, F.; Jiang, M.F. Characterization of gene expression related to milk fat synthesis in the mammary tissue of lactating yaks. J. Dairy Res. 2017, 84, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Zhao, Z.; Yu, X.; Li, J.; Lu, C.; Yang, R. Bovine lipid metabolism related gene GPAM: Molecular characterization, function identification, and association analysis with fat deposition traits. Gene 2017, 609, 9–18. [Google Scholar] [CrossRef] [PubMed]


| Gene (Accession no.) | Primer Sequence | Probe Sequence | Amplicon Size, bp |
|---|---|---|---|
| GPAT1 (NC_010456.4) | TaqMan Gene Expression Assay ID: Ss03373682_m1 (Applied Biosystems) | (FAM) | 104 |
| GPAT2 (NC_010445.3) | F: GGAAATACCGCCCTTTTGTG R: GCAGAAGCCTAGATCCATTATGC | CACCCCTAAGAGCTGG (NED) | 98 |
| RPL27 (NM_001097479) | F: CGCTACTCCGGACGCAAA R: GGTCTGAGGTGCCATCATCA | CGGTCATCGTAAAGAA (VIC) | 58 |
| RPS29 (NM_001001633) | F: CGGAAATACGGCCTCAATATG R: GCCAATATCCTTCGCGTACTG | CCGCCAGTGCTTC (VIC) | 60 |
| Gene/Tissue | GPAT1 | GPAT2 | ||
|---|---|---|---|---|
| Slaughter Traits | m. longissimus lumborum | Subcutaneous Fat | Liver Tissue | Liver Tissue |
| Slaughter performance (%) | −0.185 | 0.083 | −0.088 | 0.174 |
| Loin weight (kg) | −0.277 * | 0.023 | −0.115 | 0.187 |
| Loin weight without skin and backfat (kg) | −0.192 | 0.034 | −0.265 * | 0.033 |
| Ham weight without skin and backfat (kg) | −0.146 | −0.081 | −0.234 | −0.094 |
| Carcass length (cm) | −0.058 | 0.042 | 0.059 | 0.045 |
| Mean backfat thickness from five measurements (cm) | 0.087 | 0.024 | 0.150 | 0.068 |
| Loin eye width (cm) | 0.030 | 0.257 * | −0.056 | −0.112 |
| Loin eye height (cm) | −0.212 * | −0.080 | −0.074 | −0.034 |
| Loin eye area (cm2) | −0.140 | 0.004 | −0.220 | −0.076 |
| Lean meat content (%) | −0.022 | −0.072 | −0.228 * | −0.219 |
| Weight of meat from primal cuts (kg) | −0.177 | −0.015 | −0.261 * | −0.082 |
| Gene/Tissue | GPAT1 | GPAT2 | ||
|---|---|---|---|---|
| Meat Texture Parameters | m. longissimus lumborum | Subcutaneous Fat | Liver Tissue | Liver Tissue |
| WB_R_FIR | 0.032 | −0.048 | 0.049 | 0.003 |
| WB_R_TUG | -0.081 | −0.047 | 0.050 | −0.022 |
| WB_C_FIR | −0.270 * | −0.089 | −0.038 | −0.002 |
| WB_R_TUG | −0.091 | −0.115 | 0.111 | −0.126 |
| TPA_ hardness | 0.012 | −0.059 | 0.035 | −0.115 |
| TPA_ springiness | 0.049 | −0.408 * | 0.292 * | 0.503 * |
| TPA_ cohesiveness | 0.033 | −0.358 * | 0.229 * | 0.772 ** |
| TPA_ chewiness | 0.035 | −0.156 | 0.116 | −0.006 |
| TPA_ resilience | 0.080 | −0.334 * | 0.229 * | 0.833 ** |
| Gene/Tissue | GPAT1 | GPAT2 | ||
|---|---|---|---|---|
| Meat Quality Parameters | m. longissimus lumborum | Subcutaneous Fat | Liver Tissue | Liver Tissue |
| Intramuscular fat content (IMF; %) | 0.053 | 0.066 | 0.285 * | 0.165 |
| Water holding capacity (%) | 0.196 | 0.112 | −0.207 | −0.030 |
| Meat color (L *) | −0.080 | 0.074 | 0.163 | 0.260 |
| Meat color (a *) | 0.109 | −0.305 * | 0.132 | 0.306 * |
| Meat color (b *) | 0.073 | −0.120 | 0.183 | 0.246 |
| 45 min post-mortem pH in MLD | −0.151 | 0.083 | 0.148 | 0.323 * |
| 24 h post-mortem pH in MLD | 0.028 | 0.043 | −0.187 | 0.072 |
| 45 min post-mortem pH in SEMI | −0.228 | −0.202 | 0.205 | 0.047 |
| 24 h post-mortem pH in SEMI | −0.058 | 0.090 | 0.025 | −0.289 |
| Gene/Tissue. | GPAT1 | GPAT2 | ||
|---|---|---|---|---|
| Fattening Traits | m. longissimus lumborum | Subcutaneous Fat | Liver Tissue | Liver Tissue |
| Daily gain in performance test (g) | −0.070 | −0.057 | 0.072 | −0.310 |
| Daily live gain (g) | −0.055 | −0.231 | −0.010 | −0.302 * |
| Feed conversion ratio | 0.032 | 0.178 | −0.034 | 0.077 |
| Daily feed intake (kg) | −0.046 | 0.066 | 0.044 | −0.284 |
| Age at slaughter (days) | −0.003 | 0.246 * | 0.004 | 0.363 * |
| The number of days of control fattening (days) | 0.010 | 0.066 | −0.053 | 0.424 * |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mitka, I.; Ropka-Molik, K.; Tyra, M. Functional Analysis of Genes Involved in Glycerolipids Biosynthesis (GPAT1 and GPAT2) in Pigs. Animals 2019, 9, 308. https://doi.org/10.3390/ani9060308
Mitka I, Ropka-Molik K, Tyra M. Functional Analysis of Genes Involved in Glycerolipids Biosynthesis (GPAT1 and GPAT2) in Pigs. Animals. 2019; 9(6):308. https://doi.org/10.3390/ani9060308
Chicago/Turabian StyleMitka, Ilona, Katarzyna Ropka-Molik, and Mirosław Tyra. 2019. "Functional Analysis of Genes Involved in Glycerolipids Biosynthesis (GPAT1 and GPAT2) in Pigs" Animals 9, no. 6: 308. https://doi.org/10.3390/ani9060308
APA StyleMitka, I., Ropka-Molik, K., & Tyra, M. (2019). Functional Analysis of Genes Involved in Glycerolipids Biosynthesis (GPAT1 and GPAT2) in Pigs. Animals, 9(6), 308. https://doi.org/10.3390/ani9060308





