Evaluation of a Multi-Gene Methylation Blood-Test for the Detection of Colorectal Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Recruitment and Ethics
2.2. Clinical Specimens
2.3. DNA Isolation and Bisulfite Treatment
2.4. MethylLight Droplet Digital PCR (ML-ddPCR) Protocol
2.5. Calculation of the LoD and LoB
2.6. Statistical Analysis
3. Results
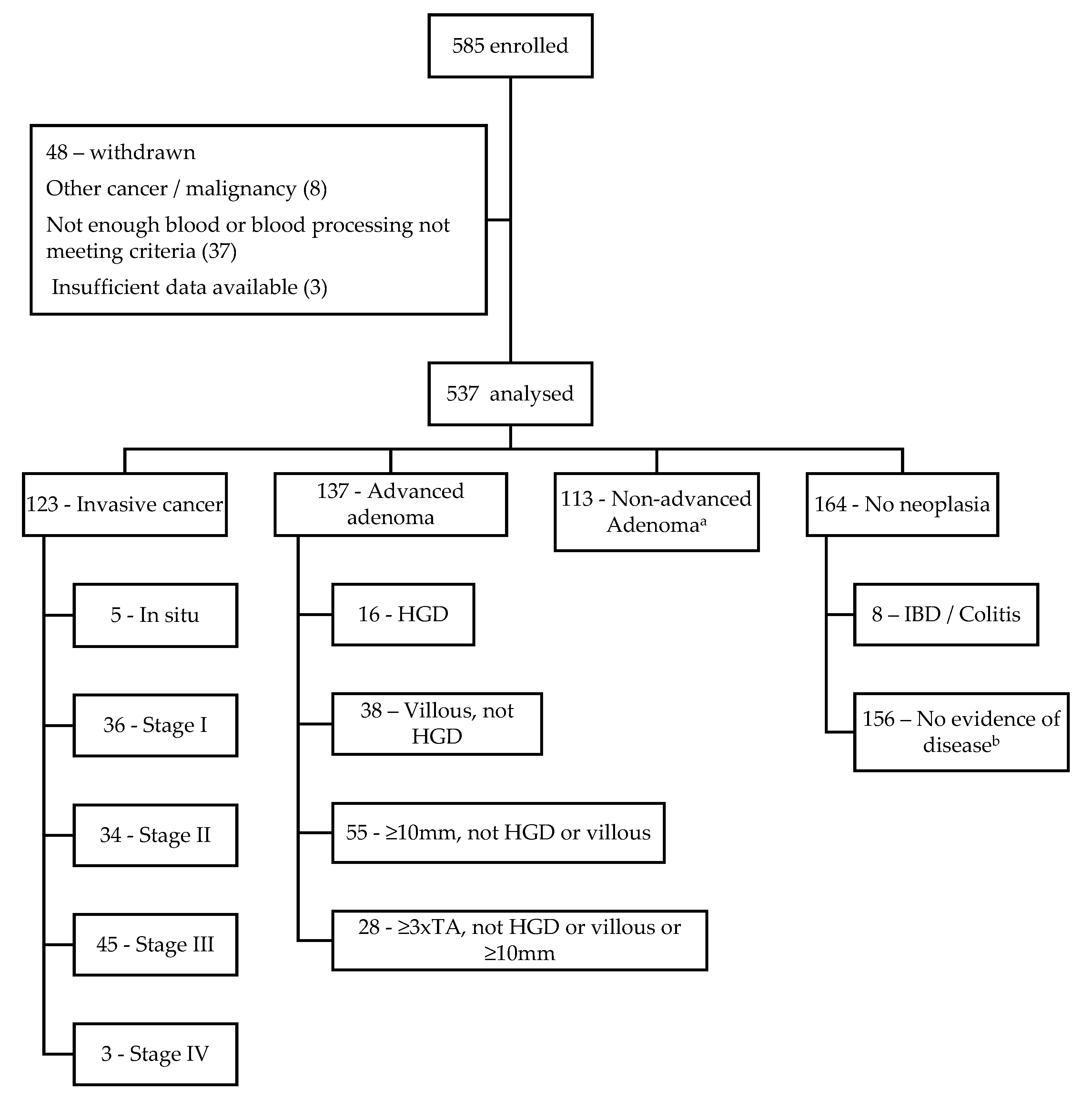
3.1. Population
3.2. Sensitivity and Specificity for CRC Using ctDNA
3.3. Sensitivity for Detection of Adenomas and Colitis
3.4. Association of Cancer-Specific Variables and Confounding Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Global Cancer Observatory: International Agency for Research on Cancer. World Health Organisation. Available online: https://gco.iarc.fr/today/data/factsheets/cancers/10_8_9-Colorectum-fact-sheet.pdf (accessed on 23 September 2021).
- Elmunzer, B.J.; Hayward, R.A.; Schoenfeld, P.S.; Saini, S.D.; Deshpande, A.; Waljee, A.K. Effect of flexible sigmoidoscopy-based screening on incidence and mortality of colorectal cancer: A systematic review and meta- analysis of randomized controlled trials. PLoS Med. 2012, 9, e1001352. [Google Scholar] [CrossRef]
- Kronborg, O.; Fenger, C.; Olsen, J.; Jørgensen, O.D.; Søndergaard, O. Randomised study of screening for colorectal cancer with faecal-occult-blood test. Lancet 1996, 348, 1467–1471. [Google Scholar] [CrossRef]
- Chiu, H.-M.; Jen, G.H.-H.; Wang, Y.-W.; Fann, J.C.-Y.; Hsu, C.-Y.; Jeng, Y.-C.; Yen, A.M.-F.; Chiu, S.Y.-H.; Chen, S.L.-S.; Hsu, W.-F.; et al. Long-term effectiveness of faecal immunochemical test screening for proximal and distal colorectal cancers. Gut 2021, 70, 2321–2329. [Google Scholar] [CrossRef] [PubMed]
- Mandel, J.S.; Church, T.R.; Bond, J.H.; Ederer, F.; Geisser, M.S.; Mongin, S.J.; Snover, D.C.; Schuman, L.M. The effect of fecal occult-blood screening on the incidence of colorectal cancer. N. Engl. J. Med. 2000, 343, 1603–1607. [Google Scholar] [CrossRef] [PubMed]
- Australian Institute of Health and Welfare. National Bowel Cancer Screening Program: Monitoring Report 2020; Cancer Series no. 128; Cat. no. CAN 133; AIHW: Canberra, Australia, 2020. Available online: https://www.aihw.gov.au/reports/cancer-screening/national-bowel-cancer-screening-monitoring-2020/summary (accessed on 11 August 2023).
- Osborne, J.; Wilson, C.; Moore, V.; Gregory, T.; Flight, I.; Young, G. Samples preference for colorectal cancer screening tests: Blood or stool? Open J. Prev. Med. 2012, 2, 326–331. [Google Scholar] [CrossRef]
- Symonds, E.L.; Pedersen, S.; Cole, S.R.; Massolino, J.; Byrne, D.; Guy, J.; Backhouse, P.; Fraser, R.J.; LaPointe, L.; Young, G.P. Improving participation in colorectal cancer screening: A randomised controlled trial of sequential offers of faecal then blood based non-invasive tests. Asian Pac. J. Cancer Prev. 2015, 16, 8455–8460. [Google Scholar] [CrossRef]
- Song, L.; Jia, J.; Peng, X.; Xiao, W.; Li, Y. The performance of the SEPT9 gene methylation assay and a comparison with other colorectal screening tests: A meta-analysis. Sci. Rep. 2017, 7, 3032. [Google Scholar] [CrossRef]
- Ladabaum, U.; Alvarez-Osorio, L.; Rosch, T.; Brueggenjuergen, B. Cost-effectiveness of colorectal cancer screening in Germany: Current endoscopic and faecal testing strategies versus plasma methylated Septin 9 DNA. Endosc. Int. Open 2014, 2, E96–E104. [Google Scholar] [CrossRef]
- Murry, D.; Baker, R.; Gaur, S.; Young, G.; Pedersen, S. Validation of a circulating tumour-derived DNA blood test for detection of methylated BCAT1 and IKZF1 DNA. J. Appl. Lab. Med. 2017, 2, 165–175. [Google Scholar] [CrossRef]
- Locke, W.; Guanzon, D.; Ma, C.; Liew, Y.; Duesing, K.; Fung, K.; Ross, J. DNA methylation cancer biomarkers: Translation to the clinic. Front. Genet. 2019, 10, 1150. [Google Scholar] [CrossRef]
- Wei, B.; Wu, F.; Xing, W.; Sun, H.; Yan, C.; Zhao, C.; Wang, D.; Chen, X.; Chen, Y.; Li, M.; et al. A panel of DNA methylation biomarkers for detection and improving diagnostic efficiency of lung cancer. Sci. Rep. 2021, 11, 16782. [Google Scholar] [CrossRef] [PubMed]
- Kubiliūtė, R.; Žukauskaitė, K.; Žalimas, A.; Ulys, A.; Sabaliauskaitė, R.; Bakavičius, A.; Želvys, A.; Jankevičius, F.; Jarmalaitė, S. Clinical significance of novel DNA methylation biomarkers for renal clear cell carcinoma. J. Cancer Res. Clin. Oncol. 2022, 148, 361–375. [Google Scholar] [CrossRef] [PubMed]
- Henriksen, S.D.; Thorlacius-Ussing, O. Cell-Free DNA Methylation as Blood-Based Biomarkers for Pancreatic Adenocarcinoma-A Literature Update. Epigenomes 2021, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Petit, J.; Carroll, G.; Gould, T.; Pockney, P.; Dun, M.; Scott, R.J. Cell-free DNA as a diagnostic blood-based biomarker for colorectal cancer: A systematic review. J. Surg. Res. 2019, 236, 184–197. [Google Scholar] [CrossRef]
- Laugsand, E.A.; Brenne, S.S.; Skorpen, F. DNA methylation markers detected in blood, stool, urine, and tissue in colorectal cancer: A systematic review of paired samples. Int. J. Color. Dis. 2021, 36, 239–251. [Google Scholar] [CrossRef]
- Petit, J.; Carroll, G.; Zhao, J.; Roper, E.; Pockney, P.; Scott, R.J. Evaluation of epigenetic methylation biomarkers for the detection of colorectal cancer using droplet digital PCR. Sci. Rep. 2023, 13, 8883. [Google Scholar] [CrossRef]
- Edge, S.; Byrd, D.; Compton, C.; Fritz, A.; Greene, F.; Trotti, A. AJCC Cancer Staging Manual, 7th ed.; Springer: New York, NY, USA, 2010. [Google Scholar]
- St. John, J.; Ee, H.; Canfell, K.; Chetcuti, A.; Emery, J.; Grogan, P.; Macrae, F.; Jenkins, M.; Mulqueeney, R.; Cancer Council Australia Colorectal Cancer Guidelines Working Party. Clinical question: Colorectal cancer screening test accuracy. In Clinical Practice Guidelines for the Prevention, Early Detection and Management of Colorectal Cancer; Cancer Council Australia: Sydney, Australia, 2017; Available online: https://wiki.cancer.org.au/australia/Guidelines:Colorectal_cancer (accessed on 20 December 2022).
- Jones, M.J.; Goodman, S.J.; Kobor, M.S. DNA Methylation and healthy human aging. Aging Cell 2015, 14, 924–932. [Google Scholar] [CrossRef]
- Selby, K.; Levine, E.H.; Doan, C.; Gies, A.; Brenner, H.; Quesenberry, C.; Lee, J.K.; Corley, D.A. Effect of Sex, Age, and Positivity Threshold on Fecal Immunochemical Test Accuracy: A Systematic Review and Meta-analysis. Gastroenterology 2019, 157, 1494–1505. [Google Scholar] [CrossRef]
- Lee, K.; Pausova, Z. Cigarette smoking and DNA methylation. Front. Genet. 2013, 4, 132. [Google Scholar] [CrossRef]
- De Klerk, C.M.; Vendrig, L.M.; Bossuyt, P.M.; Dekker, E. Participant-Related Risk Factors for False-Positive and False-Negative Fecal Immunochemical Tests in Colorectal Cancer Screening: Systematic Review and Meta-Analysis. Am. J. Gastroenterol. 2018, 113, 1778–1787. [Google Scholar] [CrossRef]
- Alexander, J.C.; Silverman, N.A.; Chretien, P.B. Effect of Age and Cigarette Smoking on Carcinoembryonic Antigen Levels. JAMA 1976, 235, 1975–1979. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.S.; Chen, C.Y.; Huang, L.K.; Wang, W.S.; Yang, S.H. Prognostic value of postoperative serum carcinoembryonic antigen levels in colorectal cancer patients who smoke. PLoS ONE 2020, 15, e0233687. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.Y.; Choe, E.K.; Park, K.J.; Lee, Y. Factors requiring adjustment in the interpretation of serum carcinoembryonic antigen: A Cross-sectional study of 18,131 healthy non-smokers. Gastroenterol. Res. Pract. 2017, 2017, 9858931. [Google Scholar] [CrossRef] [PubMed]
| Target Gene | ||
|---|---|---|
| ACTB | Forward | TGGTGATGGAGGAGGTTTAGTAAG |
| Reverse | ACCAATAAAACCTACTCCTCCCTTA | |
| Probe | VIC/ACCACCACCCAACACACAATAACAAACA/MGBNFQ | |
| IKZF1 | Forward | TGCGCGTTTCGTTTTTTGTATCG |
| Reverse | GATCCCTACTCGACCTACCCCGC | |
| Probe | FAM/CGACCGCCTCCCGAATCGC/MGBNFQ | |
| NPY | Forward | +C+G+AGGTTTTTTTTGTCGC |
| Reverse | ATAC+T+A+T+CGAACGAACG | |
| Probe | FAM/CAAAAAACGA+A+T+C+G+C+GACAA/3IABkFQ | |
| SDC2 | Forward | AAATTA+A+T+A+AGTGAGAGGGCGTC |
| Reverse | GAC+T+C+A+AACTCGAAAACTCG | |
| Probe | FAM/CGTAGGAGGAGGAAG+C+G+A+G+C/3IABkFQ | |
| SEPT9 | Forward | +C+G+T+CGTTGTTTTTCG |
| Reverse | CCCACCTTCGAAATCCG | |
| Probe | FAM/CGTTAACCGCGAAATCCG/MGBNFQ |
| No. Cases | Age (Years) | Gender | BMI | CCI | NSAID Use | Immunosuppression | Smoking Status | cfDNA | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n (%) | Median (Min-Max) | Women | Men | Median (Min-Max) | Median (Min-Max) | No | Yes | No | Yes | Non-Smoker | Ex-Smoker | Smoker | Median (Range) | Mean (SD) | |
| n (%) | n (%) | n (%) | n (%) | ||||||||||||
| Cancer | 123 (23) | 70 (37–92) | 47 (38) | 76 (62) | 28 (20–54) | 5 (2–9) | 95 (77) | 28 (23) | 115 (93) | 8 (7) | 58 (47) | 53 (43) | 12 (10) | 6.17 (56.1) | 7.96 (7.02) |
| Stage 0 | 5 | 65 (58–86) | 2 (40) | 3 (60) | 26 (21–32) | 4 (4–7) | 3 (60) | 2 (40) | 5 (100) | 0 | 2 (40) | 3 (60) | 0 | 4.37 (6.9) | 5.21 (2.73) |
| Stage I | 36 | 68 (37–85) | 17 (47) | 19 (53) | 27 (20–49) | 5 (2–9) | 29 (81) | 7 (19) | 34 (94) | 2 (6) | 16 (44) | 16 (44) | 4 (11) | 6.45 (14.6) | 6.32 (3.24) |
| Stage II | 34 | 72 (48–91) | 12 (35) | 22 (65) | 30 (20–46) | 5 (2–9) | 24 (71) | 10 (29) | 32 (94) | 2 (6) | 18 (53) | 10 (29) | 6 (18) | 6.90 (54.7) | 9.0 (9.14) |
| Stage III | 45 | 71 (48–92) | 15 (33) | 30 (67) | 31 (20–54) | 5 (2–9) | 36 (80) | 9 (20) | 41 (91) | 4 (9) | 20 (44) | 24 (53) | 1 (2) | 6.56 (37.6) | 8.43 (7.16) |
| Stage IV | 3 | 69 (56–87) | 1 (33) | 2 (67) | 29 (25–29) | 8 (3–8) | 3 (100) | 0 | 3 (100) | 0 | 2 (67) | 0 | 1 (33) | 6.0 (22.4) | 13.35 (12.89) |
| AA | 137 (25) | 64 (47–85) | 48 (35) | 89 (65) | 29 (19–43) | 3 (0–9) | 112 (82) | 25 (18) | 135 (98) | 2 (2) | 62 (45) | 42 (31) | 33 (24) | 6.07 (32.5) | 7.04 (4.42) |
| NAA | 113 (21) | 64 (49–79) | 57 (50) | 56 (50) | 29 (19–41) | 2 (0–8) | 101 (89) | 12 (11) | 109 (96) | 4 (4) | 50 (44) | 34 (30) | 29 (26) | 5.83 (21.2) | 6.65 (3.55) |
| No neoplasia a | 164 (31) | 62 (45–77) | 91 (55) | 73 (45) | 28 (17–42) | 2 (0–6) | 137 (83) | 27 (17) | 157 (96) | 7 (4) | 80 (49) | 56 (34) | 28 (17) | 5.74 (21.9) | 6.34 (2.97) |
| IBD/Colitis | 8 | 58 (47–69) | 6 (75) | 2 (25) | 24 (20–36) | 1 (0–3) | 8 (100) | 0 | 8 (100) | 0 | 3 (38) | 5 (62) | 0 | 5.0 (5.1) | 4.83 (1.62) |
| NED b | 156 | 62 (45–77) | 85 (54) | 71 (46) | 28 (17–42) | 2 (0–6) | 129 (83) | 27 (17) | 149 (95) | 7 (5) | 77 (49) | 51 (33) | 28 (18) | 5.84 (21.9) | 6.41 (3.0) |
| Study Cohort Overall | 537 (100) | 64 (37–92) | 243 (45) | 294 (55) | 29 (17–54) | 3 (0–9) | 445 (83) | 92 (17) | 516 (96) | 21 (4) | 250 (47) | 185 (34) | 102 (19) | 6.02 (56.4) | 6.95 (4.68) |
| p-value c | <0.001 | 0.08 | 0.885 | <0.001 | 0.076 | 0.11 | <0.05 | 0.154 | |||||||
| Cancer n (%) | ||
|---|---|---|
| T-Stage | In Situ | 5 (4) |
| T1 | 10 (8) | |
| T2 | 33 (27) | |
| T3 | 61 (50) | |
| T4 | 14 (11) | |
| Nodal status | Negative | 75 (61) |
| Positive | 48 (39) | |
| Metastatic disease | Negative | 120 (98) |
| Positive | 3 (2) | |
| LVI | Negative | 84 (68) |
| Positive | 39 (32) | |
| Site of Cancer | Right | 49 (40) |
| Left | 73 (59) | |
| Synchronous | 1 (1) | |
| Total | 123 |
| Analysis Method (LoD) | Status | Cancer | Total | p-Value | |
|---|---|---|---|---|---|
| No Cancer | Cancer | ||||
| n (%) | |||||
| Model 1a | Negative | 396 (96) | 78 (63) | 474 | <0.001 |
| Positive | 18 (4) | 45 (37) | 63 | ||
| Model 1b | Negative | 366 (88) | 72 (58) | 438 | <0.001 |
| Positive | 48 (12) | 51 (42) | 99 | ||
| Model 2 | Negative | 382 (92) | 69 (56) | 451 | <0.001 |
| Positive | 32 (8) | 54 (44) | 86 | ||
| Model 3 | Negative | 353 (85) | 61 (50) | 414 | <0.001 |
| Positive | 61 (15) | 62 (50) | 123 | ||
| Model 4 | Negative | 409 (99) | 89 (72) | 498 | <0.001 |
| Positive | 5 (1) | 34 (28) | 39 | ||
| Total | 414 | 123 | 537 | ||
| Analysis Method (LoD) | Status | Stage | Total | p-Value | ||||
|---|---|---|---|---|---|---|---|---|
| In Situ | 1 | 2 | 3 | 4 | ||||
| N (%) | ||||||||
| Model 1a | Negative | 5 (100) | 25 (69) | 24 (71) | 23 (51) | 1 (33) | 78 | 0.074 |
| Positive | 0 | 11 (31) | 10 (29) | 22 (49) | 2 (67) | 45 | ||
| Model 1b | Negative | 4 (80) | 20 (56) | 21 (62) | 26 (58) | 1 (33) | 72 | 0.749 |
| Positive | 1 (20) | 16 (44) | 13 (38) | 19 (42) | 2 (67) | 51 | ||
| Model 2 | Negative | 5 (100) | 21 (58) | 22 (65) | 21 (47) | 0 | 69 | 0.026 |
| Positive | 0 | 15 (42) | 12 (35) | 24 (53) | 3 (100) | 54 | ||
| Model 3 | Negative | 4 (80) | 18 (50) | 19 (56) | 19 (42) | 1 (33) | 61 | 0.469 |
| Positive | 1 (20) | 18 (50) | 15 (44) | 26 (58) | 2 (67) | 62 | ||
| Model 4 | Negative | 5 (100) | 27 (75) | 26 (76) | 30 (67) | 1 (33) | 89 | 0.244 |
| Positive | 0 | 9 (25) | 8 (24) | 15 (33) | 2 (67) | 34 | ||
| Total | 5 | 36 | 34 | 45 | 3 | 123 | ||
| Analysis Method (LoD) | Status | Principle Diagnosis | Total | p-Value | ||||
|---|---|---|---|---|---|---|---|---|
| Cancer | AA | NAA | IBD/Colitis | NED a | ||||
| N (%) | ||||||||
| Model 1a | Negative | 78 (63) | 134 (98) | 105 (93) | 8 (100) | 149 (95) | 474 | <0.001 |
| Positive | 45 (37) | 3 (2) | 8 (7) | 0 | 7 (5) | 63 | ||
| Model 1b | Negative | 72 (58) | 118 (86) | 101 (89) | 7 (87) | 140 (90) | 438 | <0.001 |
| Positive | 51 (42) | 19 (14) | 12 (11) | 1 (13) | 16 (10) | 99 | ||
| Model 2 | Negative | 69 (56) | 128 (93) | 102 (90) | 8 (100) | 144 (92) | 451 | <0.001 |
| Positive | 54 (44) | 9 (7) | 11 (10) | 0 | 12 (8) | 86 | ||
| Model 3 | Negative | 61 (50) | 117 (85) | 96 (85) | 7 (87) | 133 (85) | 414 | <0.001 |
| Positive | 62 (50) | 20 (15) | 17 (15) | 1 (13) | 23 (15) | 123 | ||
| Model 4 | Negative | 89 (72) | 135 (98) | 110 (97) | 8 (100) | 156 (100) | 498 | <0.001 |
| Positive | 34 (28) | 2 (2) | 3 (3) | 0 | 0 | 39 | ||
| Total | 123 | 137 | 113 | 8 | 156 | 537 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petit, J.; Carroll, G.; Williams, H.; Pockney, P.; Scott, R.J. Evaluation of a Multi-Gene Methylation Blood-Test for the Detection of Colorectal Cancer. Med. Sci. 2023, 11, 60. https://doi.org/10.3390/medsci11030060
Petit J, Carroll G, Williams H, Pockney P, Scott RJ. Evaluation of a Multi-Gene Methylation Blood-Test for the Detection of Colorectal Cancer. Medical Sciences. 2023; 11(3):60. https://doi.org/10.3390/medsci11030060
Chicago/Turabian StylePetit, Joel, Georgia Carroll, Henry Williams, Peter Pockney, and Rodney J. Scott. 2023. "Evaluation of a Multi-Gene Methylation Blood-Test for the Detection of Colorectal Cancer" Medical Sciences 11, no. 3: 60. https://doi.org/10.3390/medsci11030060
APA StylePetit, J., Carroll, G., Williams, H., Pockney, P., & Scott, R. J. (2023). Evaluation of a Multi-Gene Methylation Blood-Test for the Detection of Colorectal Cancer. Medical Sciences, 11(3), 60. https://doi.org/10.3390/medsci11030060







