Whole Organ Engineering: Approaches, Challenges, and Future Directions
Abstract
:Featured Application
Abstract
1. Introduction
2. Review Methodology
2.1. Literature Search
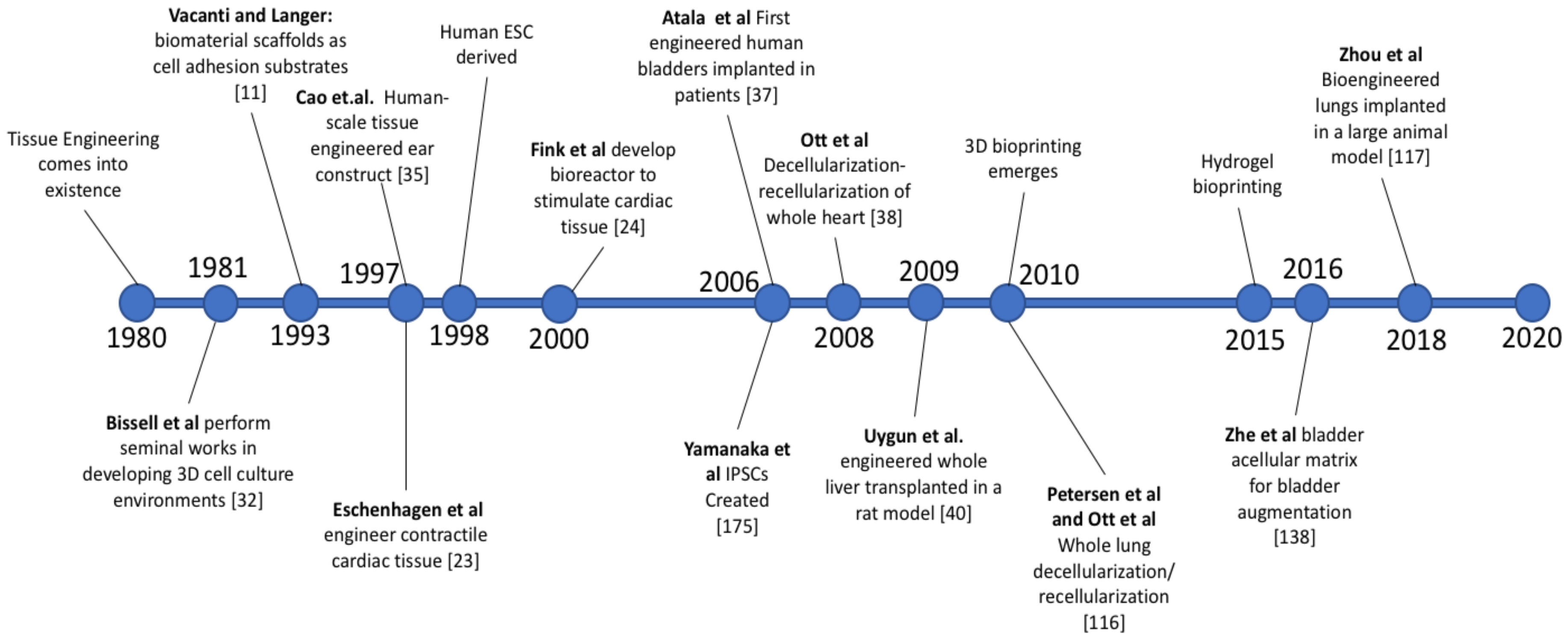
2.2. History of Whole Organ Engineering
2.3. Approaches to Engineering Whole Organs
2.4. Recent Developments and Accomplishments
2.5. Supporting Technological Advancements
2.6. Major Challenges and Barriers to Market
3. History of Whole Organ Engineering
3.1. Origin and Foundational Technologies
3.2. Extracellular Matrix
3.3. Growing Human Tissue In Vivo
3.4. Autologous Engineered Tissues for Organ Resuscitation
3.5. First Engineered Beating Heart
3.6. Early Success in Animal Studies for Liver Engineering
3.7. Academic and Industry Pioneers
4. Approaches to Engineering Whole Organs
4.1. High-Level Strategies
4.2. Building Organs from Cadaveric Organs (De-Cell/Re-Cell)
4.3. Building Organs from Synthetic Biomaterials
4.4. Building Organs from Biohybrid Materials
4.5. Three-Dimensional Bioprinting
4.6. Organoids
5. Recent Developments and Accomplishments
5.1. Heart
5.2. Liver
5.3. Kidney
5.4. Lungs
5.5. Pancreas
5.6. Bladder
5.7. Skin
5.8. Gastrointestinal
| Year | Title | Number of Citations | Summary and Significance |
|---|---|---|---|
| 2010 | Preparation of Cardiac Extracellular Matrix from an Intact Porcine Heart (Wainwright) [97] | 201 | In this study, a whole porcine decellularized heart was obtained in less than ten hours using pulsatile retrograde aortic perfusion of various enzymes, detergents, and acid solutions. This decellularized heart not only maintained the ECM components and mechanical properties of a native heart but also supported the differentiation of chicken cardiomyocytes into sarcomeres. This study marked the first full-scale decellularization of a heart close to human proportions and paved the way to using decellularized hearts in preclinical and clinical studies for cardiac therapies. Additionally, the perfusion method used for decellularization of whole organs is superior to previous methods in maintaining the native ECM compositions and architecture and efficiently removing cells. As a result, this method has been adapted for decellularizing other organ types, advancing the entire field of whole organ engineering as a result. |
| 2011 | The Use of Whole Organ Decellularization for the Generation of a Vascularized Liver Organoid (Baptista) [63] | 393 | In this study, whole livers were engineered with functional vasculature by first using perfusion decellularization of detergents through the vascular network of the liver followed by perfusion of human fetal liver and endothelial cells through this vascular network. This resulted in the creation of whole livers in vitro seeded with human cells containing vasculature with fully formed endothelium and differentiated hepatic and epithelial tissues surrounding them. This study represents a significant advancement in whole organ engineering since many approaches lack the creation of functional vasculature, which is essential to support cell viability and tissue function, and struggle to both adequately decellularize large organs and recellularize them with the appropriate number of cells to create a functional engineered organ. This method overcomes those challenges by using the existing vascular network of the organ to both remove the cells, recellularize the organ, and form a new vascular network to support the organ. The influence of this study on the field of whole organ engineering is immense as it opens the door to clinical translation of engineered whole organs for organ repair and transplantation. |
| 2012 | Decellularization Methods of Porcine Kidneys for Whole Organ Engineering using a High-Throughput System (Sullivan) [153] | 178 | In this study, whole porcine kidneys were decellularized with a high-throughput system that pumped detergents through the renal vasculature. The porcine cells were successfully cleared while maintaining the complex microvasculature of the kidneys as well as the native ECM structure and composition. The decellularized kidneys were then evaluated for cytotoxicity and the kidneys perfused with 0.5% SDS showed no significant difference in cell viability from tissue plastic controls. The ability to efficiently and reproducibly decellularize organs with complex microvasculature like the kidney at a clinically relevant scale is a huge step towards clinical translation of engineered whole organs. Additionally, the low cytotoxicity and maintenance of ECM composition and morphology of this method allows for the application of a number of different recellularization strategies using pluripotent stem cells, adult stem cells, and somatic cells. The ability of this system to simultaneously decellularize 24 human-sized whole organs at once gives it the production efficiency to meet the high unmet demand for whole organ transplantation. Overall, this study marks the first step towards mass production of clinically relevant ECM scaffolds for whole organ engineering. |
| 2013 | Perfusion-Decellularized Pancreas as a Natural 3D Scaffold for Pancreatic Tissue and Whole Organ Engineering (Goh) [124] | 151 | In this study, the unmet need for a regenerative medicine therapy to address type I diabetes is addressed by creating a decellularized pancreas whole organ and reseeding it with pancreatic cells. The pancreas was perfused with 0.5% SDS through the pancreatic vasculature to remove the cellular components. These scaffolds were successfully implanted in vivo without killing surrounding cells or inducing the foreign body response. The scaffolds were also seeded in vitro with beta islet and acinar cells using retrograde perfusion of the hepatic portal vein and the pancreatic duct, respectively. Both cell types maintained high viability and showed appropriate localization in the scaffold during a five-day culture period. Additionally, the beta cells maintained insulin production capabilities and the acinar cells maintained amylase production over the entire culture period. This study is the first successful attempt at creating a tissue-engineered whole pancreas that produces insulin and provides a unique opportunity for the millions of people afflicted with type 1 diabetes. This treatment could eventually replace insulin injections and pumps, which are expensive and inconvenient, as a treatment for type 1 diabetes by instead providing an engineered whole organ transplant that would function as an autologous native pancreas. |
| 2014 | Recellularization of Well-Preserved Acellular Kidney Scaffold Using Embryonic Stem Cells (Bonandrini) [154] | 96 | Though leveraging the native ECM structure and biochemistry of cadaveric whole organs via decellularization and recellularization is a promising approach to engineering whole organs, one of its limitations is the time needed to perfuse and decellularize the organs. In this publication, Bonandrini et al. presented an accelerated perfusion decellularization protocol that was capable of using an ionic detergent solution of SDS to decellularize rat kidneys in just 17 h, with sufficient preservation of native ECM for infused murine embryonic stem cells to uniformly distribute, attach, and begin to differentiate. This relatively short time to decellularize rat kidneys is significant due to the need for improved manufacturing efficiencies for engineered whole organs to be clinically relevant. However, this protocol is limited due to a lack of replication in human-scale organs and its specificity to kidneys. Different tissues require varying decellularization protocols, and only using an SDS as the decellularization solution may not translate to other organs due to the documented harshness of ionic detergents. |
| 2015 | Whole-Organ Tissue Engineering: Decellularization and Recellularization of Three-Dimensional Matrix Liver Scaffolds (Sabetkish) [155] | 51 | In this study, rat and sheep livers were decellularized through perfusion of either Triton X-100 + SDS or SDS-only detergents and then recellularized either by in vivo implantation into rats or in vitro through perfusion of cell suspension media through the hepatic vein of the decellularized constructs. The results of the study showed that decellularization using the Triton X-100 + SDS detergent showed superior removal of cells, maintenance of native ECM architecture, and retention of mechanical properties relative to native tissue. These decellularized whole organs were successfully reseeded with hepatocytes in vivo and in vitro. The in vitro reseeding resulted in a native liver-like cellular architecture infiltrating both the regions surrounding the vessels and the parenchyma. This study represents a significant advancement over previous de-cell-re-cell studies of whole livers, which were only able to recellularize the vasculature or areas immediately surrounding the vasculature, whereas this study developed a technique for decellularization that enabled cellular infiltration throughout the entire thickness of the construct. This is a major step towards clinical viability of engineered whole organs since many rely on perfusion recellularization approaches but have had limited success with seeding the entire organ due to the distorted morphology of the ECM resulting from using an SDS only method of decellularization. |
| 2016 | Bioengineering Human Myocardium on Native Extracellular Matrix (Guyette) [98] | 121 | In this study, human hearts were decellularized using perfusion-decellularization to obtain human acellular cardiac scaffolds and whole heart scaffolds. Scaffolds derived from 2D and 3D slices of acellular human myocardium were seeded with iPSC derived cardiomyocytes in static culture conditions and formed contractile cardiac tissue after 4–7 and 7–10 days, respectively. Human whole heart scaffolds were partially seeded with iPSC-derived cardiomyocytes by injection into different regions of the left ventricle followed by coronary artery perfusion of media and left ventricle wall mechanical stimulation in a custom bioreactor. The resulting ventricle tissue showed 50% cellular repopulation and maintained 90% cell viability over the culture period. The tissue also demonstrated visible contractile function when electrically stimulated and produced measurable spontaneous contractions and ventricular pressures of 2.4 mmHg. While this study did not produce a fully functional engineered heart, it is the closest anyone has come to producing an engineered whole heart at the clinically relevant scale. Additionally, this study used iPSCs to recellularize the whole heart matrix, which can allow for the creation of a patient-specific organ by obtaining iPSCs from the transplant patients own tissues. The ability to create a patient-specific organ for an organ with such complexity and high metabolic demands as the heart makes this study is a major milestone in the field of whole organ engineering. More work is needed to develop this technique to produce engineered whole human hearts that are clinically relevant for transplantation. |
| 2017 | Bioengineering of Functional Human Induced Pluripotent Stem Cell-Derived Intestinal Grafts (Kitano) [156] | 39 | In 2017, Kitano and colleagues published their research on the successful engineering of intestinal grafts populated with human iPSCs, which, after 28 days of culturing, demonstrated intestine-specific functionality. Sections of intestine were perfusion decellularized and then recellularized using spheroids comprising human iPSCs, with these spheroids adhering to the scaffold and forming a functional epithelial barrier capable of transferring nutrients across itself. These functional intestine grafts were then implanted into mice, after which the epithelial layer formed further matured and continued to transfer glucose and fatty acid nutrients. The functionalization of an implanted engineered graft is highly promising; however, this protocol is suited towards relatively simpler organs comprised of thinner tissues and may not be able to be translated for engineering more complex whole organs with thicker, more solid tissue compositions. |
| 2018 | Bioengineering Human Lung Grafts on Porcine Matrix (Zhou) [117] | 28 | In this study, human-scale tissue-engineered lungs were created by perfusion decellularization of porcine whole lungs with SDS followed by Triton X-100. The vasculature of the lungs was then seeded with HUVECs by perfusion of cell suspension media through the pulmonary artery and vein. Epithelial cells were seeded into the bronchi in media suspension at a pressure of 80 mmHg followed by a period of perfusion culture through the pulmonary artery for 6 days to allow for endothelialization and epithelialization of the decellularized lung scaffold. The engineered lungs were then transplanted into adult pigs and evaluated for gas exchange and perfusion in vivo. The tissue-engineered lungs not only developed endothelialized vasculature and capillaries as well as a complete epithelium in the bronchial tubes, but the endothelial and epithelial cells rebuilt functional alveoli in the decellularized ECM allowing for gas exchange to occur. While the capacity for gas exchange of the engineered lungs was about 50% of that of the native lung in the porcine model, this study is the first to successfully create a whole bioartificial lung at a clinically relevant scale that recapitulates the function of the native lung in vivo. With further research, this technique could potentially produce clinically relevant bioartificial lungs for human transplant. |
| 2019 | Vascular Bioengineering of Scaffolds Derived from Human Discarded Transplant Kidneys using Human Pluripotent Stem Cell-Derived Endothelium (Leuning) [157] | 7 | Leuning et al., demonstrated that focusing on preservation of GAGs when decellularizing kidneys can result in improved reendothelialization after infusing the kidneys with cells. They found that GAG preservation resulted in enhanced reendothelialization when coupled with preloading of growth factors, as the growth factors utilize GAGs to exert their effects. In addition, they developed a novel perfusion recellularization system that simultaneously infused acellular kidney scaffolds via the arteries and veins, which resulted in significantly improved endothelial covering of the vasculature by the infused endothelial cells derived from iPSCs. The methods presented in this publication were successfully used on both mouse and human cadaveric kidneys. As adequate endothelialization is vital for vasculature to function properly in organs, Leuning and colleagues’ work is a notable contribution to whole organ engineering. Their work in retaining specific ECM components during decellularization and developing a novel recellularization method, if translatable to other organs, would be a very useful protocol. |
| 2020 | Fast, Robust and Effective Decellularization of Whole Human Livers using Mild Detergents and Pressure Controlled Perfusion (Willemse) [158] | 4 | This study developed an improved technique for perfusion decellularization that keeps a constant pressure of 120 mmHg in the perfused detergent solution and uses only TX-100 instead of SDS or a combination of SDS and TX-100. The researchers first conducted a preliminary study with porcine livers to show the decellularization properties of different detergent combinations when perfused through the liver vasculature. The preliminary study showed that while SDS is more effective at clearing cells from the tissue, it is a harsh solvent and partially destroys the ECM and GAGs of the livers in the process, which makes it less effective as a biomimetic scaffold. In contrast, the TX-100 does not damage the ECM and GAGs but does not remove cells efficiently enough in oscillating flow conditions. The pressure-controlled TX-100-only method overcame both of the issues with the previous methods. The constant pressure enabled the TX-100 to more efficiently clear cells from the tissue, cutting down the decellularization time to 20 h compared to 96 h for the oscillating flow TX-100 protocol, and the use of TX-100-only preserved the ECM and GAGs of the liver tissue. To prove the effectiveness of this procedure, the human liver tissue and whole porcine livers were perfusion recellularized with hepatic cells and the resulting engineered tissues/organs showed high cell viability and significant cellular infiltration of the parenchyma. This improved method for decellularization of whole organs will improve the quality and resulting clinical viability of future tissue-engineered whole organs that utilize this method, not just liver. |
6. Supporting Technological Advances
6.1. Cell Transplantation
6.2. Immuno-Therapy
6.3. Three-Dimensional Cell Cultures
6.4. Stem Cell Technologies
6.5. Genetic Engineering
7. Major Challenges and Barriers to Market
7.1. Incorporation into Host Tissue
7.2. Achieving Sufficient Organ Function Post-Implantation
7.3. Cost of Large-Scale Manufacturing
7.4. Regulatory Approval
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
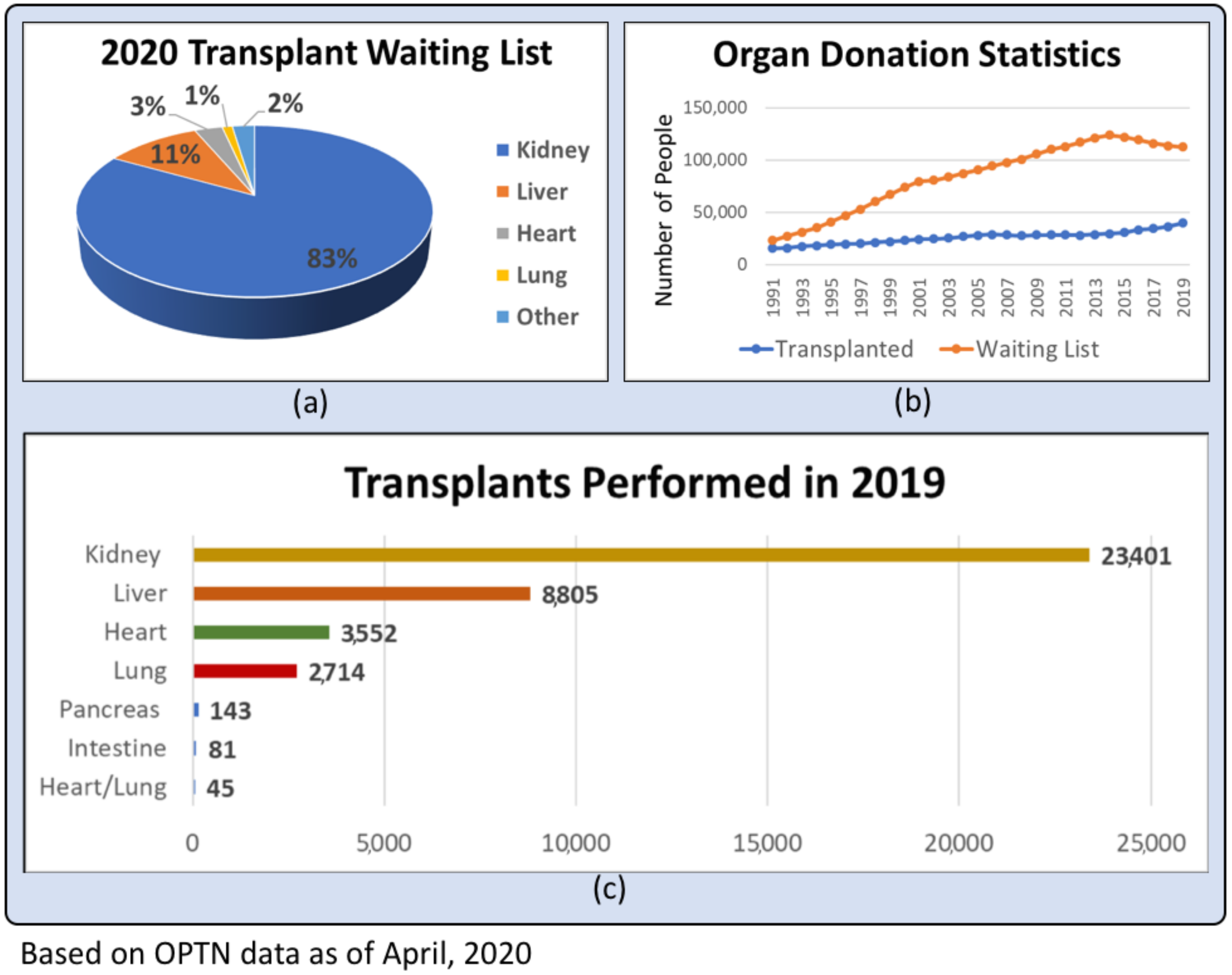
- Centers for Disease Control and Prevention. Heart Failure | cdc.gov. 2019. Available online: https://www.cdc.gov/heartdisease/heart_failure.htm (accessed on 23 April 2020).
- Chronic Kidney Disease in the United States. 2019. Available online: https://www.cdc.gov/kidneydisease/publications-resources/2019-national-facts.html (accessed on 23 April 2020).
- Kidney Disease Statistics for the United States | NIDDK. Available online: https://www.niddk.nih.gov/health-information/health-statistics/kidney-disease (accessed on 23 April 2020).
- Grinyó, J.M. Why is organ transplantation clinically important? Cold Spring Harb. Perspect. Med. 2013, 3, a014985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- U.S. Department of Health & Human Service. Organ Donation Statistics | Organ Donor. Available online: https://www.organdonor.gov/statistics-stories/statistics.html (accessed on 20 April 2020).
- Global Observatory on Donation and Transplantation (GODT). International Report on Organ Donation and Transplantation Activities—2017, no. October; World Health Organization: Geneva, Switzerland, 2019; pp. 1–52. [Google Scholar]
- Matching Donors and Recipients | Organ Donor. Available online: https://www.organdonor.gov/about/process/matching.html (accessed on 23 April 2020).
- Ingulli, E. Mechanism of cellular rejection in transplantation. Pediatr. Nephrol. 2010, 25, 61–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duncan, M.D.; Wilkes, D.S. Transplant-related Immunosuppression A Review of Immunosuppression and Pulmonary Infections. Proc. Am. Thorac. Soc. 2005, 2, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Langer, R.; Vacanti, J. Tissue engineering. Science 1993, 260, 920–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vacanti, J.P.; Morse, M.A.; Saltzman, W.M.; Domb, A.J.; Perez-Atayde, A.; Langer, R. Selective cell transplantation using bioabsorbable artificial polymers as matrices. J. Pediatr. Surg. 1988, 23, 3–9. [Google Scholar] [CrossRef]
- Vacanti, J.P. Beyond Transplantation. Arch. Surg. 1988, 123, 545. [Google Scholar] [CrossRef]
- Lee, E.J.; Kasper, F.K.; Mikos, A.G. Biomaterials for tissue engineering. In Annals of Biomedical Engineering; Kluwer Academic Publishers: Berlin/Heidelberg, Germany, 2014; Volume 42, pp. 323–337. [Google Scholar]
- Dhandayuthapani, B.; Yoshida, Y.; Maekawa, T.; Kumar, D.S. Polymeric scaffolds in tissue engineering application: A review. Int. J. Polym. Sci. 2011, 2011, 290602. [Google Scholar] [CrossRef]
- Lanza, R.; Langer, R.; Vacanti, J. Principles of Tissue Engineering, 2nd ed.; Academic Press: Cambridge, MA, USA, 2000. [Google Scholar]
- ter Horst, B.; Chouhan, G.; Moiemen, N.S.; Grover, L.M. Advances in keratinocyte delivery in burn wound care. Adv. Drug Deliv. Rev. 2018, 123, 18–32. [Google Scholar] [CrossRef]
- Chen, F.M.; Liu, X. Advancing biomaterials of human origin for tissue engineering. Prog. Polym. Sci. 2016, 53, 86–168. [Google Scholar] [CrossRef] [Green Version]
- Dolcimascolo, A.; Calabrese, G.; Conoci, S.; Parenti, R. Innovative Biomaterials for Tissue Engineering. In Biomaterial-Supported Tissue Reconstruction or Regeneration; IntechOpen: London, UK, 2019. [Google Scholar]
- Sun, J.; Tan, H. Alginate-based biomaterials for regenerative medicine applications. Materials 2013, 6, 1285–1309. [Google Scholar] [CrossRef]
- Sarker, B.; Boccaccini, A.R. Alginate Utilization in Tissue Engineering and Cell Therapy. In Alginates and Their Biomedical Applications; Springer: Singapore, 2018; pp. 121–155. [Google Scholar]
- Li, X.; Chen, S.; Zhang, B.; Li, M.; Diao, K.; Zhang, Z.; Li, J.; Xu, Y.; Wang, X.; Chen, H. In situ injectable nano-composite hydrogel composed of curcumin, N,O-carboxymethyl chitosan and oxidized alginate for wound healing application. Int. J. Pharm. 2012, 437, 110–119. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Monaco, E.; Maki, A.; De Lima, A.S.; Kong, H.J.; Hurley, W.L.; Wheeler, M.B. Morphologic and transcriptomic comparison of adipose- and bone-marrow-derived porcine stem cells cultured in alginate hydrogels. Cell Tissue Res. 2010, 341, 359–370. [Google Scholar] [CrossRef] [PubMed]
- Eschenhagen, T.; Fink, C.; Remmers, U.; Scholz, H.; Wattchow, J.; Weil, J.; Zimmermann, W.; Dohmen, H.H.; Schäfer, H.; Bishopric, N.; et al. Three-dimensional reconstitution of embryonic cardiomyocytes in a collagen matrix: A new heart muscle model system. FASEB J. 1997, 11, 683–694. [Google Scholar] [CrossRef]
- Fink, C.; Ergün, S.; Kralisch, D.; Remmers, U.; Weil, J.; Eschenhagen, T. Chronic stretch of engineered heart tissue induces hypertrophy and functional improvement. FASEB J. 2000, 14, 669–679. [Google Scholar] [CrossRef] [Green Version]
- Mooney, D.; Hansen, L.; Vacanti, J.; Langer, R.; Farmer, S.; Ingber, D. Switching from differentiation to growth in hepatocytes: Control by extracellular matrix. J. Cell. Physiol. 1992, 151, 497–505. [Google Scholar] [CrossRef]
- Kim, B.S.; Mooney, D.J. Engineering smooth muscle tissue with a predefined structure. J. Biomed. Mater. Res. 1998, 41, 322–332. [Google Scholar] [CrossRef]
- Badylak, S.F.; Arnoczky, S.; Plouhar, P.; Haut, R.; Mendenhall, V.; Clarke, R.; Horvath, C. Naturally occurring extracellular matrix as a scaffold for musculoskeletal repair. Clin. Orthop. Relat. Res. 1999, 367, S333–S343. [Google Scholar] [CrossRef] [PubMed]
- Van Amerongen, M.J.; Harmsen, M.C.; Petersen, A.H.; Kors, G.; Van Luyn, M.J.A. The enzymatic degradation of scaffolds and their replacement by vascularized extracellular matrix in the murine myocardium. Biomaterials 2006, 27, 2247–2257. [Google Scholar] [CrossRef]
- Petreaca, M.; Martins-Green, M. Cell–Extracellular Matrix Interactions in Repair and Regeneration. In Principles of Regenerative Medicine; Academic Press: Cambridge, MA, USA, 2019; pp. 15–35. [Google Scholar]
- Ethier, C.R.; Simmons, C.A. Introductory Biomechanics; Cambridge University Press: Cambridge, UK, 2007; ISBN 9780511809217. [Google Scholar]
- Bertini, E.; Pepe, G. Collagen type VI and related disorders: Bethlem myopathy and Ullrich scleroatonic muscular dystrophy. Eur. J. Paediatr. Neurol. 2002, 6, 193–198. [Google Scholar] [CrossRef]
- Bissell, M.J.; Hall, H.G.; Parry, G. How does the extracellular matrix direct gene expression? J. Theor. Biol. 1982, 99, 31–68. [Google Scholar] [CrossRef]
- Bissell, M.J.; Barcellos-Hoff, M.H. The influence of extracellular matrix on gene expression: Is structure the message? J. Cell Sci. 1987, 343, 327–343. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caldeira, J.; Sousa, A.; Sousa, D.M.; Barros, D. Extracellular Matrix Constitution and Function for Tissue Regeneration and Repair; Elsevier Ltd.: Amsterdam, The Netherlands, 2018; ISBN 9780081008522. [Google Scholar]
- Cao, Y.; Vacanti, J.P.; Paige, K.T.; Upton, J.; Vacanti, C.A. Transplantation of chondrocytes utilizing a polymer-cell construct to produce tissue-engineered cartilage in the shape of a human ear. Plast. Reconstr. Surg. 1997, 100, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Oberpenning, F.; Meng, J.; Yoo, J.J.; Atala, A. De Novo Reconstitution of a Functional Mammalian Urinary Bladder by Tissue Engineering. Nat. Biotechnol. 1999, 17, 149–155. [Google Scholar] [CrossRef] [PubMed]
- Atala, A.; Bauer, S.B.; Soker, S.; Yoo, J.J.; Retik, A.B. Tissue-engineered autologous bladders for patients needing cystoplasty. Lancet 2006, 367, 1241–1246. [Google Scholar] [CrossRef]
- Ott, H.C.; Matthiesen, T.S.; Goh, S.K.; Black, L.D.; Kren, S.M.; Netoff, T.I.; Taylor, D.A. Perfusion-decellularized matrix: Using nature’s platform to engineer a bioartificial heart. Nat. Med. 2008, 14, 213–221. [Google Scholar] [CrossRef]
- Martinez-Hernandez, A.; Amenta, P.S. The extracellular matrix in hepatic regeneration. FASEB J. 1995, 9, 1401–1410. [Google Scholar]
- Uygun, B.E.; Soto-Gutierrez, A.; Yagi, H.; Izamis, M.L.; Guzzardi, M.A.; Shulman, C.; Milwid, J.; Kobayashi, N.; Tilles, A.; Berthiaume, F.; et al. Organ reengineering through development of a transplantable recellularized liver graft using decellularized liver matrix. Nat. Med. 2010, 16, 814–820. [Google Scholar] [CrossRef]
- Jaramillo, M.; Yeh, H.; Yarmush, M.L.; Uygun, B.E. Decellularized human liver extracellular matrix (hDLM)-mediated hepatic differentiation of human induced pluripotent stem cells (hIPSCs). J. Tissue Eng. Regen. Med. 2018, 12, e1962–e1973. [Google Scholar] [CrossRef]
- Devalliere, J.; Chen, Y.; Dooley, K.; Yarmush, M.L.; Uygun, B.E. Improving functional re-endothelialization of acellular liver scaffold using REDV cell-binding domain. Acta Biomater. 2018, 78, 151–164. [Google Scholar] [CrossRef]
- Soto-Gutierrez, A.; Zhang, L.; Medberry, C.; Fukumitsu, K.; Faulk, D.; Jiang, H.; Reing, J.; Gramignoli, R.; Komori, J.; Ross, M.; et al. A whole-organ regenerative medicine approach for liver replacement. Tissue Eng. Part C Methods 2011, 17, 677–686. [Google Scholar] [CrossRef]
- Miromatrix Medical Inc. Available online: https://www.miromatrix.com/ (accessed on 2 June 2020).
- Technology Platform | TRS—Tissue Regeneration Systems. Available online: https://tissuesys.com/technology/ (accessed on 2 June 2020).
- Rokit Healthcare, Inc. Aging is Disease. Available online: http://rokithealthcare.com/ (accessed on 2 June 2020).
- Abolbashari, M.; Agcaoili, S.M.; Lee, M.K.; Ko, I.K.; Aboushwareb, T.; Jackson, J.D.; Yoo, J.J.; Atala, A. Repopulation of porcine kidney scaffold using porcine primary renal cells. Acta Biomater. 2016, 29, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Ott, H.C.; Clippinger, B.; Conrad, C.; Schuetz, C.; Pomerantseva, I.; Ikonomou, L.; Kotton, D.; Vacanti, J.P. Regeneration and orthotopic transplantation of a bioartificial lung. Nat. Med. 2010, 16, 927–933. [Google Scholar] [CrossRef]
- Gilpin, S.E.; Guyette, J.P.; Gonzalez, G.; Ren, X.; Asara, J.M.; Mathisen, D.J.; Vacanti, J.P.; Ott, H.C. Perfusion decellularization of human and porcine lungs: Bringing the matrix to clinical scale. J. Heart Lung Transplant. 2014, 33, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Chia, H.N.; Wu, B.M. Recent advances in 3D printing of biomaterials. J. Biol. Eng. 2015, 9, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishop, E.S.; Mostafa, S.; Pakvasa, M.; Luu, H.H.; Lee, M.J.; Wolf, J.M.; Ameer, G.A.; He, T.C.; Reid, R.R. 3-D bioprinting technologies in tissue engineering and regenerative medicine: Current and future trends. Genes Dis. 2017, 4, 185–195. [Google Scholar] [CrossRef]
- Kang, H.W.; Lee, S.J.; Ko, I.K.; Kengla, C.; Yoo, J.J.; Atala, A. A 3D bioprinting system to produce human-scale tissue constructs with structural integrity. Nat. Biotechnol. 2016, 34, 312–319. [Google Scholar] [CrossRef]
- Nakamura, M.; Iwanaga, S.; Henmi, C.; Arai, K.; Nishiyama, Y. Biomatrices and biomaterials for future developments of bioprinting and biofabrication. Biofabrication 2010, 2, 014110. [Google Scholar] [CrossRef]
- Hinton, T.J.; Jallerat, Q.; Palchesko, R.N.; Park, J.H.; Grodzicki, M.S.; Shue, H.J.; Ramadan, M.H.; Hudson, A.R.; Feinberg, A.W. Three-dimensional printing of complex biological structures by freeform reversible embedding of suspended hydrogels. Sci. Adv. 2015, 1, e1500758. [Google Scholar] [CrossRef] [Green Version]
- Grigoryan, B.; Paulsen, S.J.; Corbett, D.C.; Sazer, D.W.; Fortin, C.L.; Zaita, A.J.; Greenfield, P.T.; Calafat, N.J.; Gounley, J.P.; Ta, A.H.; et al. Multivascular networks and functional intravascular topologies within biocompatible hydrogels. Science 2019, 364, 458–464. [Google Scholar] [CrossRef]
- Mandrycky, C.; Phong, K.; Zheng, Y. Tissue engineering toward organ-specific regeneration and disease modeling. MRS Commun. 2017, 7, 332–347. [Google Scholar] [CrossRef] [Green Version]
- Mazza, G.; Rombouts, K.; Rennie Hall, A.; Urbani, L.; Vinh Luong, T.; Al-Akkad, W.; Longato, L.; Brown, D.; Maghsoudlou, P.; Dhillon, A.P.; et al. Decellularized human liver as a natural 3D-scaffold for liver bioengineering and transplantation. Sci. Rep. 2015, 5, 13079. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.; Hudson, A.R.; Shiwarski, D.J.; Tashman, J.W.; Hinton, T.J.; Yerneni, S.; Bliley, J.M.; Campbell, P.G.; Feinberg, A.W. 3D bioprinting of collagen to rebuild components of the human heart. Science 2019, 365, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Gungor-Ozkerim, P.S.; Inci, I.; Zhang, Y.S.; Khademhosseini, A.; Dokmeci, M.R. Bioinks for 3D bioprinting: An overview. Biomater. Sci. 2018, 6, 915–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Traphagen, S.; Yelick, P.C. Reclaiming a natural beauty: Whole-organ engineering with natural extracellular materials. Regen. Med. 2009, 747–758. [Google Scholar] [CrossRef] [Green Version]
- Londono, R.; Badylak, S.F. Biologic Scaffolds for Regenerative Medicine: Mechanisms of In vivo Remodeling. Ann. Biomed. Eng. 2015, 43, 577–592. [Google Scholar] [CrossRef]
- Gilpin, S.E.; Ren, X.; Okamoto, T.; Guyette, J.P.; Mou, H.; Rajagopal, J.; Mathisen, D.J.; Vacanti, J.P.; Ott, H.C. Enhanced lung epithelial specification of human induced pluripotent stem cells on decellularized lung matrix. Ann. Thorac. Surg. 2014, 98, 1721–1729. [Google Scholar] [CrossRef] [Green Version]
- Baptista, P.M.; Siddiqui, M.M.; Lozier, G.; Rodriguez, S.R.; Atala, A.; Soker, S. The use of whole organ decellularization for the generation of a vascularized liver organoid. Hepatology 2011, 53, 604–617. [Google Scholar] [CrossRef]
- Huling, J.C.; Atala, A.; Yoo, J.J. Decellularized Whole Organ Scaffolds for the Regeneration of Kidneys; Elsevier Inc.: Amsterdam, The Netherlands, 2015; ISBN 9780128004388. [Google Scholar]
- Faulk, D.M.; Wildemann, J.D.; Badylak, S.F. Decellularization and cell seeding of whole liver biologic scaffolds composed of extracellular matrix. J. Clin. Exp. Hepatol. 2015, 5, 69–80. [Google Scholar] [CrossRef] [Green Version]
- Jank, B.J.; Goverman, J.; Guyette, J.P.; Charest, J.M.; Randolph, M.; Gaudette, G.R.; Gershlak, J.R.; Purschke, M.; Javorsky, E.; Nazarian, R.M.; et al. Creation of a Bioengineered Skin Flap Scaffold with a Perfusable Vascular Pedicle. Tissue Eng. Part A 2017, 23, 696–707. [Google Scholar] [CrossRef]
- Obata, T.; Tsuchiya, T.; Akita, S.; Kawahara, T.; Matsumoto, K.; Miyazaki, T.; Masumoto, H.; Kobayashi, E.; Niklason, L.E.; Nagayasu, T. Utilization of Natural Detergent Potassium Laurate for Decellularization in Lung Bioengineering. Tissue Eng. Part C Methods 2019, 25, 459–471. [Google Scholar] [CrossRef]
- Faulk, D.M.; Johnson, S.A.; Zhang, L.; Badylak, S.F. Role of the extracellular matrix in whole organ engineering. J. Cell. Physiol. 2014, 229, 984–989. [Google Scholar] [CrossRef] [PubMed]
- Yagi, H.; Soto-Gutierrez, A.; Kitagawa, Y. Whole-organ re-engineering: A regenerative medicine approach in digestive surgery for organ replacement. Surg. Today 2013, 43, 587–594. [Google Scholar] [CrossRef] [Green Version]
- Keane, T.J.; DeWard, A.; Londono, R.; Saldin, L.T.; Castleton, A.A.; Carey, L.; Nieponice, A.; Lagasse, E.; Badylak, S.F. Tissue-Specific Effects of Esophageal Extracellular Matrix. Tissue Eng. Part A 2015, 21, 2293–2300. [Google Scholar] [CrossRef]
- Macchiarini, P.; Jungebluth, P.; Go, T.; Asnaghi, M.A.; Rees, L.E.; Cogan, T.A.; Dodson, A.; Martorell, J.; Bellini, S.; Parnigotto, P.P.; et al. Clinical transplantation of a tissue-engineered airway. Lancet 2008, 372, 2023–2030. [Google Scholar] [CrossRef]
- Bracaglia, L.G.; Fisher, J.P. Extracellular Matrix-Based Biohybrid Materials for Engineering Compliant, Matrix-Dense Tissues. Adv. Healthc. Mater. 2015, 4, 2475–2487. [Google Scholar] [CrossRef] [Green Version]
- Heydarkhan-Hagvall, S.; Schenke-Layland, K.; Dhanasopon, A.P.; Rofail, F.; Smith, H.; Wu, B.M.; Shemin, R.; Beygui, R.E.; MacLellan, W.R. Three-dimensional electrospun ECM-based hybrid scaffolds for cardiovascular tissue engineering. Biomaterials 2008, 29, 2907–2914. [Google Scholar] [CrossRef] [Green Version]
- He, W.; Yong, T.; Teo, W.E.; Ma, Z.; Ramakrishna, S. Fabrication and endothelialization of collagen-blended biodegradable polymer nanofibers: Potential vascular graft for blood vessel tissue engineering. Tissue Eng. 2005, 11, 1574–1588. [Google Scholar] [CrossRef] [PubMed]
- Barnes, C.P.; Sell, S.A.; Boland, E.D.; Simpson, D.G.; Bowlin, G.L. Nanofiber technology: Designing the next generation of tissue engineering scaffolds. Adv. Drug Deliv. Rev. 2007, 59, 1413–1433. [Google Scholar] [CrossRef] [PubMed]
- Kumbar, S.G.; James, R.; Nukavarapu, S.P.; Laurencin, C.T. Electrospun nanofiber scaffolds: Engineering soft tissues. Biomed. Mater. 2008, 3, 034002. [Google Scholar] [CrossRef] [Green Version]
- Ramanathan, G.; Sobhana, L.; Sobhanadhas, S.; Felciya, G.; Jeyakumar, S.; Devi, V.; Sivagnanam, U.T.; Fardim, P. Fabrication of biohybrid cellulose acetate-collagen bilayer matrices as nanofibrous spongy dressing material for wound healing application. Biomacromolecules 2020, 21, 2512–2524. [Google Scholar] [CrossRef]
- Li, X.; Guo, Y.; Ziegler, K.R.; Model, L.S.; Eghbalieh, S.D.D.; Brenes, R.A.; Kim, S.T.; Shu, C.; Dardik, A. Current usage and future directions for the bovine pericardial patch. Ann. Vasc. Surg. 2011, 25, 561–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.C.; Boland, E.D.; Williams, S.K.; Hoying, J.B. Direct-write bioprinting three-dimensional biohybrid systems for future regenerative therapies. J. Biomed. Mater. Res. Part B Appl. Biomater. 2011, 98, 160–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J. Bioactive modification of poly(ethylene glycol) hydrogels for tissue engineering. Biomaterials 2010, 31, 4639–4656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, J.P.; Sprangers, A.J.; Byce, J.R.; Su, J.; Squirrell, J.M.; Messersmith, P.B.; Eliceiri, K.W.; Ogle, B.M. ECM-incorporated hydrogels cross-linked via native chemical ligation to engineer stem cell microenvironments. Biomacromolecules 2013, 14, 3102–3111. [Google Scholar] [CrossRef] [Green Version]
- Jammalamadaka, U.; Tappa, K. Recent advances in biomaterials for 3D printing and tissue engineering. J. Funct. Biomater. 2018, 9, 22. [Google Scholar] [CrossRef] [Green Version]
- Do, A.V.; Khorsand, B.; Geary, S.M.; Salem, A.K. 3D Printing of Scaffolds for Tissue Regeneration Applications. Adv. Healthc. Mater. 2015, 4, 1742–1762. [Google Scholar] [CrossRef] [Green Version]
- Ozbolat, I.T. Bioprinting scale-up tissue and organ constructs for transplantation. Trends Biotechnol. 2015, 33, 395–400. [Google Scholar] [CrossRef]
- Kodama, H. Automatic method for fabricating a three-dimensional plastic model with photo-hardening polymer. Rev. Sci. Instrum. 1981, 52, 1770–1773. [Google Scholar] [CrossRef]
- Hull, C.W. Arcadia, and Calif. United States Patent (19) no. 19, 8 August 1984. [Google Scholar]
- Deckard, C.R. Method and Apparatus for Producing Parts by Selective Sintering. U.S. Patent 4,863,538, 5 September 1989. [Google Scholar]
- Crump, S. Apparatus and Method for Creating Three-Dimensional Objects. U.S. Patent 5,121,329, 9 June 1992. [Google Scholar]
- Ventola, C.L. Medical Applications for 3D Printing: Current and Projected Uses. Pharm. Ther. 2014, 39, 704–711. [Google Scholar]
- Sudarmadji, N.; Tan, J.Y.; Leong, K.F.; Chua, C.K.; Loh, Y.T. Investigation of the mechanical properties and porosity relationships in selective laser-sintered polyhedral for functionally graded scaffolds. Acta Biomater. 2011, 7, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.M.; Adewunmi, A.; Schek, R.M.; Flanagan, C.L.; Krebsbach, P.H.; Feinberg, S.E.; Hollister, S.J.; Das, S. Bone tissue engineering using polycaprolactone scaffolds fabricated via selective laser sintering. Biomaterials 2005, 26, 4817–4827. [Google Scholar] [CrossRef] [PubMed]
- Noor, N.; Shapira, A.; Edri, R.; Gal, I.; Wertheim, L.; Dvir, T. 3D Printing of Personalized Thick and Perfusable Cardiac Patches and Hearts. Adv. Sci. 2019, 6, 1900344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miri, A.K.; Nieto, D.; Iglesias, L.; Goodarzi Hosseinabadi, H.; Maharjan, S.; Ruiz-Esparza, G.U.; Khoshakhlagh, P.; Manbachi, A.; Dokmeci, M.R.; Chen, S.; et al. Microfluidics-Enabled Multimaterial Maskless Stereolithographic Bioprinting. Adv. Mater. 2018, 30, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Cristobal, A.; van den Toorn, H.W.P.; van de Wetering, M.; Clevers, H.; Heck, A.J.R.; Mohammed, S. Personalized Proteome Profiles of Healthy and Tumor Human Colon Organoids Reveal Both Individual Diversity and Basic Features of Colorectal Cancer. Cell Rep. 2017, 18, 263–274. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Kim, H.; Ko, U.H.; Oh, Y.; Lim, A.; Sohn, J.W.; Shin, J.H.; Kim, H.; Han, Y.M. Islet-like organoids derived from human pluripotent stem cells efficiently function in the glucose responsiveness in vitro and in vivo. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Hohwieler, M.; Illing, A.; Hermann, P.C.; Mayer, T.; Stockmann, M.; Perkhofer, L.; Eiseler, T.; Antony, J.S.; Müller, M.; Renz, S.; et al. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut 2017, 66, 473–486. [Google Scholar] [CrossRef] [Green Version]
- Wainwright, J.M.; Czajka, C.A.; Patel, U.B.; Freytes, D.O.; Tobita, K.; Gilbert, T.W.; Badylak, S.F. Preparation of cardiac extracellular matrix from an intact porcine heart. Tissue Eng. Part C Methods 2010, 16, 525–532. [Google Scholar] [CrossRef]
- Guyette, J.P.; Charest, J.M.; Mills, R.W.; Jank, B.J.; Moser, P.T.; Gilpin, S.E.; Gershlak, J.R.; Okamoto, T.; Gonzalez, G.; Milan, D.J.; et al. Bioengineering Human Myocardium on Native Extracellular Matrix. Circ. Res. 2016, 118, 56–72. [Google Scholar] [CrossRef] [Green Version]
- Taylor, D.A.; Frazier, O.H.; Elgalad, A.; Hochman-Mendez, C.; Sampaio, L.C. Building a Total Bioartificial Heart: Harnessing Nature to Overcome the Current Hurdles. Artif. Organs 2018, 42, 970–982. [Google Scholar] [CrossRef]
- Sánchez, P.L.; Fernández-Santos, M.E.; Espinosa, M.A.; González-Nicolas, M.A.; Acebes, J.R.; Costanza, S.; Moscoso, I.; Rodríguez, H.; García, J.; Romero, J.; et al. Data from acellular human heart matrix. Data Brief 2016, 8, 211–219. [Google Scholar] [CrossRef] [Green Version]
- Ko, I.K.; Peng, L.; Peloso, A.; Smith, C.J.; Dhal, A.; Deegan, D.B.; Zimmerman, C.; Clouse, C.; Zhao, W.; Shupe, T.D.; et al. Bioengineered transplantable porcine livers with re-endothelialized vasculature. Biomaterials 2015, 40, 72–79. [Google Scholar] [CrossRef]
- Hussein, K.H.; Park, K.M.; Kang, K.S.; Woo, H.M. Heparin-gelatin mixture improves vascular reconstruction efficiency and hepatic function in bioengineered livers. Acta Biomater. 2016, 38, 82–93. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Mizuno, S. The discovery of Hepatocyte Growth Factor (HGF) and its significance for cell biology, life sciences and clinical medicine. Proc. Jpn. Acad. Ser. B 2010, 86, 588–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, S.A. Sustained In Vivo Perfusion of a Re-Endothelialized Tissue Engineered Porcine Liver. Int. J. Transplant. Res. Med. 2017, 3, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Schumacher, J.D.; Guo, G.L. Regulation of Hepatic Stellate Cells and Fibrogenesis by Fibroblast Growth Factors. BioMed Res. Int. 2016, 2016, 8323747. [Google Scholar] [CrossRef] [Green Version]
- Orlando, G.; Booth, C.; Wang, Z.; Totonelli, G.; Ross, C.L.; Moran, E.; Salvatori, M.; Maghsoudlou, P.; Turmaine, M.; Delario, G.; et al. Discarded human kidneys as a source of ECM scaffold for kidney regeneration technologies. Biomaterials 2013, 34, 5915–5925. [Google Scholar] [CrossRef] [PubMed]
- Ross, E.A.; Williams, M.J.; Hamazaki, T.; Terada, N.; Clapp, W.L.; Adin, C.; Ellison, G.W.; Jorgensen, M.; Batich, C.D. Embryonic stem cells proliferate and differentiate when seeded into kidney scaffolds. J. Am. Soc. Nephrol. 2009, 20, 2338–2347. [Google Scholar] [CrossRef] [Green Version]
- Song, J.J.; Guyette, J.P.; Gilpin, S.E.; Gonzalez, G.; Vacanti, J.P.; Ott, H.C. Regeneration and experimental orthotopic transplantation of a bioengineered kidney. Nat. Med. 2013, 19, 646–651. [Google Scholar] [CrossRef]
- Kajbafzadeh, A.M.; Khorramirouz, R.; Nabavizadeh, B.; Ladi Seyedian, S.S.; Akbarzadeh, A.; Heidari, R.; Masoumi, A.; Azizi, B.; Seyed Hossein Beigi, R. Whole organ sheep kidney tissue engineering and in vivo transplantation: Effects of perfusion-based decellularization on vascular integrity. Mater. Sci. Eng. C 2019, 98, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Katari, R.; Peloso, A.; Zambon, J.P.; Soker, S.; Stratta, R.J.; Atala, A.; Orlando, G. Renal bioengineering with scaffolds generated from human kidneys. Nephron Exp. Nephrol. 2014, 126, 119–124. [Google Scholar] [CrossRef]
- Ciampi, O.; Bonandrini, B.; Derosas, M.; Conti, S.; Rizzo, P.; Benedetti, V.; Figliuzzi, M.; Remuzzi, A.; Benigni, A.; Remuzzi, G.; et al. Engineering the vasculature of decellularized rat kidney scaffolds using human induced pluripotent stem cell-derived endothelial cells. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Anil Kumar, P.R.; Yoo, J.J.; Zahran, F.; Atala, A.; Lee, S.J. A Photo-Crosslinkable Kidney ECM-Derived Bioink Accelerates Renal Tissue Formation. Adv. Healthc. Mater. 2019, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Denizet, G.; Calame, P.; Lihoreau, T.; Kleinclauss, F.; Aubry, S. 3D multi-tissue printing for kidney transplantation. Quant. Imaging Med. Surg. 2019, 9, 101. [Google Scholar] [CrossRef] [PubMed]
- Homan, K.A.; Kolesky, D.B.; Skylar-Scott, M.A.; Herrmann, J.; Obuobi, H.; Moisan, A.; Lewis, J.A. Bioprinting of 3D Convoluted Renal Proximal Tubules on Perfusable Chips. Sci. Rep. 2016, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Carreno-Galeano, G.; Ali, M.; Jackson, J.; Yoo, J.; Lee, S.J.; Atala, A. 3D Bioprinted Renal Tissue Constructs Using a Novel Kidney ECM-Derived Bioink. J. Urol. 2020, 203, e775. [Google Scholar] [CrossRef]
- Petersen, T.H.; Calle, E.A.; Zhao, L.; Lee, E.J.; Gui, L.; Raredon, M.S.B.; Gavrilov, K.; Yi, T.; Zhuang, Z.W.; Breuer, C.; et al. Tissue-engineered lungs for in vivo implantation. Science 2010, 329, 538–541. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.; Kitano, K.; Ren, X.; Rajab, T.K.; Wu, M.; Gilpin, S.E.; Wu, T.; Baugh, L.; Black, L.D.; Mathisen, D.J.; et al. Bioengineering Human Lung Grafts on Porcine Matrix. Ann. Surg. 2018, 267, 590–598. [Google Scholar] [CrossRef]
- Urbano, J.J.; Da Palma, R.K.; De Lima, F.M.; Fratini, P.; Guimaraes, L.L.; Uriarte, J.J.; Alvarenga, L.H.; Miglino, M.A.; Vieira, R.D.P.; Prates, R.A.; et al. Effects of two different decellularization routes on the mechanical properties of decellularized lungs. PLoS ONE 2017, 12, e0178696. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, J.D.; Anfang, R.; Anandappa, A.; Costa, J.; Javidfar, J.; Wobma, H.M.; Singh, G.; Freytes, D.O.; Bacchetta, M.D.; Sonett, J.R.; et al. Decellularization of human and porcine lung tissues for pulmonary tissue engineering. Ann. Thorac. Surg. 2013, 96, 1046–1056. [Google Scholar] [CrossRef] [Green Version]
- Balestrini, J.L.; Gard, A.L.; Liu, A.; Leiby, K.L.; Schwan, J.; Kunkemoeller, B.; Calle, E.A.; Sivarapatna, A.; Lin, T.; Dimitrievska, S.; et al. Production of decellularized porcine lung scaffolds for use in tissue engineering. Integr. Biol. 2015, 7, 1598–1610. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Engler, A.J.; Raredon, M.S.; Le, A.; Baevova, P.; Yoder, M.C.; Niklason, L.E. Epac agonist improves barrier function in iPSC-derived endothelial colony forming cells for whole organ tissue engineering. Biomaterials 2019, 200, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Nichols, J.E.; La Francesca, S.; Niles, J.A.; Vega, S.P.; Argueta, L.B.; Frank, L.; Christiani, D.C.; Pyles, R.B.; Himes, B.E.; Zhang, R.; et al. Production and transplantation of bioengineered lung into a large-animal model. Sci. Transl. Med. 2018, 10, eaao3926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Citro, A.; Ott, H.C. Can We Re-Engineer the Endocrine Pancreas? Curr. Diab. Rep. 2018, 18, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Goh, S.K.; Bertera, S.; Olsen, P.; Candiello, J.E.; Halfter, W.; Uechi, G.; Balasubramani, M.; Johnson, S.A.; Sicari, B.M.; Kollar, E.; et al. Perfusion-decellularized pancreas as a natural 3D scaffold for pancreatic tissue and whole organ engineering. Biomaterials 2013, 34, 6760–6772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirmalek-Sani, S.H.; Orlando, G.; McQuilling, J.P.; Pareta, R.; Mack, D.L.; Salvatori, M.; Farney, A.C.; Stratta, R.J.; Atala, A.; Opara, E.C.; et al. Porcine pancreas extracellular matrix as a platform for endocrine pancreas bioengineering. Biomaterials 2013, 34, 5488–5495. [Google Scholar] [CrossRef] [Green Version]
- Peloso, A.; Urbani, L.; Cravedi, P.; Katari, R.; Maghsoudlou, P.; Fallas, M.E.A.; Sordi, V.; Citro, A.; Purroy, C.; Niu, G.; et al. The human pancreas as a source of protolerogenic extracellular matrix scaffold for a new-generation bioartificial endocrine pancreas. Ann. Surg. 2016, 264, 169–179. [Google Scholar] [CrossRef] [Green Version]
- Hashemi, J.; Pasalar, P.; Soleimani, M.; Khorramirouz, R.; Fendereski, K.; Enderami, S.E.; Kajbafzadeh, A.M. Application of a novel bioreactor for in vivo engineering of pancreas tissue. J. Cell. Physiol. 2018, 233, 3805–3816. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, J.; Pasalar, P.; Soleimani, M.; Arefian, E.; Khorramirouz, R.; Akbarzadeh, A.; Ghorbani, F.; Enderami, S.E.; Kajbafzadeh, A.M. Decellularized pancreas matrix scaffolds for tissue engineering using ductal or arterial catheterization. Cells Tissues Organs 2018, 205, 72–84. [Google Scholar] [CrossRef] [PubMed]
- Pagliuca, F.W.; Millman, J.R.; Gürtler, M.; Segel, M.; Van Dervort, A.; Ryu, J.H.; Peterson, Q.P.; Greiner, D.; Melton, D.A. Generation of functional human pancreatic β cells in vitro. Cell 2014, 159, 428–439. [Google Scholar] [CrossRef] [Green Version]
- Vegas, A.J.; Veiseh, O.; Gürtler, M.; Millman, J.R.; Pagliuca, F.W.; Bader, A.R.; Doloff, J.C.; Li, J.; Chen, M.; Olejnik, K.; et al. Long-term glycemic control using polymer-encapsulated human stem cell-derived beta cells in immune-competent mice. Nat. Med. 2016, 22, 306–311. [Google Scholar] [CrossRef] [PubMed]
- Deuse, T.; Hu, X.; Gravina, A.; Wang, D.; Tediashvili, G.; De, C.; Thayer, W.O.; Wahl, A.; Garcia, J.V.; Reichenspurner, H.; et al. Hypoimmunogenic derivatives of induced pluripotent stem cells evade immune rejection in fully immunocompetent allogeneic recipients. Nat. Biotechnol. 2019, 37, 252–258. [Google Scholar] [CrossRef]
- Lanza, R.; Russell, D.W.; Nagy, A. Engineering universal cells that evade immune detection. Nat. Rev. Immunol. 2019, 19, 723–733. [Google Scholar] [CrossRef]
- Piechota, H.J.; Dahms, S.E.; Nunes, L.S.; Dahiya, R.; Lue, T.F.; Tanagho, E.A. In vitro functional properties of the rat bladder regenerated by the bladder acellular matrix graft. J. Urol. 1998, 159, 1717–1724. [Google Scholar] [CrossRef] [PubMed]
- Olsen, L.; Bowald, S.; Busch, C.; Carlsten, J.; Eriksson, I. Urethral reconstruction with a new synthetic absorbable device: An experimental study. Scand. J. Urol. Nephrol. 1992, 26, 323–326. [Google Scholar] [CrossRef] [PubMed]
- Eberli, D.; Filho, L.F.; Atala, A.; Yoo, J.J. Composite scaffolds for the engineering of hollow organs and tissues. Methods 2009, 47, 109–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, L.; Murphy, S.V.; Yang, B.; Xu, Y.; Zhang, Y.; Atala, A. Bladder acellular matrix and its application in bladder augmentation. Tissue Eng. Part B Rev. 2014, 20, 163–172. [Google Scholar] [CrossRef] [PubMed]
- Joseph, D.B.; Borer, J.G.; De Filippo, R.E.; Hodges, S.J.; McLorie, G.A. Autologous cell seeded biodegradable scaffold for augmentation cystoplasty: Phase II study in children and adolescents with spina bifida. J. Urol. 2014, 191, 1389–1395. [Google Scholar] [CrossRef]
- Zhe, Z.; Jun, D.; Yang, Z.; Mingxi, X.; Ke, Z.; Ming, Z.; Zhong, W.; Mujun, L. Bladder Acellular Matrix Grafts Seeded with Adipose-Derived Stem Cells and Incubated Intraperitoneally Promote the Regeneration of Bladder Smooth Muscle and Nerve in a Rat Model of Bladder Augmentation. Stem Cells Dev. 2016, 25, 405–414. [Google Scholar] [CrossRef]
- Moreno-Manzano, V.; Mellado-López, M.; Morera-Esteve, M.J.; Alastrue-Agudo, A.; Bisbal-Velasco, V.; Forteza-Vila, J.; Serrano-Aroca, Á.; Vera-Donoso, C.D. Human adipose-derived mesenchymal stem cells accelerate decellularized neobladder regeneration. Regen. Biomater. 2020, 7, 161–169. [Google Scholar] [CrossRef]
- Sander, E.A.; Lynch, K.A.; Boyce, S.T. Development of the Mechanical Properties of Engineered Skin Substitutes after Grafting to Full-Thickness Wounds. J. Biomech. Eng. 2014, 136, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Itoh, M.; Umegaki-Arao, N.; Guo, Z.; Liu, L.; Higgins, C.A.; Christiano, A.M. Generation of 3D Skin Equivalents Fully Reconstituted from Human Induced Pluripotent Stem Cells (iPSCs). PLoS ONE 2013, 8, e77673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baltazar, T.; Merola, J.; Catarino, C.; Xie, C.B.; Kirkiles-Smith, N.C.; Lee, V.; Hotta, S.; Dai, G.; Xu, X.; Ferreira, F.C.; et al. Three Dimensional Bioprinting of a Vascularized and Perfusable Skin Graft Using Human Keratinocytes, Fibroblasts, Pericytes, and Endothelial Cells. Tissue Eng. Part A 2020, 26, 227–238. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Huang, S.; Yao, B.; Xie, J.; Wu, X.; Fu, X. 3D bioprinting matrices with controlled pore structure and release function guide in vitro self-organization of sweat gland. Sci. Rep. 2016, 6, 1–8. [Google Scholar] [CrossRef]
- Parnigotto, P.P.; Marzaro, M.; Artusi, T.; Perrino, G.; Conconi, M.T. Short bowel syndrome: Experimental approach to increase intestinal surface in rats by gastric homologous acellular matrix. J. Pediatr. Surg. 2000, 35, 1304–1308. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.K.; Badylak, S.F. Small bowel tissue engineering using small intestinal submucosa as a scaffold. J. Surg. Res. 2001, 99, 352–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maemura, T.; Shin, M.; Kinoshita, M.; Majima, T.; Ishihara, M.; Saitoh, D.; Ichikura, T. A tissue-engineered stomach shows presence of proton pump and G-cells in a rat model, resulting in improved anemia following total gastrectomy. Artif. Organs 2008, 32, 234–239. [Google Scholar] [CrossRef]
- Grikscheit, T.C.; Siddique, A.; Ochoa, E.R.; Srinivasan, A.; Alsberg, E.; Hodin, R.A.; Vacanti, J.P. Tissue-engineered small intestine improves recovery after massive small bowel resection. Ann. Surg. 2004, 240, 748–754. [Google Scholar] [CrossRef]
- Grant, C.N.; Mojica, S.G.; Sala, F.G.; Ryan Hill, J.; Levin, D.E.; Speer, A.L.; Barthel, E.R.; Shimada, H.; Zachos, N.C.; Grikscheit, T.C. Human and mouse tissue-engineered small intestine both demonstrate digestive and absorptive function. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 308, G664–G677. [Google Scholar] [CrossRef] [Green Version]
- Levin, D.E.; Barthel, E.R.; Speer, A.L.; Sala, F.G.; Hou, X.; Torashima, Y.; Grikscheit, T.C. Human tissue-engineered small intestine forms from postnatal progenitor cells. J. Pediatr. Surg. 2013, 48, 129–137. [Google Scholar] [CrossRef]
- Nakatsu, H.; Ueno, T.; Oga, A.; Nakao, M.; Nishimura, T.; Kobayashi, S.; Masaaki, O. Influence of mesenchymal stem cells on stomach tissue engineering using small intestinal submucosa. J. Tissue Eng. Regen. Med. 2015, 9, 296–304. [Google Scholar] [CrossRef] [Green Version]
- Kim, I.G.; Wu, Y.; Park, S.A.; Cho, H.; Choi, J.J.; Kwon, S.K.; Shin, J.W.; Chung, E.J. Tissue-engineered esophagus via bioreactor cultivation for circumferential esophageal reconstruction. Tissue Eng. Part A 2019, 25, 1478–1492. [Google Scholar] [CrossRef] [PubMed]
- Dadhich, P.; Bohl, J.L.; Tamburrini, R.; Zakhem, E.; Scott, C.; Kock, N.; Mitchell, E.; Gilliam, J.; Bitar, K.N. BioSphincters to treat Fecal Incontinence in Nonhuman Primates. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, D.C.; Mirmalek-Sani, S.H.; Deegan, D.B.; Baptista, P.M.; Aboushwareb, T.; Atala, A.; Yoo, J.J. Decellularization methods of porcine kidneys for whole organ engineering using a high-throughput system. Biomaterials 2012, 33, 7756–7764. [Google Scholar] [CrossRef] [PubMed]
- Bonandrini, B.; Figliuzzi, M.; Papadimou, E.; Morigi, M.; Perico, N.; Casiraghi, F.; Dipl, C.; Sangalli, F.; Conti, S.; Benigni, A.; et al. Recellularization of well-preserved acellular kidney scaffold using embryonic stem cells. Tissue Eng. Part A 2014, 20, 1486–1498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabetkish, S.; Kajbafzadeh, A.M.; Sabetkish, N.; Khorramirouz, R.; Akbarzadeh, A.; Seyedian, S.L.; Pasalar, P.; Orangian, S.; Hossein Beigi, R.S.; Aryan, Z.; et al. Whole-organ tissue engineering: Decellularization and recellularization of three-dimensional matrix liver scaffolds. J. Biomed. Mater. Res. Part A 2015, 103, 1498–1508. [Google Scholar] [CrossRef]
- Kitano, K.; Schwartz, D.M.; Zhou, H.; Gilpin, S.E.; Wojtkiewicz, G.R.; Ren, X.; Sommer, C.A.; Capilla, A.V.; Mathisen, D.J.; Goldstein, A.M.; et al. Bioengineering of functional human induced pluripotent stem cell-derived intestinal grafts. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Leuning, D.G.; Witjas, F.M.R.; Maanaoui, M.; de Graaf, A.M.A.; Lievers, E.; Geuens, T.; Avramut, C.M.; Wiersma, L.E.; van den Berg, C.W.; Sol, W.M.P.J.; et al. Vascular bioengineering of scaffolds derived from human discarded transplant kidneys using human pluripotent stem cell-derived endothelium. Am. J. Transplant. 2019, 19, 1328–1343. [Google Scholar] [CrossRef] [Green Version]
- Willemse, J.; Verstegen, M.M.A.; Vermeulen, A.; Schurink, I.J.; Roest, H.P.; van der Laan, L.J.W.; de Jonge, J. Fast, robust and effective decellularization of whole human livers using mild detergents and pressure controlled perfusion. Mater. Sci. Eng. C 2020, 108, 110200. [Google Scholar] [CrossRef]
- Anazawa, T.; Okajima, H.; Masui, T.; Uemoto, S. Current state and future evolution of pancreatic islet transplantation. Ann. Gastroenterol. Surg. 2019, 3, 34–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Den Bos, E.J.; Van Der Giessen, W.J.; Duncker, D.J. Cell transplantation for cardiac regeneration: Where do we stand? Neth. Heart J. 2008, 16, 88–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Wu, J.; Liu, G.H. First stem cell transplantation to regenerate human lung. Protein Cell 2018, 9, 244–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Foggia, V.; Makwana, P.; Ali, R.R.; Sowden, J.C. Induced pluripotent stem cell therapies for degenerative disease of the outer retina: Disease modeling and cell replacement. J. Ocul. Pharmacol. Ther. 2016, 32, 240–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foster, E.D.; Bridges, N.D.; Feurer, I.D.; Eggerman, T.L.; Hunsicker, L.G.; Alejandro, R. Improved Health-Related Quality of Life in a Phase 3 Islet Transplantation Trial in Type 1 Diabetes Complicated by Severe Hypoglycemia. Diabetes Care 2018, 41, 1001–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jain, A.; Bansal, R. Applications of regenerative medicine in organ transplantation. J. Pharm. Bioallied Sci. 2015, 7, 188–194. [Google Scholar] [CrossRef]
- Boehler, R.M.; Graham, J.G.; Shea, L.D. Tissue engineering tools for modulation of the immune response. Biotechniques 2011, 51, 239–254. [Google Scholar] [CrossRef] [Green Version]
- Andorko, J.I.; Jewell, C.M. Designing biomaterials with immunomodulatory properties for tissue engineering and regenerative medicine. Bioeng. Transl. Med. 2017, 2, 139–155. [Google Scholar] [CrossRef] [Green Version]
- Von Der Mark, K.; Gauss, V.; Von Der Mark, H.; Müller, P. Relationship between cell shape and type of collagen synthesised as chondrocytes lose their cartilage phenotype in culture. Nature 1977, 267, 531–532. [Google Scholar] [CrossRef]
- Benya, P.D.; Shaffer, J.D. Dedifferentiated chondrocytes reexpress the differentiated collagen phenotype when cultured in agarose gels. Cell 1982, 30, 215–224. [Google Scholar] [CrossRef]
- Ravi, M.; Paramesh, V.; Kaviya, S.R.; Anuradha, E.; Paul Solomon, F.D. 3D cell culture systems: Advantages and applications. J. Cell. Physiol. 2015, 230, 16–26. [Google Scholar] [CrossRef]
- Bissell, M.J. The Differentiated State of Normal and Malignant Cells or How to Define a “Normal” Cell in Culture. In International Review of Cytology; Academic Press: Cambridge, MA, USA, 1981; Volume 70, ISBN 0123644704. [Google Scholar]
- Hirschi, K.K.; Li, S.; Roy, K. Induced Pluripotent Stem Cells for Regenerative Medicine. Annu. Rev. Biomed. Eng. 2014, 16, 277–294. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.T.; Jacobs, K.; Spits, C. Human pluripotent stem cells in regenerative medicine: Where do we stand? Reproduction 2018, 156, R143–R153. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.A. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Studer. Stem Cell in Disease Review. Ethn. Dis. 2010, 20, 1–26. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.C.; Leu, S.; Sun, C.K.; Yen, C.H.; Kao, Y.H.; Chang, L.T.; Tsai, T.H.; Chua, S.; Fu, M.; Ko, S.F.; et al. Early combined treatment with sildenafil and adipose-derived mesenchymal stem cells preserves heart function in rat dilated cardiomyopathy. J. Transl. Med. 2010, 8, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yañez, R.; Lamana, M.L.; García-Castro, J.; Colmenero, I.; Ramírez, M.; Bueren, J.A. Adipose Tissue-Derived Mesenchymal Stem Cells Have In Vivo Immunosuppressive Properties Applicable for the Control of the Graft-versus-Host Disease. Stem Cells 2006, 24, 2582–2591. [Google Scholar] [CrossRef] [PubMed]
- Orabi, H.; Lin, G.; Ferretti, L.; Lin, C.S.; Lue, T.F. Scaffoldless Tissue Engineering of Stem Cell Derived Cavernous Tissue for Treatment of Erectile Function. J. Sex. Med. 2012, 9, 1522–1534. [Google Scholar] [CrossRef]
- Urrutia, D.N.; Caviedes, P.; Mardones, R.; Minguell, J.J.; Vega-Letter, A.M.; Jofre, C.M. Comparative study of the neural differentiation capacity of mesenchymal stromal cells from different tissue sources: An approach for their use in neural regeneration therapies. PLoS ONE 2019, 14, e0213032. [Google Scholar] [CrossRef]
- Dos Santos, A.; Balayan, A.; Funderburgh, M.L.; Ngo, J.; Funderburgh, J.L.; Deng, S.D. Differentiation capacity of human mesenchymal stem cells into keratocyte lineage. Investig. Ophthalmol. Vis. Sci. 2019, 60, 3013–3023. [Google Scholar] [CrossRef] [Green Version]
- Ben-David, U.; Benvenisty, N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat. Rev. Cancer 2011, 11, 268–277. [Google Scholar] [CrossRef]
- Yasuda, S.; Kusakawa, S.; Kuroda, T.; Miura, T.; Tano, K.; Takada, N.; Matsuyama, S.; Matsuyama, A.; Nasu, M.; Umezawa, A.; et al. Tumorigenicity-associated characteristics of human iPS cell lines. PLoS ONE 2018, 13, e0205022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papapetrou, E.P. Patient-derived induced pluripotent stem cells in cancer research and precision oncology. Nat. Med. 2016, 22, 1392–1401. [Google Scholar] [CrossRef] [PubMed]
- Blum, B.; Benvenisty, N. The Tumorigenicity of Human Embryonic Stem Cells. Adv. Cancer Res. 2008, 100, 133–158. [Google Scholar] [PubMed]
- Baker, M. Unregulated stem cell transplant causes tumours. Nat. Rep. Stem Cells 2009. [Google Scholar] [CrossRef]
- Liu, Z.; Tang, Y.; Lü, S.; Zhou, J.; Du, Z.; Duan, C.; Li, Z.; Wang, C. The tumourigenicity of iPS cells and their differentiated derivates. J. Cell. Mol. Med. 2013, 17, 782–791. [Google Scholar] [CrossRef]
- Xu, H.; Wang, B.; Ono, M.; Kagita, A.; Fujii, K.; Sasakawa, N.; Ueda, T.; Gee, P.; Nishikawa, M.; Nomura, M.; et al. Targeted Disruption of HLA Genes via CRISPR-Cas9 Generates iPSCs with Enhanced Immune Compatibility. Cell Stem Cell 2019, 24, 566–578. [Google Scholar] [CrossRef] [Green Version]
- Elliott, M.J.; De Coppi, P.; Speggiorin, S.; Roebuck, D.; Butler, C.R.; Samuel, E.; Crowley, C.; McLaren, C.; Fierens, A.; Vondrys, D.; et al. Stem-cell-based, tissue engineered tracheal replacement in a child: A 2-year follow-up study. Lancet 2012, 380, 994–1000. [Google Scholar] [CrossRef] [Green Version]
- Griffith, L.G.; Swartz, M.A. Capturing complex 3D tissue physiology in vitro. Nat. Rev. Mol. Cell Biol. 2006, 7, 211–224. [Google Scholar] [CrossRef]
- Maby-El Hajjami, H.; Petrova, T.V. Developmental and pathological lymphangiogenesis: From models to human disease. Histochem. Cell Biol. 2008, 130, 1063–1078. [Google Scholar] [CrossRef] [Green Version]
- Li, G.N.; Hoffman-Kim, D. Tissue-engineered platforms of axon guidance. Tissue Eng. Part B Rev. 2008, 14, 33–51. [Google Scholar] [CrossRef]
- Zhang, H.; Wei, Y.T.; Tsang, K.S.; Sun, C.R.; Li, J.; Huang, H.; Cui, F.Z.; An, Y.H. Implantation of neural stem cells embedded in hyaluronic acid and collagen composite conduit promotes regeneration in a rabbit facial nerve injury model. J. Transl. Med. 2008, 6, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichihara, S.; Inada, Y.; Nakamura, T. Artificial nerve tubes and their application for repair of peripheral nerve injury: An update of current concepts. Injury 2008, 39, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Tiruvannamalai-Annamalai, R.; Armant, D.R.; Matthew, H.W.T. A glycosaminoglycan based, modular tissue scaffold system for rapid assembly of perfusable, high cell density, engineered tissues. PLoS ONE 2014, 9, e84287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakayama, K.H.; Batchelder, C.A.; Lee, C.I.; Tarantal, A.F. Renal tissue engineering with decellularized rhesus monkey kidneys: Age-related differences. Tissue Eng. Part A 2011, 17, 2891–2901. [Google Scholar] [CrossRef]
- Kropp, C.; Massai, D.; Zweigerdt, R. Progress and challenges in large-scale expansion of human pluripotent stem cells. Process Biochem. 2017, 59, 244–254. [Google Scholar] [CrossRef] [Green Version]
- Tirziu, D.; Giordano, F.J.; Simons, M. Cell communications in the heart. Circulation 2010, 122, 928–937. [Google Scholar] [CrossRef] [Green Version]
- Park, K.-M.; Hussein, K.H.; Hong, S.-H.; Ahn, C.; Yang, S.-R.; Park, S.-M.; Kweon, O.-K.; Kim, B.-M.; Woo, H.-M. Decellularized Liver Extracellular Matrix as Promising Tools for Transplantable Bioengineered Liver Promotes Hepatic Lineage Commitments of Induced Pluripotent Stem Cells. Tissue Eng. Part A 2016, 22, 449–460. [Google Scholar] [CrossRef]
- FDA. Tissue & Tissue Products. Available online: https://www.fda.gov/vaccines-blood-biologics/tissue-tissue-products (accessed on 19 April 2020).
- FDA. CFR—Code of Federal Regulations Title 21. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfcfr/CFRSearch.cfm?CFRPart=1270 (accessed on 19 April 2020).
- FDA. Premarket Approval (PMA). Available online: https://www.fda.gov/medical-devices/premarket-submissions/premarket-approval-pma (accessed on 19 April 2020).


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sohn, S.; Buskirk, M.V.; Buckenmeyer, M.J.; Londono, R.; Faulk, D. Whole Organ Engineering: Approaches, Challenges, and Future Directions. Appl. Sci. 2020, 10, 4277. https://doi.org/10.3390/app10124277
Sohn S, Buskirk MV, Buckenmeyer MJ, Londono R, Faulk D. Whole Organ Engineering: Approaches, Challenges, and Future Directions. Applied Sciences. 2020; 10(12):4277. https://doi.org/10.3390/app10124277
Chicago/Turabian StyleSohn, Sogu, Maxwell Van Buskirk, Michael J. Buckenmeyer, Ricardo Londono, and Denver Faulk. 2020. "Whole Organ Engineering: Approaches, Challenges, and Future Directions" Applied Sciences 10, no. 12: 4277. https://doi.org/10.3390/app10124277
APA StyleSohn, S., Buskirk, M. V., Buckenmeyer, M. J., Londono, R., & Faulk, D. (2020). Whole Organ Engineering: Approaches, Challenges, and Future Directions. Applied Sciences, 10(12), 4277. https://doi.org/10.3390/app10124277




