Microbiota in Waterlogged Archaeological Wood: Use of Next-Generation Sequencing to Evaluate the Risk of Biodegradation
Abstract
:1. Introduction
2. Materials and Methods
Bioinformatic Analyses
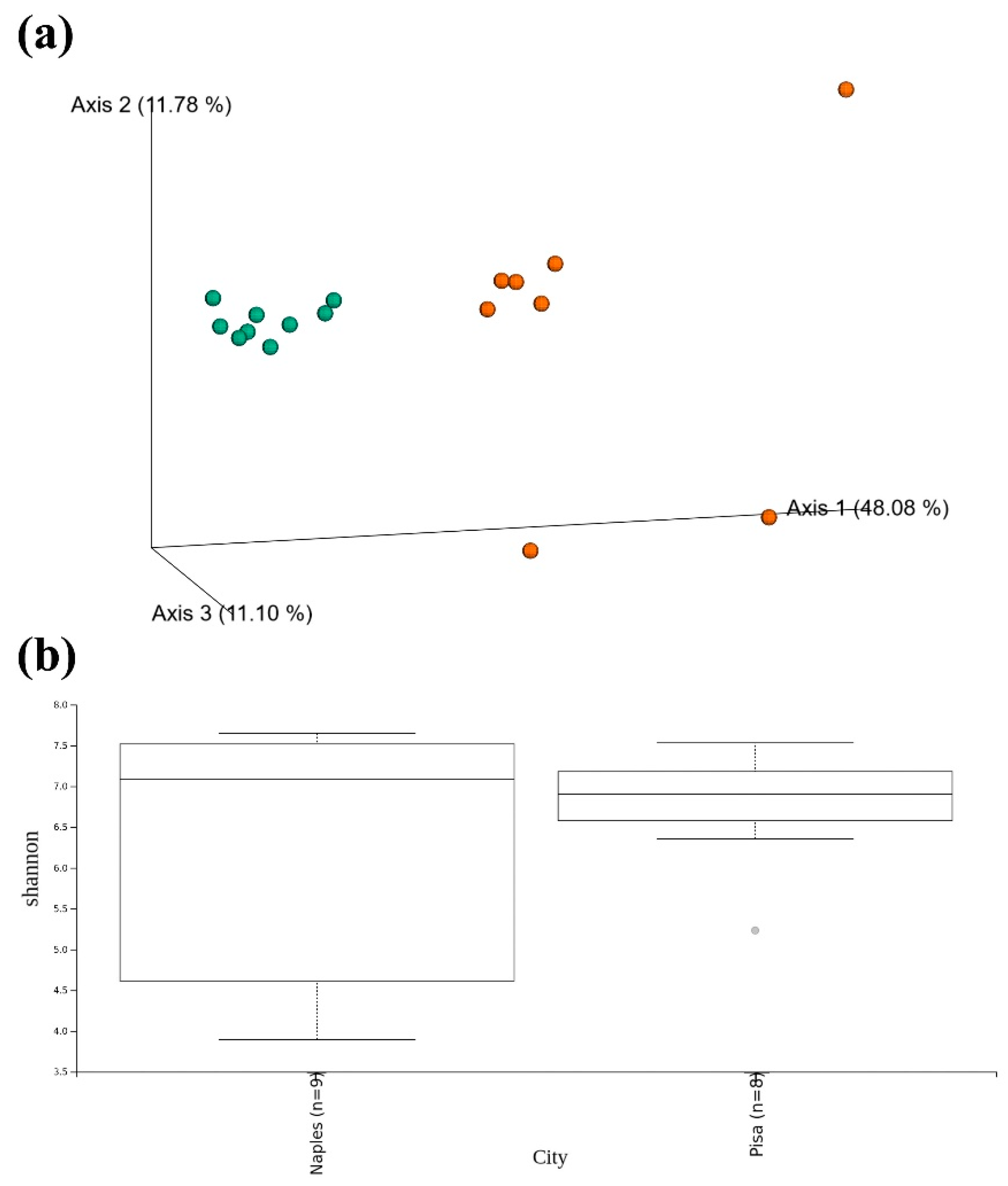
3. Results and Discussion
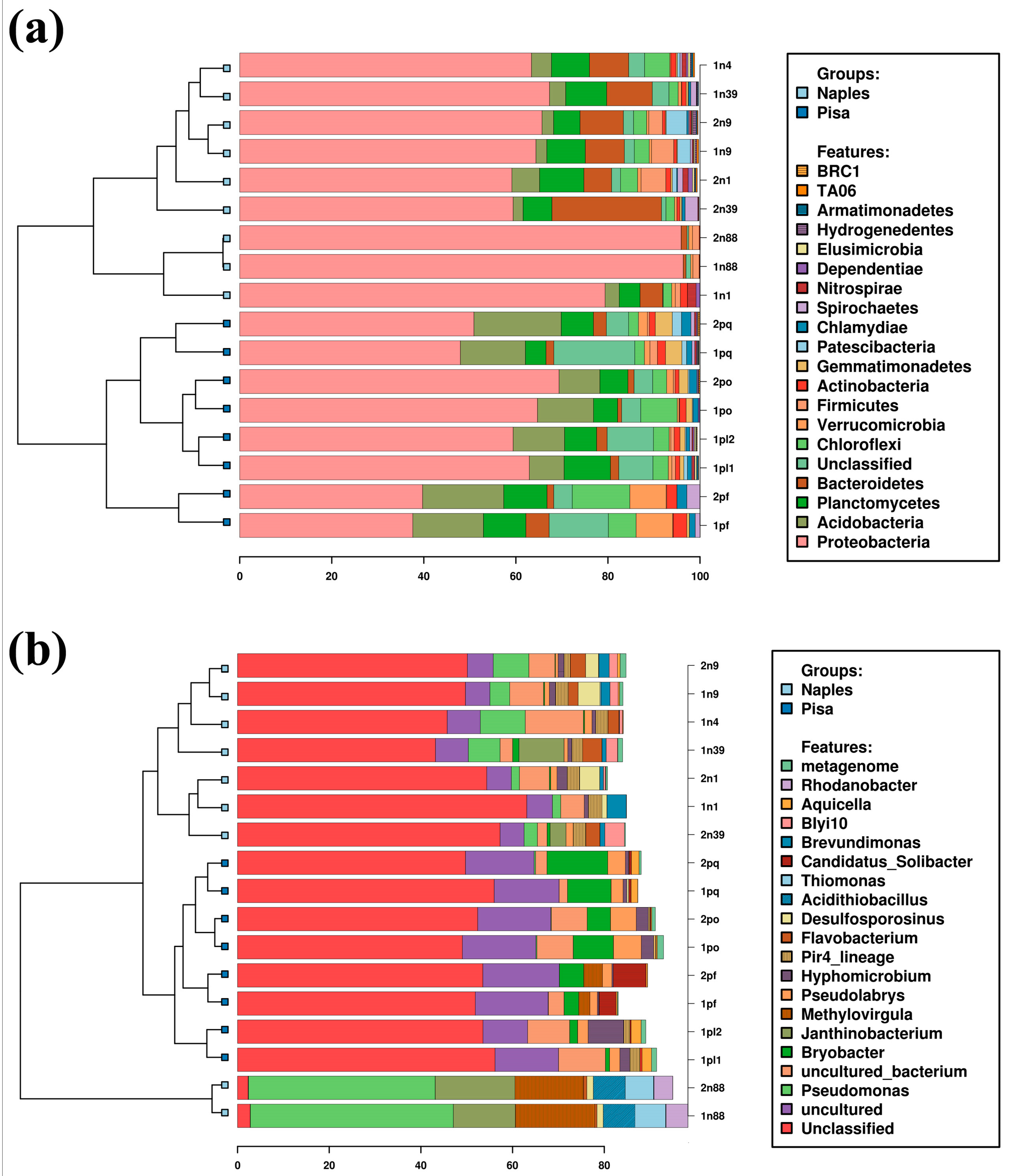
3.1. Bacterial Community
3.2. Fungal Community
3.3. Microbiota and Wood Decay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rowell, R.M.; Barbour, R.J. Archaeological Wood; American Chemical Society: Washington, DC, USA, 1989; Volume 225, ISBN 0-8412-1623-1. [Google Scholar]
- EN 16873:2016. Conservation of Cultural Heritage. Guidelines for the Management of Waterlogged Wood on Archaeological Terrestrial Sites; European Committee for Standardization: Brussels, Belgium, 2016. [Google Scholar]
- Grattan, D.W.; Clarke, R.W. Conservation of waterlogged wood. In Conservation of Marine Archaeological Objects; Pearson, C., Ed.; Butterworth-Heinemann: London, UK, 1987; pp. 164–206. [Google Scholar]
- Barbour, R.J. Treatments for waterlogged and dry archaeological wood. In Archaeological Wood; RowellR, M., Barbour, R.J., Eds.; Advances in Chemistry; 1989; Volume 225, pp. 177–192. ISBN ISBN 0-8412-1623-1. [Google Scholar]
- Eaton, R.A.; Hale, M.D.C. Wood: Decay, Pests and Protection; Chapman and Hall Ltd: Washington, DC, USA, 1993. [Google Scholar]
- Blanchette, R.A.; Nilsson, T.; Daniel, G.; Abad, A. Biological degradation of wood. In Archaeological Wood. Properties, Chemistry, and Preservation; Rowell, R.M., Barbour, J.R., Eds.; American Chemical Society: Washington, DC, USA, 1989; pp. 141–174. [Google Scholar]
- Björdal, C.G.; Nilsson, T. Waterlogged archaeological wood—A substrate for white rot fungi during drainage of wetlands. Int. Biodeterior. Biodegrad. 2002, 50, 17–23. [Google Scholar] [CrossRef]
- Blanchette, R.A. A review of microbial deterioration found in archaeological wood from different environments. Int. Biodeterior. Biodegrad. 2000, 46, 189–204. [Google Scholar] [CrossRef]
- Björdal, C.G. Microbial degradation of waterlogged archaeological wood. J. Cult. Herit. 2012, 13, S118–S122. [Google Scholar] [CrossRef]
- Björdal, C.G.; Nilsson, T. Observations on microbial growth during conservation treatment of waterlogged archaeological wood. Stud. Conserv. 2001, 46, 211–220. [Google Scholar]
- Kim, Y.S.; Singh, A.P.; Nilsson, T. Bacteria as important degraders in waterlogged archaeological woods. Holzforschung 1996, 50, 389–392. [Google Scholar] [CrossRef]
- Björdal, C.G. Evaluation of microbial degradation of shipwrecks in the Baltic Sea. Int. Biodeterior. Biodegrad. 2012, 70, 126–140. [Google Scholar] [CrossRef]
- Singh, A.P. A review of microbial decay types found in wooden objects of cultural heritage recovered from buried and waterlogged environments. J. Cult. Herit. 2012, 13, S16–S20. [Google Scholar] [CrossRef]
- Preston, J.; Watts, J.E.M.; Jones, M. Novel bacterial community associated with 500-year-old unpreserved archaeological wood from King Henry VIII’s Tudor Warship the Mary Rose. Appl. Environ. Microbiol. 2012, 78, 8822–8828. [Google Scholar] [CrossRef] [Green Version]
- Giampaola, D.; Carsana, V.; Boetto, G.; Crema, F.; Florio, C.; Panza, D.; Bartolini, M.; Capretti, C.; Galotta, G.; Giachi, G.; et al. La scoperta del porto di” Neapolis”: Dalla ricostruzione topografica allo scavo e al recupero dei relitti. Archaeol. Marit. Mediterr. 2005, 2, 1000–1045. [Google Scholar]
- Anagnost, S.E. Light microscopic diagnosis of wood decay. IAWA J. 1998, 19, 141–167. [Google Scholar] [CrossRef]
- Nilsson, T.; Björdal, C.G.; Fällman, E. Culturing erosion bacteria: Procedures for obtaining purer cultures and pure strains. Int. Biodeterior. Biodegrad. 2008, 61, 17–23. [Google Scholar] [CrossRef]
- Nilsson, T.; Björdal, C.G. Culturing wood-degrading erosion bacteria. Int. Biodeterior. Biodegrad. 2008, 61, 3–10. [Google Scholar] [CrossRef]
- Nilsson, T.; Björdal, C.G. The use of kapok fibres for enrichment cultures of lignocellulose-degrading bacteria. Int. Biodeterior. Biodegrad. 2008, 61, 11–16. [Google Scholar] [CrossRef]
- Schwarze, F.W.M.R. Wood decay under the microscope. Fungal Biol. Rev. 2007, 21, 133–170. [Google Scholar] [CrossRef]
- Kretschmar, E.I.; Gelbrich, J.; Militz, H.; Lamersdorf, N. Studying bacterial wood decay under low oxygen conditions-results of microcosm experiments. Int. Biodeterior. Biodegrad. 2008, 61, 69–84. [Google Scholar] [CrossRef]
- Helms, A.C.; Camillo Martiny, A.; Hofman-Bang, H.J.; Ahring, B.K.; Kilstrup, M. Identification of bacterial cultures from archaeological wood using molecular biological techniques. Int. Biodeterior. Biodegrad. 2004, 53, 79–88. [Google Scholar] [CrossRef] [Green Version]
- Landy, E.T.; Mitchell, J.I.; Hotchkiss, S.; Eaton, R.A. Bacterial diversity associated with archaeological waterlogged wood: Ribosomal RNA clone libraries and denaturing gradient gel electrophoresis (DGGE). Int. Biodeterior. Biodegrad. 2008, 61, 106–116. [Google Scholar] [CrossRef]
- Palla, F.; Barresi, G.; Di Carlo, E. Identification of bacterial taxa in archaeological waterlogged wood. Conserv. Sci. Cult. Herit. 2014, 14, 247–254. [Google Scholar]
- Palla, F.; Mancuso, F.P.; Billeci, N. Multiple approaches to identify bacteria in archaeological waterlogged wood. J. Cult. Herit. 2013, 14, e61–e64. [Google Scholar] [CrossRef] [Green Version]
- Pang, K.L.; Eaton, R.A.; Mitchell, J.I. Molecular detection of bacteria in the seventeenth century Swedish warship Vasa. In Proceedings of the 9th ICOM Group on Wet Organic Archeaological Materials Conference, Copenhagen, Denmark, 15–17 January 2004; Hoffmann, P., Straetkvern, K., Spriggs, J.A., Gregory, D.J., Eds.; Verlag H.M. Hauschild GmbH: Bremerhaven, Germany, 2004; pp. 243–259. [Google Scholar]
- Li, Q.; Cao, L.; Wang, W.; Tan, H.; Jin, T.; Wang, G.; Lin, G.; Xu, R. Analysis of the bacterial communities in the waterlogged wooden cultural relics of the Xiaobaijiao No. 1 shipwreck via high-throughput sequencing technology. Holzforschung 2018, 72, 609–619. [Google Scholar] [CrossRef]
- Liu, Z.; Fu, T.; Hu, C.; Shen, D.; Macchioni, N.; Sozzi, L.; Chen, Y.; Liu, J.; Tian, X.; Ge, Q.; et al. Microbial community analysis and biodeterioration of waterlogged archaeological wood from the Nanhai No. 1 shipwreck during storage. Sci. Rep. 2018, 8, 7170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, S.; Lagane, F.; Seguin-Orlando, A.; Schubert, M.; Leroy, T.; Guichoux, E.; Chancerel, E.; Bech-Hebelstrup, I.; Bernard, V.; Billard, C.; et al. High-Throughput DNA sequencing of ancient wood. Mol. Ecol. 2018, 27, 1138–1154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Wei, Y.; Jia, H.; Xiao, L.; Gong, D. A new perspective on studying burial environment before archaeological excavation: Analyzing bacterial community distribution by high-throughput sequencing. Sci. Rep. 2017, 7, 41691. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Jia, Y.; Wang, M.; Yu, H.; Jing, Y.; Hu, C.; Zhang, F.; Sun, M.; Liu, Z.; Chen, Y.; et al. Bacterial and biodeterioration analysis of the waterlogged wooden lacquer plates from the Nanhai No. 1 Shipwreck. Appl. Sci. 2019, 9, 653. [Google Scholar] [CrossRef] [Green Version]
- Di Donato, V.; Ruello, M.R.; Liuzza, V.; Carsana, V.; Giampaola, D.; Di Vito, M.A.; Morhange, C.; Cinque, A.; Russo Ermolli, E. Development and decline of the ancient harbor of Neapolis. Geoarchaeology 2018, 33, 542–557. [Google Scholar] [CrossRef]
- Giachi, G.; Lazzeri, S.; Lippi, M.M.; Macchioni, N.; Paci, S. The wood of “C” and “F” Roman ships found in the ancient harbour of Pisa (Tuscany, Italy): The utilisation of different timbers and the probable geographical area which supplied them. J. Cult. Herit. 2003, 4, 269–283. [Google Scholar] [CrossRef]
- Bruni, S. Le navi antiche di Pisa. In Proceedings of the Il mare degli Etruschi: Atti del convegno promosso dalla Quinta Commissione consiliare Attività culturali e turismo del Consiglio regionale della Toscana, Orbetello, Italy, 18–20 September 2009; Tipografia Consiglio regionale della Toscana: Orbetello, Italy, 2010; pp. 63–84. [Google Scholar]
- Camilli, A. Il contesto delle navi antiche di Pisa. Un breve punto della situazione. J. Fasti Online 2005, 31, 1–7. [Google Scholar]
- Cragnolini, E.; Sarra, A. Sperimentazione di Metodi di Impregnazione e Liofilizzazione di Legni Archeologici Provenienti da Ambiente Saturo D’acqua; Istituto Centrale per il Restauro: Rome, Italy, 2008. [Google Scholar]
- Jensen, P.; Gregory, D.J. Selected physical parameters to characterize the state of preservation of waterlogged archaeological wood: A practical guide for their determination. J. Archaeol. Sci. 2006, 33, 551–559. [Google Scholar] [CrossRef]
- Antonelli, F.; Galotta, G.; Sidoti, G.; Zikeli, F.; Nisi, R.; Davidde Petriaggi, B.; Romagnoli, M. Cellulose and Lignin Nano-Scale Consolidants for Waterlogged Archaeological Wood. Front. Chem. 2020, 8, 32. [Google Scholar] [CrossRef]
- Italian Technical Standard. UNI11205 Beni Culturali—Legno di Interesse Archeologico ed Archeobotanico—Linee Guida per la Caratterizzazione; Italian Technical Standard: Milan, Italy, 2007. [Google Scholar]
- Romagnoli, M.; Galotta, G.; Antonelli, F.; Sidoti, G.; Humar, M.; Kržišnik, D.; Čufar, K.; Davidde Petriaggi, B. Micro-morphological, physical and thermogravimetric analyses of waterlogged archaeological wood from the prehistoric village of Gran Carro (Lake Bolsena-Italy). J. Cult. Herit. 2018, 33, 30–38. [Google Scholar] [CrossRef]
- Capretti, C.; Macchioni, N.; Pizzo, B.; Galotta, G.; Giachi, G.; Giampaola, D. The characterization of waterlogged archaeological wood: The three roman ships found in Naples (Italy). Archaeometry 2008, 50, 855–876. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Antonelli, F.; Esposito, A.; Calvo, L.; Licursi, V.; Tisseyre, P.; Ricci, S.; Romagnoli, M.; Piazza, S.; Guerrieri, F. Characterization of black patina from the Tiber River embankments using next-generation sequencing. PLoS ONE 2020, 15, e0227639. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581. [Google Scholar] [CrossRef] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT: Iterative refinement and additional methods. In Multiple Sequence Alignment Methods; Humana press: Totowa, NJ, USA, 2014; pp. 131–146. [Google Scholar]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2018, 47, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewski, M.; Proietti, C.; Ellis, J.J.; Hasan, S.; Brion, M.J.; Berger, B.; Krause, L. Calypso: A user-friendly web-server for mining and visualizing microbiome–environment interactions | Bioinformatics | Oxford Academic. Bioinformatics 2017, 33, 782–783. [Google Scholar] [PubMed] [Green Version]
- Björdal, C.G.; Daniel, G.; Nilsson, T. Depth of burial, an important factor in controlling bacterial decay of waterlogged archaeological poles. Int. Biodeterior. Biodegrad. 2000, 45, 15–26. [Google Scholar] [CrossRef]
- Blanchette, R.A.; Jurgens, J.A. Characterization of wood destroying microorganisms in archaeological woods from marine environments. In Proceedings of the International Academy of Wood Science Annual Meeting, Chile, 14–17 September 2005; p. 14. [Google Scholar]
- Shui Yang, J.; Ren Ni, J.; Li Yuan, H.; Wang, E. Biodegradation of three different wood chips by Pseudomonas sp. PKE117. Int. Biodeterior. Biodegrad. 2007, 60, 90–95. [Google Scholar] [CrossRef]
- Zimmermann, W. Degradation of lignin by bacteria. J. Biotechnol. 1990, 13, 119–130. [Google Scholar] [CrossRef]
- Stern, I.J.; Wang, C.H.; Gilmour, C.M. Comparative catabolism of carbohydrates in Pseudomonas species. J. Bacteriol. 1960, 79, 601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palleroni, N.J.; Doudoroff, M. Metabolism of carbohydrates by Pseudomonas saccharophila III: Oxidation of d-Arabinose1. J. Bacteriol. 1957, 74, 180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wood, W.A.; Schwerdt, R.F. Carbohydrate oxidation by Pseudomonas fluorescens I. The mechanism of glucose and gluconate oxidation. J. Biol. Chem. 1953, 201, 501–511. [Google Scholar]
- Dumova, V.A.; Kruglov, Y.V. A cellulose-decomposing bacterial association. Microbiology 2009, 78, 234–239. [Google Scholar] [CrossRef]
- Bugg, T.D.; Ahmad, M.; Hardiman, E.M.; Singh, R. The emerging role for bacteria in lignin degradation and bio-product formation. Curr. Opin. Biotechnol. 2011, 22, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chang, H.; Fan, C.; Chen, W.; Lee, M. Screening, isolation and characterization of cellulose biotransformation bacteria from specific soils. Int. Conf. Environ. Ind. Innov. 2011, 12, 216–220. [Google Scholar]
- Lopatina, A.; Krylenkov, V.; Severinov, K. Activity and bacterial diversity of snow around Russian Antarctic stations. Res. Microbiol. 2013, 164, 949–958. [Google Scholar] [CrossRef] [PubMed]
- Shivaji, S.; Kumari, K.; Kishore, K.H.; Pindi, P.K.; Rao, P.S.; Radha Srinivas, T.N.; Asthana, R.; Ravindra, R. Vertical distribution of bacteria in a lake sediment from Antarctica by culture-independent and culture-dependent approaches. Res. Microbiol. 2011, 162, 191–203. [Google Scholar] [CrossRef]
- Pearce, D.A.; Gast, C.J.; Lawley, B.; Ellis-Evans, J.C. Bacterioplankton community diversity in a maritime Antarctic lake, determined by culture-dependent and culture-independent techniques. FEMS Microbiol. Ecol. 2003, 45, 59–70. [Google Scholar] [CrossRef] [Green Version]
- Ravindran, A.; Yang, S.S. Production and characterization of cellulase from Janthinobacterium sp. AR-129 and Pedobacter sp. AR-138. Taiwan J. Agric. Chem. Food Sci. 2016, 54, 124–134. [Google Scholar]
- Bernardet, J.F.; Bowman, J.P. The Genus Flavobacterium. In The Prokaryotes; Springer: New York, NY, USA, 2006; pp. 481–531. [Google Scholar]
- Kuo, I.; Saw, J.; Kapan, D.D.; Christensen, S.; Kaneshiro, K.Y.; Donachie, S.P. Flavobacterium akiainvivens sp. nov., from decaying wood of Wikstroemia oahuensis, Hawai’i, and emended description of the genus Flavobacterium. Int. J. Syst. Evol. Microbiol. 2013, 63, 3280–3286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, L.; Yi, L.; Zheng, T. Advance in the research of marine algicidal functional bacteria and their algicidal mechanism. Adv. Earth Sci. 2013, 28, 243–252. [Google Scholar]
- White, D.C.; Sutton, S.D.; Ringelberg, D.B. The genus Sphingomonas: Physiology and ecology. Curr. Opin. Biotechnol. 1996, 7, 301–306. [Google Scholar] [CrossRef]
- Nishiyama, M.; Senoo, K.; Wada, H.; Matsumoto, S. Identification of soil micro-habitats for growth, death and survival of a bacterium, Î3-1,2,3,4,5,6-hexachlorocyclohexane-assimilating Sphingomonas paucimobilis, by fractionation of soil. FEMS Microbiol. Lett. 1992, 101, 145–150. [Google Scholar] [CrossRef] [Green Version]
- Yang, D.C.; Im, W.T.; Kim, M.K.; Ohta, H.; Lee, S.T. Sphingomonas soli sp. nov., a beta-glucosidase-producing bacterium in the family Sphingomonadaceae in the 4 subgroup of the Proteobacteria. Int. J. Syst. Evol. Microbiol. 2006, 56, 703–707. [Google Scholar] [CrossRef] [Green Version]
- Baraniecki, C.; Aislabie, J.; Foght, J. Characterization of Sphingomonas sp. Ant 17, an aromatic hydrocarbon-degrading bacterium isolated from Antarctic soil. Microb. Ecol. 2002, 43, 44–54. [Google Scholar] [CrossRef]
- Masai, E.; Katayama, Y.; Nishikawa, S.; Fukuda, M. Characterization of Sphingomonas paucimobilis SYK-6 genes involved in degradation of lignin-related compounds. J. Ind. Microbiol. Biotechnol. 1999, 23, 364–373. [Google Scholar] [CrossRef]
- Hoppe, B.; Krüger, D.; Kahl, T.; Arnstadt, T.; Buscot, F.; Bauhus, J.; Wubet, T. A pyrosequencing insight into sprawling bacterial diversity and community dynamics in decaying deadwood logs of Fagus sylvatica and Picea abies. Sci. Rep. 2015, 5, 9456. [Google Scholar] [CrossRef] [Green Version]
- Kielak, A.M.; Scheublin, T.R.; Mendes, L.W.; van Veen, J.A.; Kuramae, E.E. Bacterial community succession in pine-wood decomposition. Front. Microbiol. 2016, 7, 231. [Google Scholar] [CrossRef] [Green Version]
- Vorob’ev, A.V.; de Boer, W.; Folman, L.B.; Bodelier, P.L.E.; Doronina, N.V.; Suzina, N.E.; Trotsenko, Y.A.; Dedysh, S.N. Methylovirgula ligni gen. nov., sp. nov., an obligately acidophilic, facultatively methylotrophic bacterium with a highly divergent mxaF gene. Int. J. Syst. Evol. Microbiol. 2009, 59, 2538–2545. [Google Scholar] [CrossRef] [Green Version]
- Dedysh, S.N. Bryobacter. In Bergey’s manual of systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2019; pp. 1–5. [Google Scholar]
- Kulichevskaya, I.S.; Suzina, N.E.; Liesack, W.; Dedysh, S.N. Bryobacter aggregatus gen. nov., sp. nov., a peat-inhabiting, aerobic chemo-organotroph from subdivision 3 of the Acidobacteria. Int. J. Syst. Evol. Microbiol. 2010, 60, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Kampfer, P.; Young, C.C.; Arun, A.B.; Shen, F.T.; Jäckel, U.; Rosselló-Mora, R.; Lai, W.A.; Rekha, P.D. Pseudolabrys taiwanensis gen. nov., sp. nov., an alphaproteobacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 2006, 56, 2469–2472. [Google Scholar] [CrossRef] [Green Version]
- Leifson, E. Hyphomicrobium neptunium sp. n. Antonie Van Leeuwenhoek 1964, 30, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthy, S.; Sass, H.; Langner, H.; Schumann, P.; Kroppenstedt, R.M.; Spring, S.; Overmann, J.; Rosenzweig, R.F. Desulfosporosinus lacus sp. nov., a sulfate-reducing bacterium isolated from pristine freshwater lake sediments. Int. J. Syst. Evol. Microbiol. 2006, 56, 2729–2736. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, W.J.; Bowman, J.P.; Mee, B.J.; Franzmann, P.D. Desulfosporosinus meridiei sp. nov., a spore-forming sulfate-reducing bacterium isolated from gasolene-contaminated groundwater. Int. J. Syst. Evol. Microbiol. 2001, 51, 133–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kostka, J.E.; Green, S.J.; Rishishwar, L.; Prakash, O.; Katz, L.S.; Mariño-Ramírez, L.; Jordan, I.K.; Munk, C.; Ivanova, N.; Mikhailova, N.; et al. Genome sequences for six Rhodanobacter strains, isolated from soils and the terrestrial subsurface, with variable denitrification capabilities. J. Bacteriol. 2012, 194, 4461–4462. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Sanchez, A.; Ariza, C.; Ubeda, J.; Martin-Sanchez, P.; Jurado, V.; Bastian, F.; Alabouvette, C.; Saiz-Jimenez, C. Free-living amoebae in sediments from the Lascaux Cave in France. Int. J. Speleol. 2013, 42, 9–13. [Google Scholar] [CrossRef]
- Pagnier, I.; Raoult, D.; La Scola, B. Isolation and characterization of Reyranella massiliensis gen. nov., sp. nov. from freshwater samples by using an amoeba co-culture procedure. Int. J. Syst. Evol. Microbiol. 2011, 61, 2151–2154. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Kim, D.U.; Lee, S.; Park, S.; Yoon, J.H.; Seong, C.N.; Ka, J.O. Reyranella terrae sp. nov., isolated from an agricultural soil, and emended description of the genus Reyranella. Int. J. Syst. Evol. Microbiol. 2017, 67, 2031–2035. [Google Scholar] [CrossRef]
- Yoon, J.H.; Kang, S.J.; Park, S.; Oh, T.K. Devosia insulae sp. nov., isolated from soil, and emended description of the genus Devosia. Int. J. Syst. Evol. Microbiol. 2007, 57, 1310–1314. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.U.; Lee, H.; Kim, H.; Kim, S.G.; Ka, J.O. Dongia soli sp. nov., isolated from soil from Dokdo, Korea. Antonie Van Leeuwenhoek 2016, 109, 1397–1402. [Google Scholar] [CrossRef] [PubMed]
- Baik, K.S.; Hwang, Y.M.; Choi, J.S.; Kwon, J.; Seong, C.N. Dongia rigui sp. nov., isolated from freshwater of a large wetland in Korea. Antonie Van Leeuwenhoek 2013, 104, 1143–1150. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Yokota, A. Curvibacter fontana sp. nov., a microaerobic bacteria isolated from well water. J. Gen. Appl. Microbiol. 2010, 56, 267–271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramírez, P.; Guiliani, N.; Valenzuela, L.; Beard, S.; Jerez, C.A. Differential protein expression during growth of Acidithiobacillus ferrooxidans on ferrous iron, sulfur compounds, or metal sulfides. Appl. Environ. Microbiol. 2004, 70, 4491–4498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hallberg, K.B.; González-Toril, E.; Johnson, D.B. Acidithiobacillus ferrivorans, sp. nov.; facultatively anaerobic, psychrotolerant iron-, and sulfur-oxidizing acidophiles isolated from metal mine-impacted environments. Extremophiles 2010, 14, 9–19. [Google Scholar] [CrossRef]
- Chen, X.G.; Geng, A.L.; Yan, R.; Gould, W.D.; Ng, Y.L.; Liang, D.T. Isolation and characterization of sulphur-oxidizing Thiomonas sp. and its potential application in biological deodorization. Lett. Appl. Microbiol. 2004, 39, 495–503. [Google Scholar] [CrossRef]
- Zecchin, S.; Colombo, M.; Cavalca, L. Exposure to different arsenic species drives the establishment of iron- and sulfur-oxidizing bacteria on rice root iron plaques. World J. Microbiol. Biotechnol. 2019, 35, 117. [Google Scholar] [CrossRef]
- De Bont, J.A.M.; Van Dijken, J.P.; Harder, W. Dimethyl sulphoxide and dimethyl sulphide as a carbon, sulphur and energy source for growth of Hyphomicrobium S. Microbiology 1981, 127, 315–323. [Google Scholar] [CrossRef] [Green Version]
- Van Den Heuvel, R.N.; Van Der Biezen, E.; Jetten, M.S.M.; Hefting, M.M.; Kartal, B. Denitrification at pH 4 by a soil-derived Rhodanobacter-dominated community. Environ. Microbiol. 2010, 12, 3264–3271. [Google Scholar] [CrossRef]
- Green, S.J.; Prakash, O.; Jasrotia, P.; Overholt, W.A.; Cardenas, E.; Hubbard, D.; Tiedje, J.M.; Watson, D.B.; Schadt, C.W.; Brooks, S.C.; et al. Denitrifying bacteria from the genus Rhodanobacter dominate bacterial communities in the highly contaminated subsurface of a nuclear legacy waste site. Appl. Environ. Microbiol. 2012, 78, 1039–1047. [Google Scholar] [CrossRef] [Green Version]
- Nilsson, R.H.; Ryberg, M.; Kristiansson, E.; Abarenkov, K.; Larsson, K.H.; Köljalg, U. Taxonomic reliability of DNA sequences in public sequences databases: A fungal perspective. PLoS ONE 2006, 1. [Google Scholar] [CrossRef] [PubMed]
- Balajee, S.A.; Borman, A.M.; Brandt, M.E.; Cano, J.; Cuenca-Estrella, M.; Dannaoui, E.; Guarro, J.; Haase, G.; Kibbler, C.C.; Meyer, W.; et al. Sequence-based identification of Aspergillus, Fusarium, and Mucorales species in the clinical mycology laboratory: Where are we and where should we go from here? J. Clin. Microbiol. 2009, 47, 877–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seifert, K.A. Progress towards DNA barcoding of fungi. Mol. Ecol. Resour. 2009, 9, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Réblová, M.; Seifert, K.A.; Fournier, J.; Štěpánek, V. Phylogenetic classification of Pleurothecium and Pleurotheciella gen. nov. and its dactylaria-like anamorph (Sordariomycetes) based on nuclear ribosomal and protein-coding genes. Mycologia 2012, 104, 1299–1314. [Google Scholar] [CrossRef]
- Luo, Z.L.; Hyde, K.D.; Bhat, D.J.; Jeewon, R.; Maharachchikumbura, S.S.N.; Bao, D.F.; Li, W.L.; Su, X.J.; Yang, X.Y.; Su, H.Y. Morphological and molecular taxonomy of novel species Pleurotheciaceae from freshwater habitats in Yunnan, China. Mycol. Prog. 2018, 17, 511–530. [Google Scholar] [CrossRef]
- Gramaje, D.; Mostert, L.; Armengol Fortí, J. Characterization of Cadophora luteo-olivacea and C. melinii isolates obtained from grapevines and environmental samples from grapevine nurseries in Spain. Phytopathol. Mediterr. 2011, 50, 112–126. [Google Scholar]
- Harrington, T.C.; McNew, D.L. Phylogenetc analysis places the Phialophora-like anamorph genus Cadophora in the Helotiales. Mycotaxon 2003, 87, e151. [Google Scholar]
- Blanchette, R.A.; Held, B.W.; Jurgens, J.A.; McNew, D.L.; Harrington, T.C.; Duncan, S.M.; Farrell, R.L. Wood-destroying soft rot fungi in the historic expedition huts of Antarctica. Appl. Environ. Microbiol. 2004, 70, 1328–1335. [Google Scholar] [CrossRef] [Green Version]
- Arenz, B.E.; Held, B.W.; Jurgens, J.A.; Farrell, R.L.; Blanchette, R.A. Fungal diversity in soils and historic wood from the Ross Sea Region of Antarctica. Soil Biol. Biochem. 2006, 38, 3057–3064. [Google Scholar] [CrossRef]
- Held, B.W.; Blanchette, R.A. Deception Island, Antarctica, harbors a diverse assemblage of wood decay fungi. Fungal Biol. 2017, 121, 145–157. [Google Scholar] [CrossRef] [Green Version]
- Blanchette, R.A.; Held, B.W.; Hellmann, L.; Millman, L.; Büntgen, U. Arctic driftwood reveals unexpectedly rich fungal diversity. Fungal Ecol. 2016, 23, 58–65. [Google Scholar] [CrossRef]
- Miller, A.N.; Huhndorf, S.M. Multi-gene phylogenies indicate ascomal wall morphology is a better predictor of phylogenetic relationships than ascospore morphology in the Sordariales (Ascomycota, Fungi). Mol. Phylogenet. Evol. 2005, 35, 60–75. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, L.E.; Havrylenko, M. The genera Arnium and Podospora from Argentina. Mycologia 2001, 93, 1221–1230. [Google Scholar] [CrossRef]
- Mirza, J.H.; Cain, R.F. Revision of the genus Podospora. Can. J. Bot. 1969, 47, 1999–2048. [Google Scholar] [CrossRef]
- Che, Y.; Araujo, A.R.; Gloer, J.B.; Scott, J.A.; Malloch, D. Communiols E−H: new polyketide metabolites from the coprophilous fungus Podospora communis. J. Nat. Prod. 2005, 68, 435–438. [Google Scholar] [CrossRef] [Green Version]
- Walsh, E.; Luo, J.; Zhang, N. Acidomelania panicicola gen. et sp. nov. from switchgrass roots in acidic New Jersey pine barrens. Mycologia 2014, 106, 856–864. [Google Scholar] [CrossRef]
- Réblová, M.; Gams, W.; Seifert, K.A. Monilochaetes and allied genera of the Glomerellales, and a reconsideration of families in the Microascales. Stud. Mycol. 2011, 68, 163–191. [Google Scholar] [CrossRef]
- Day, M.J.; Hall, J.C.; Currah, R.S. Phialide arrangement and character evolution in the helotialean anamorph genera Cadophora and Phialocephala. Mycologia 2012, 104, 371–381. [Google Scholar] [CrossRef]
- Crous, P.W.; Groenewald, J.Z.; Gams, W. Eyespot of cereals revisited: ITS phylogeny reveals new species relationships. Eur. J. Plant Pathol. 2003, 109, 841–850. [Google Scholar] [CrossRef]
- Leonhardt, S.; Büttner, E.; Gebauer, A.M.; Hofrichter, M.; Kellner, H. Draft genome sequence of the Sordariomycete Lecythophora (Coniochaeta) hoffmannii CBS 245.38. Genome Announc. 2018, 6. [Google Scholar] [CrossRef] [Green Version]
- Bugos, R.C.; Sutherland, J.B.; Adler, J.H. Phenolic compound utilization by the soft rot fungus Lecythophora hoffmannii. Appl. Environ. Microbiol. 1988, 54, 1882–1885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamed, S.A.M. In-vitro studies on wood degradation in soil by soft-rot fungi: Aspergillus niger and Penicillium chrysogenum. Int. Biodeterior. Biodegrad. 2013, 78, 98–102. [Google Scholar] [CrossRef]
- Rodríguez, A.; Carnicero, A.; Perestelo, F.; de la Fuente, G.; Milstein, O.; Falcón, M.A. Effect of Penicillium chrysogenum on lignin transformation. Appl. Environ. Microbiol. 1994, 60, 2971–2976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Launen, L.; Pinto, L.; Wiebe, C.; Kiehlmann, E.; Moore, M. The oxidation of pyrene and benzo[a]pyrene by nonbasidiomycete soil fungi. Can. J. Microbiol. 1995, 41, 477–488. [Google Scholar] [CrossRef]
- Tillett, R.; Walker, J.R.L. Metabolism of ferulic acid by a Penicillium sp. Arch. Microbiol. 1990, 154, 206–208. [Google Scholar] [CrossRef]
- Osman, M.E.; El-Shaphy, A.A.E.N.; Meligy, D.A.; Ayid, M.M. Survey for fungal decaying archaeological wood and their enzymatic activity. Int. J. Conserv. Sci. 2014, 5, 295–301. [Google Scholar]
- Zeng, G.M.; Yu, H.Y.; Huang, H.L.; Huang, D.L.; Chen, Y.N.; Huang, G.H.; Li, J.B. Laccase activities of a soil fungus Penicillium simplicissimum in relation to lignin degradation. World J. Microbiol. Biotechnol. 2006, 22, 317–324. [Google Scholar] [CrossRef]
- Yu, H.; Zeng, G.; Huang, G.; Huang, D.; Chen, Y. Lignin degradation by Penicillium simplicissimum. Environ. Sci. 2005, 26, 167–171. [Google Scholar]
- Fraaije, M.W.; Pikkemaat, M.; Van Berkel, W. Enigmatic gratuitous induction of the covalent flavoprotein vanillyl-alcohol oxidase in Penicillium simplicissimum. Appl. Environ. Microbiol. 1997, 63, 435–439. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Hu, T.J.; Zeng, G.M.; Huang, D.L.; Yin, L.; Liu, Y.; Wu, J.J.; Liu, H. Biodegradation of lignocellulose by Penicillium simplicissimum and characters of lignocellulolytic enzymes. Environ. Sci. 2013, 34, 781–788. [Google Scholar]
- Zeng, J. Developing Species Recognition and Diagnostics of Rare Opportunistic Fungi; AmsterdamIBED: Amsterdam, The Netherlands, 2007. [Google Scholar]
- Morio, F.; Le Berre, J.Y.; Garcia-Hermoso, D.; Najafzadeh, M.J.; de Hoog, S.; Benard, L.; Michau, C. Phaeohyphomycosis due to Exophiala xenobiotica as a cause of fungal arthritis in an HIV-infected patient. Med. Mycol. 2012, 50, 513–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aoyama, Y.; Aoyama, Y.; Nomura, M.; Yamanaka, S.; Ogawa, Y.; Kitajima, Y. Subcutaneous phaeohyphomycosis caused by Exophiala xenobiotica in a non-Hodgkin lymphoma patient. Med. Mycol. 2009, 47, 95–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Hoog, G.S.; Zeng, J.S.; Harrak, M.J.; Sutton, D.A. Exophiala xenobiotica sp. nov., an opportunistic black yeast inhabiting environments rich in hydrocarbons. Antonie Van Leeuwenhoek 2006, 90, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Yelle, D.J.; Ralph, J.; Lu, F.; Hammel, K.E. Evidence for cleavage of lignin by a brown rot basidiomycete. Environ. Microbiol. 2008, 10, 1844–1849. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.; Suzuki, M.R.; Hammel, K.E. Processive endoglucanase active in crystalline cellulose hydrolysis by the brown rot basidiomycete Gloeophyllum trabeum. Appl. Environ. Microbiol. 2005, 71, 2412–2417. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.J.; Kim, Y.K. Degradation of crystalline cellulose by the brown-rot basidiomycete Fomitopsis palustris. J. Microbiol. 2005, 43, 487–492. [Google Scholar]
- D’Souza, T.M.; Merritt, C.S.; Reddy, C.A. Lignin-modifying enzymes of the white rot basidiomycete Ganoderma lucidum. Appl. Environ. Microbiol. 1999, 65, 5307–5313. [Google Scholar] [CrossRef] [Green Version]
- Gold, M.H.; Wariishi, H.; Valli, K. Extracellular peroxidases involved in lignin degradation by the white rot basidiomycete Phanerochaete chrysosporium. In Biocatalysis in Agricultural Biotechnology; American Chemical Society: Washington, DC, USA, 1989; pp. 127–140. [Google Scholar]
- Sutcliffe, B.; Chariton, A.A.; Harford, A.J.; Hose, G.C.; Greenfield, P.; Midgley, D.J.; Paulsen, I.T. Diverse fungal lineages in subtropical ponds are altered by sediment-bound copper. Fungal Ecol. 2018, 34, 28–42. [Google Scholar] [CrossRef]
- Letcher, P.M.; Vélez, C.G.; Barrantes, M.E.; Powell, M.J.; Churchill, P.F.; Wakefield, W.S. Ultrastructural and molecular analyses of Rhizophydiales (Chytridiomycota) isolates from North America and Argentina. Mycol. Res. 2008, 112, 759–782. [Google Scholar] [CrossRef]
- Ohm, R.A.; de Jong, J.F.; Lugones, L.G.; Aerts, A.; Kothe, E.; Stajich, J.E.; de Vries, R.P.; Record, E.; Levasseur, A.; Baker, S.E.; et al. Genome sequence of the model mushroom Schizophyllum commune. Nat. Biotechnol. 2010, 28, 957–963. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, O.; Liese, W. Variability of wood degrading enzymes of Schizophyllum commune. Holzforschung 1980, 34, 67–72. [Google Scholar] [CrossRef]
- Ghosh, S.; Sachan, A.; Mitra, A. Degradation of ferulic acid by a white rot fungus Schizophyllum commune. World J. Microbiol. Biotechnol. 2005, 21, 385–388. [Google Scholar] [CrossRef]
- Irshad, M.; Asgher, M. Production and optimization of ligninolytic enzymes by white rot fungus Schizophyllum commune IBL-06 in solid state medium banana stalks. Afr. J. Biotechnol. 2002, 10, 18234–18242. [Google Scholar]
- Haltrich, D.; Steiner, W. Formation of xylanase by Schizophyllum commune: Effect of medium components. Enzym. Microb. Technol. 1994, 16, 229–235. [Google Scholar] [CrossRef]
- Steiner, W.; Lafferty, R.M.; Gomes, I.; Esterbauer, H. Studies on a wild strain of Schizophyllum commune: Cellulase and xylanase production and formation of the extracellular polysaccharide. Biotechnol. Bioeng. 1987, 30, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Baldrian, P.; Valášková, V. Degradation of cellulose by basidiomycetous fungi. FEMS Microbiol. Rev. 2008, 32, 501–521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ozgun, H.D.; Jacobs, D.L.; Toms, S.A. Cladophialophora bantiana. In Fungal Infections of the Central Nervous System; Springer International Publishing: Cham, Switzerland, 2019; pp. 195–204. [Google Scholar]
- Shields, G.S.; Castillo, M. Myelitis Caused by Cladophialophora bantiana. Am. J. Roentgenol. 2002, 179, 278–279. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef]
- Johnston, S.R.; Boddy, L.; Weightman, A.J. Bacteria in decomposing wood and their interactions with wood-decay fungi. FEMS Microbiol. Ecol. 2016, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Der Wal, A.; De Boer, W.; Smant, W.; Van Veen, J.A. Initial decay of woody fragments in soil is influenced by size, vertical position, nitrogen availability and soil origin. Plant Soil 2007, 301, 189–201. [Google Scholar] [CrossRef] [Green Version]
- Sun, H.; Terhonen, E.; Kasanen, R.; Asiegbu, F.O. Diversity and community structure of primary wood-inhabiting bacteria in boreal forest. Geomicrobiol. J. 2014, 31, 315–324. [Google Scholar] [CrossRef]



| Sample name | Wood Species | Provenance |
|---|---|---|
| N1 | Silver fir—Abies alba Mill. | Ancient port of Neapolis—archaeological site of Piazza Municipio, Naples, Italy |
| N4 | Silver fir—Abies alba Mill. | |
| N9 | Elm—Ulmus sp. | |
| N39 | Elm—Ulmus sp. | |
| N88 | Silver fir—Abies alba Mill. | |
| PF | Ash—Fraxinus sp. | Archaeological site of San Rossore, Pisa, Italy |
| PO | Elm—Ulmus sp. | |
| PQ | Oak—Quercus sp. | |
| PL1 | Holm oak—Quercus ilex L. | |
| PL2 | Holm oak—Quercus ilex L. |
| Protocol 1—Maxwell® RSC Plant DNA Kit | Protocol 2—Maxwell® RSC PureFood GMO and Authentication Kit |
|---|---|
| 1N1 | 2N1 |
| 1N4 | 2N4 |
| 1N9 | 2N9 |
| 1N39 | 2N39 |
| 1N88 | 2N88 |
| 1PF | 2PF |
| 1PL1 | 2PO |
| 1PL2 | 2PQ |
| 1PO | |
| 1PQ |
| Protocol 1—Maxwell® RSC Plant DNA Kit | Protocol 2—Maxwell® RSC PureFood GMO and Authentication Kit | ||||
|---|---|---|---|---|---|
| Sample | DNA Concentration (ng/µl) | 260/280 Ratio | Sample | DNA Concentration (ng/µl) | 260/280 Ratio |
| 1N1 | 33.5 | 1.73 | 2N1 | 13.2 | 1.38 |
| 1N4 | 49.8 | 1.29 | 2N4 | 8.5 | 1.63 |
| 1N9 | 19.5 | 1.36 | 2N9 | 7.3 | 2.56 |
| 1N39 | 45.7 | 1.20 | 2N39 | 5 | 3.34 |
| 1N88 | 52.9 | 1.1 | 2N88 | 4 | 22.78 |
| 1PF | 233.6 | 0.98 | 2PF | 123.3 | 1.05 |
| 1PO | 67.3 | 1.27 | 2PO | 40.1 | 40.1 |
| 1PQ | 780.4 | 0.91 | 2PQ | 35.9 | 35.9 |
| 1PL1 | 170.8 | 1.11 | |||
| 1PL2 | 551.7 | 0.96 | |||
| Sample | Dbd (g × cm−3) | MWC (%) | LWS (%) | |
|---|---|---|---|---|
| 2008 | Elm 1 | 0.20 | 441 | 63 |
| Elm 2 | 0.18 | 517 | 67 | |
| Holm oak | 0.16 | 582 | 78 | |
| Oak | 0.20 | 453 | 71 | |
| Ash | 0.14 | 657 | 78 | |
| 2018 | PF | 0.09 | 1102 | 83 |
| PO | 0.11 | 879 | 81 | |
| PQ | 0.17 | 530 | 75 | |
| PL | 0.13 | 729 | 82 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antonelli, F.; Esposito, A.; Galotta, G.; Davidde Petriaggi, B.; Piazza, S.; Romagnoli, M.; Guerrieri, F. Microbiota in Waterlogged Archaeological Wood: Use of Next-Generation Sequencing to Evaluate the Risk of Biodegradation. Appl. Sci. 2020, 10, 4636. https://doi.org/10.3390/app10134636
Antonelli F, Esposito A, Galotta G, Davidde Petriaggi B, Piazza S, Romagnoli M, Guerrieri F. Microbiota in Waterlogged Archaeological Wood: Use of Next-Generation Sequencing to Evaluate the Risk of Biodegradation. Applied Sciences. 2020; 10(13):4636. https://doi.org/10.3390/app10134636
Chicago/Turabian StyleAntonelli, Federica, Alfonso Esposito, Giulia Galotta, Barbara Davidde Petriaggi, Silvano Piazza, Manuela Romagnoli, and Francesca Guerrieri. 2020. "Microbiota in Waterlogged Archaeological Wood: Use of Next-Generation Sequencing to Evaluate the Risk of Biodegradation" Applied Sciences 10, no. 13: 4636. https://doi.org/10.3390/app10134636
APA StyleAntonelli, F., Esposito, A., Galotta, G., Davidde Petriaggi, B., Piazza, S., Romagnoli, M., & Guerrieri, F. (2020). Microbiota in Waterlogged Archaeological Wood: Use of Next-Generation Sequencing to Evaluate the Risk of Biodegradation. Applied Sciences, 10(13), 4636. https://doi.org/10.3390/app10134636







