Construction of an Enzymatically-Conjugated DNA Aptamer–Protein Hybrid Molecule for Use as a BRET-Based Biosensor
Abstract
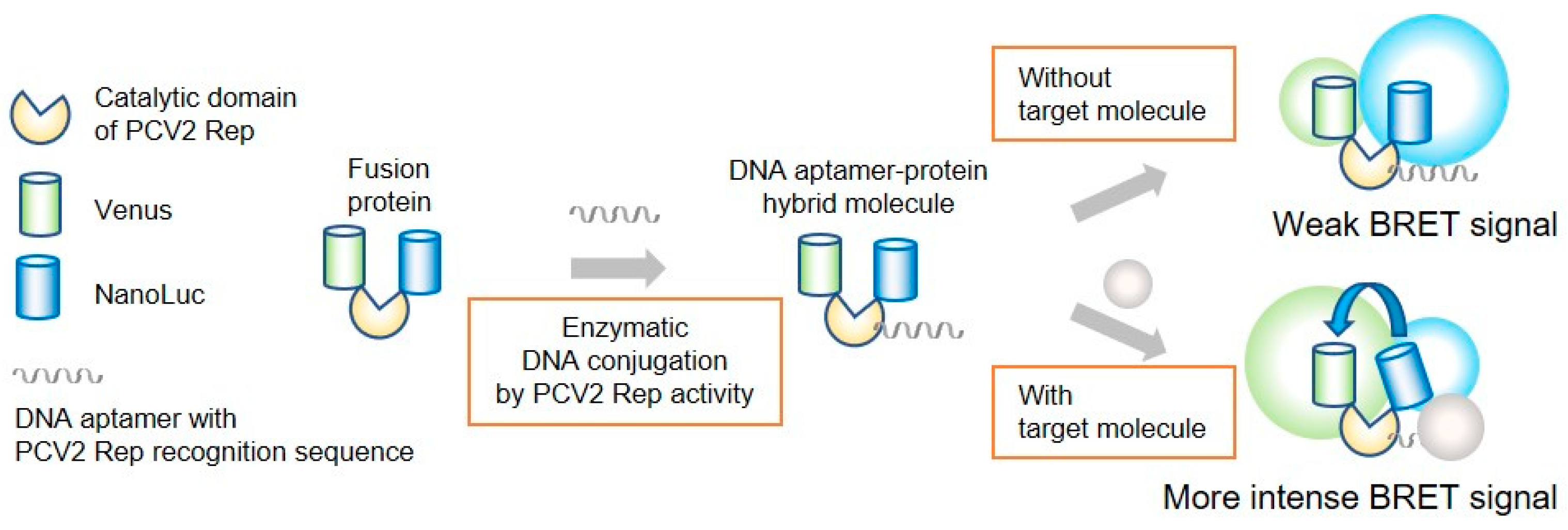
:1. Introduction
2. Materials and Methods
2.1. Construction of Plasmids
2.2. Protein Expression and Purification
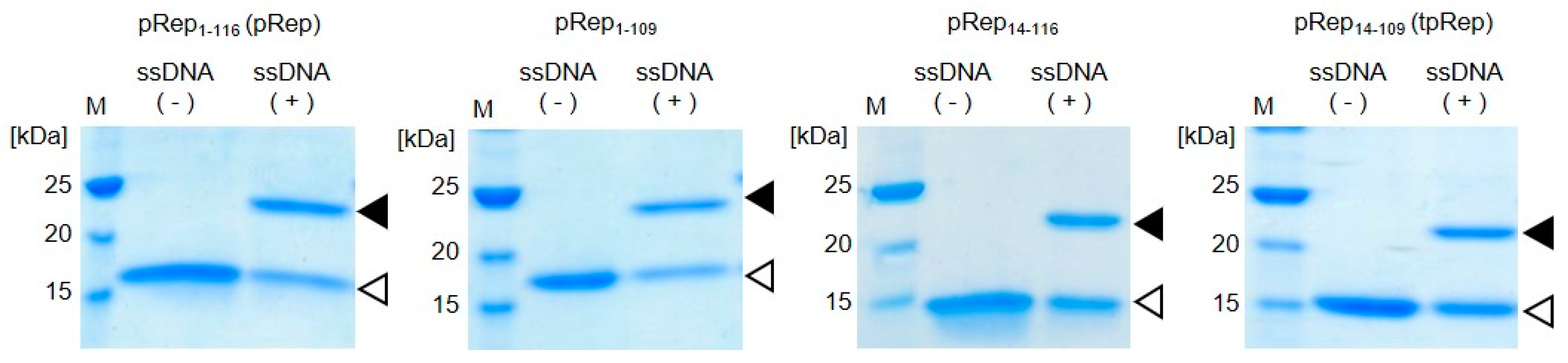
2.3. Evaluation of DNA Binding Ability of pRep
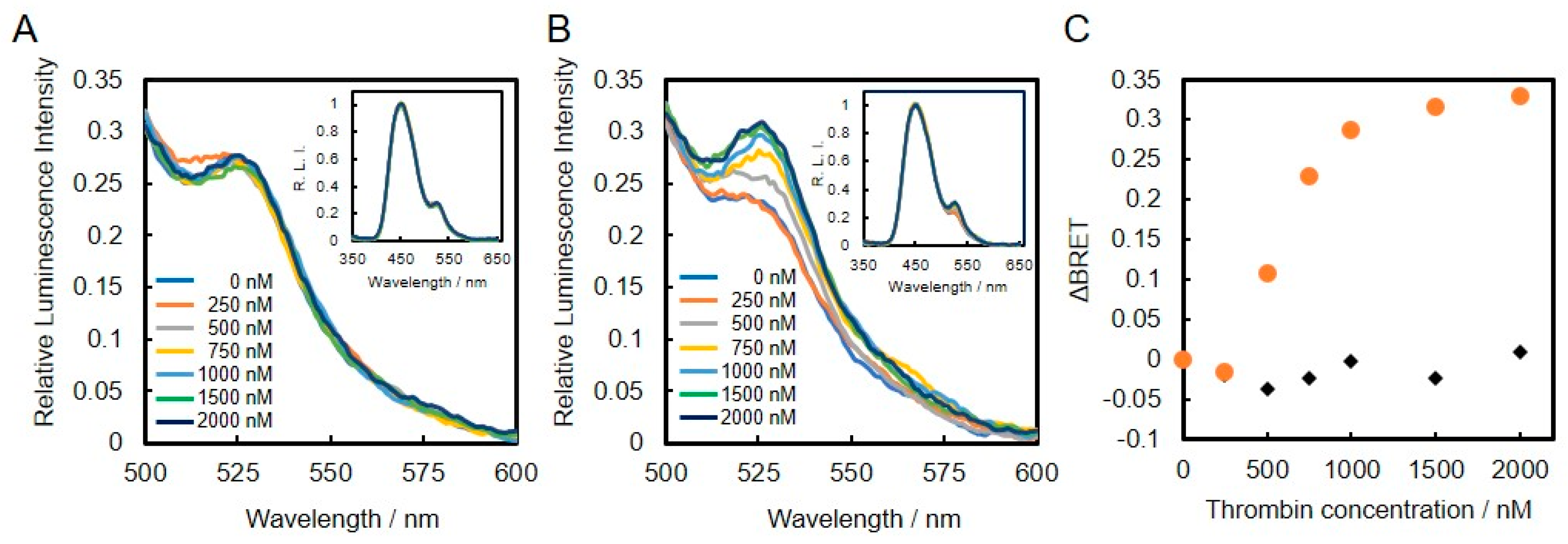
2.4. Evaluation of Emission Spectra
2.5. Homogeneous Assay with DNA Aptamer
3. Results and Discussion
3.1. Truncation of the Catalytic Domain of PCV2 Rep
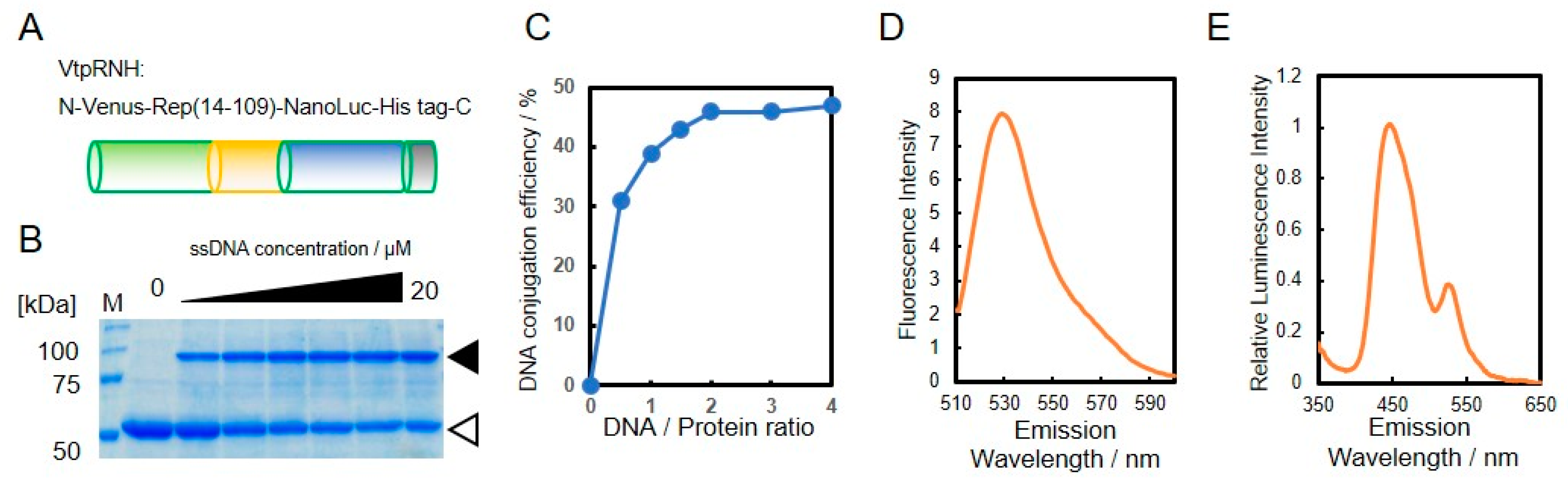
3.2. Construction of DNA-Protein Conjugates with NanoLuc and Venus for BRET-Based Biosensor
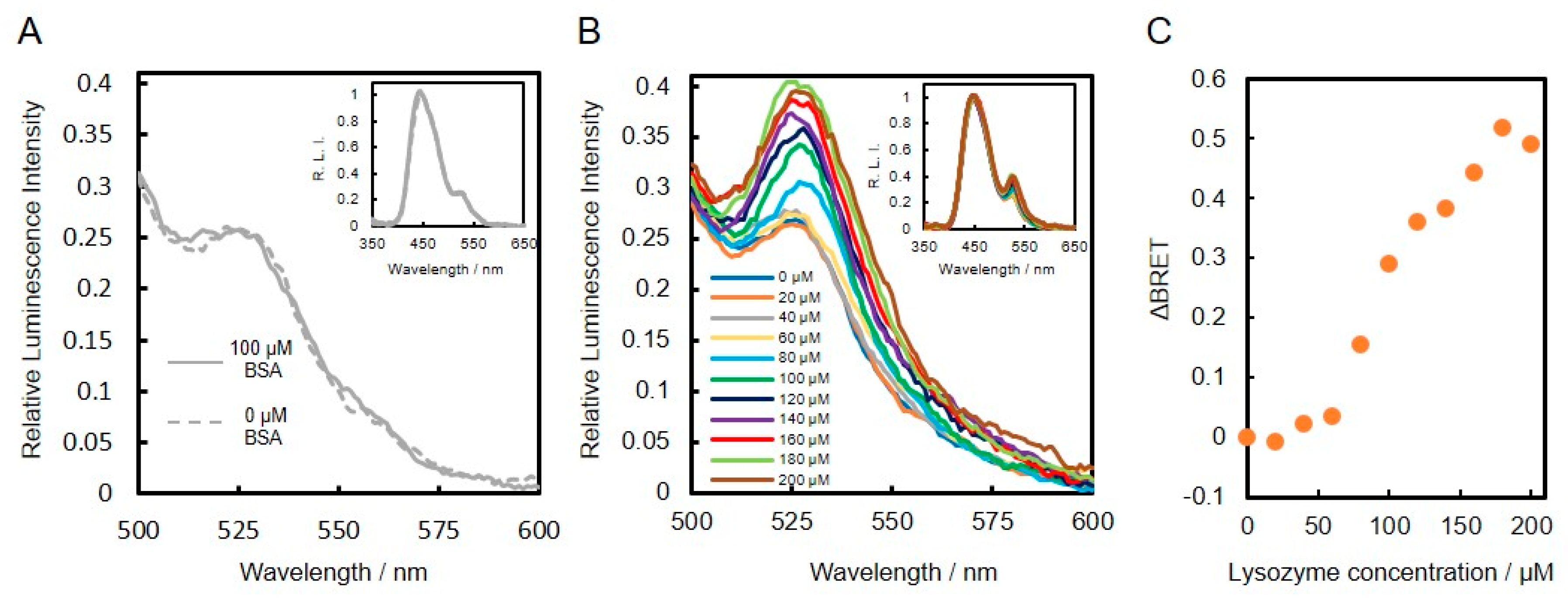
3.3. Construction and Evaluation of BRET-Based Biosensor with DNA-Protein Conjugates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zhao, D.; Kong, Y.; Zhao, S.; Xing, H. Engineering Functional DNA–Protein Conjugates for Biosensing, Biomedical, and Nanoassembly Applications. Top. Curr. Chem. 2020, 378, 41. [Google Scholar] [CrossRef]
- Sano, T.; Smith, C.; Cantor, C. Immuno-PCR: Very sensitive antigen detection by means of specific antibody-DNA conjugates. Science 1992, 258, 120–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimada, J.; Maruyama, T.; Kitaoka, M.; Kamiya, N.; Goto, M. DNA–enzyme conjugate with a weak inhibitor that can specifically detect thrombin in a homogeneous medium. Anal. Biochem. 2011, 414, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Takahara, M.; Wakabayashi, R.; Minamihata, K.; Goto, M.; Kamiya, N. Primary Amine-Clustered DNA Aptamer for DNA–Protein Conjugation Catalyzed by Microbial Transglutaminase. Bioconjug. Chem. 2017, 28, 2954–2961. [Google Scholar] [CrossRef]
- Mie, M.; Niimi, T.; Mashimo, Y.; Kobatake, E. Construction of DNA-NanoLuc luciferase conjugates for DNA aptamer-based sandwich assay using Rep protein. Biotechnol. Lett. 2019, 41, 357–362. [Google Scholar] [CrossRef]
- Wang, G.; Wang, Y.; Chen, L.; Choo, J. Nanomaterial-assisted aptamers for optical sensing. Biosens. Bioelectron. 2010, 25, 1859–1868. [Google Scholar] [CrossRef]
- Sassolas, A.; Blum, L.J.; Leca-Bouvier, B.D. Homogeneous assays using aptamers. Analyst 2011, 136, 257–274. [Google Scholar] [CrossRef]
- Citartan, M.; Gopinath, S.C.B.; Tominaga, J.; Tan, S.-C.; Tang, T.-H. Assays for aptamer-based platforms. Biosens. Bioelectron. 2012, 34, 1–11. [Google Scholar] [CrossRef]
- Gokulrangan, G.; Unruh, J.R.; Holub, D.F.; Ingram, B.; Johnson, C.K.; Wilson, G.S. DNA Aptamer-Based Bioanalysis of IgE by Fluorescence Anisotropy. Anal. Chem. 2005, 77, 1963–1970. [Google Scholar] [CrossRef]
- Rupcich, N.; Chiuman, W.; Nutiu, R.; Mei, S.; Flora, K.K.; Li, Y.; Brennan, J.D. Quenching of Fluorophore-Labeled DNA Oligonucleotides by Divalent Metal Ions: Implications for Selection, Design, and Applications of Signaling Aptamers and Signaling Deoxyribozymes. J. Am. Chem. Soc. 2006, 128, 780–790. [Google Scholar] [CrossRef] [PubMed]
- Urata, H.; Nomura, K.; Wada, S.; Akagi, M. Fluorescent-labeled single-strand ATP aptamer DNA: Chemo- and enantio-selectivity in sensing adenosine. Biochem. Biophys. Res. Commun. 2007, 360, 459–463. [Google Scholar] [CrossRef]
- Hao, L.; Zhao, Q. A fluorescein labeled aptamer switch for thrombin with fluorescence decrease response. Anal. Methods 2015, 7, 3888–3892. [Google Scholar] [CrossRef]
- Mashimo, Y.; Maeda, H.; Mie, M.; Kobatake, E. Construction of Semisynthetic DNA-Protein Conjugates with Phi X174 Gene-A* Protein. Bioconjug. Chem. 2012, 23, 1349–1355. [Google Scholar] [CrossRef] [PubMed]
- Brown, N.E.; Blumer, J.B.; Hepler, J.R. Bioluminescence Resonance Energy Transfer to Detect Protein-Protein Interactions in Live Cells. In Protein-Protein Interactions; Meyerkord, C.L., Fu, H., Eds.; Methods in Molecular Biology; Springer: New York, NY, USA, 2015; Volume 1278, pp. 457–465. ISBN 978-1-4939-2424-0. [Google Scholar]
- Yang, J.; Cumberbatch, D.; Centanni, S.; Shi, S.; Winder, D.; Webb, D.; Johnson, C.H. Coupling optogenetic stimulation with NanoLuc-based luminescence (BRET) Ca++ sensing. Nat. Commun. 2016, 7, 13268. [Google Scholar] [CrossRef] [Green Version]
- Inagaki, S.; Tsutsui, H.; Suzuki, K.; Agetsuma, M.; Arai, Y.; Jinno, Y.; Bai, G.; Daniels, M.J.; Okamura, Y.; Matsuda, T.; et al. Genetically encoded bioluminescent voltage indicator for multi-purpose use in wide range of bioimaging. Sci. Rep. 2017, 7, 42398. [Google Scholar] [CrossRef]
- Dale, N.C.; Johnstone, E.K.M.; White, C.W.; Pfleger, K.D.G. NanoBRET: The Bright Future of Proximity-Based Assays. Front. Bioeng. Biotechnol. 2019, 7, 56. [Google Scholar] [CrossRef]
- Vega-Rocha, S.; Byeon, I.-J.L.; Gronenborn, B.; Gronenborn, A.M.; Campos-Olivas, R. Solution Structure, Divalent Metal and DNA Binding of the Endonuclease Domain from the Replication Initiation Protein from Porcine Circovirus 2. J. Mol. Biol. 2007, 367, 473–487. [Google Scholar] [CrossRef] [PubMed]
- Kirby, R.; Cho, E.J.; Gehrke, B.; Bayer, T.; Park, Y.S.; Neikirk, D.P.; McDevitt, J.T.; Ellington, A.D. Aptamer-Based Sensor Arrays for the Detection and Quantitation of Proteins. Anal. Chem. 2004, 76, 4066–4075. [Google Scholar] [CrossRef] [PubMed]
- Tasset, D.M.; Kubik, M.F.; Steiner, W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J. Mol. Biol. 1997, 272, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Lovendahl, K.N.; Hayward, A.N.; Gordon, W.R. Sequence-Directed Covalent Protein–DNA Linkages in a Single Step Using HUH-Tags. J. Am. Chem. Soc. 2017, 139, 7030–7035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagai, T.; Ibata, K.; Park, E.S.; Kubota, M.; Mikoshiba, K.; Miyawaki, A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002, 20, 87–90. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.P.; Unch, J.; Binkowski, B.F.; Valley, M.P.; Butler, B.L.; Wood, M.G.; Otto, P.; Zimmerman, K.; Vidugiris, G.; Machleidt, T.; et al. Engineered Luciferase Reporter from a Deep Sea Shrimp Utilizing a Novel Imidazopyrazinone Substrate. ACS Chem. Biol. 2012, 7, 1848–1857. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mie, M.; Hirashima, R.; Mashimo, Y.; Kobatake, E. Construction of an Enzymatically-Conjugated DNA Aptamer–Protein Hybrid Molecule for Use as a BRET-Based Biosensor. Appl. Sci. 2020, 10, 7646. https://doi.org/10.3390/app10217646
Mie M, Hirashima R, Mashimo Y, Kobatake E. Construction of an Enzymatically-Conjugated DNA Aptamer–Protein Hybrid Molecule for Use as a BRET-Based Biosensor. Applied Sciences. 2020; 10(21):7646. https://doi.org/10.3390/app10217646
Chicago/Turabian StyleMie, Masayasu, Rena Hirashima, Yasumasa Mashimo, and Eiry Kobatake. 2020. "Construction of an Enzymatically-Conjugated DNA Aptamer–Protein Hybrid Molecule for Use as a BRET-Based Biosensor" Applied Sciences 10, no. 21: 7646. https://doi.org/10.3390/app10217646





