1. Introduction
Currently, due to the consumption habits in the developed countries, diabetes-related diseases have been increasing in prevalence [
1]. Diabetes mellitus is characterized by a defective use or lack of insulin in the body, resulting in an increase in sugar levels in blood. This causes a progressive damage in different parts of the human body, specially in its most sensible structures like the retinal and choroidal vascularity [
2,
3]. Among the most common consequences of this pathology, we can find the Diabetic Retinopathy [
4] and the Diabetic Macular Edema (DME). In particular, the latter is characterized by fluid accumulations in between the retinal layers, producing the progressive vision deterioration that may result in severe blindness [
5,
6].
Given that the pathology only causes vision impairment in advanced stages (where the damage may be irreversible), an early detection is critical [
7]. Additionally, the more the pathology advances, the more drastic and severe the treatment has to be for a successful management of the pathology.
Currently, to study these pathologies, eye care practitioners commonly use Optical Coherence Tomography (OCT) images [
8,
9,
10]. These images correspond to cross-sectional cuts of the retina, allowing to perceive its histological organization and internal characteristics. Using the OCT images as reference, Otani et al. [
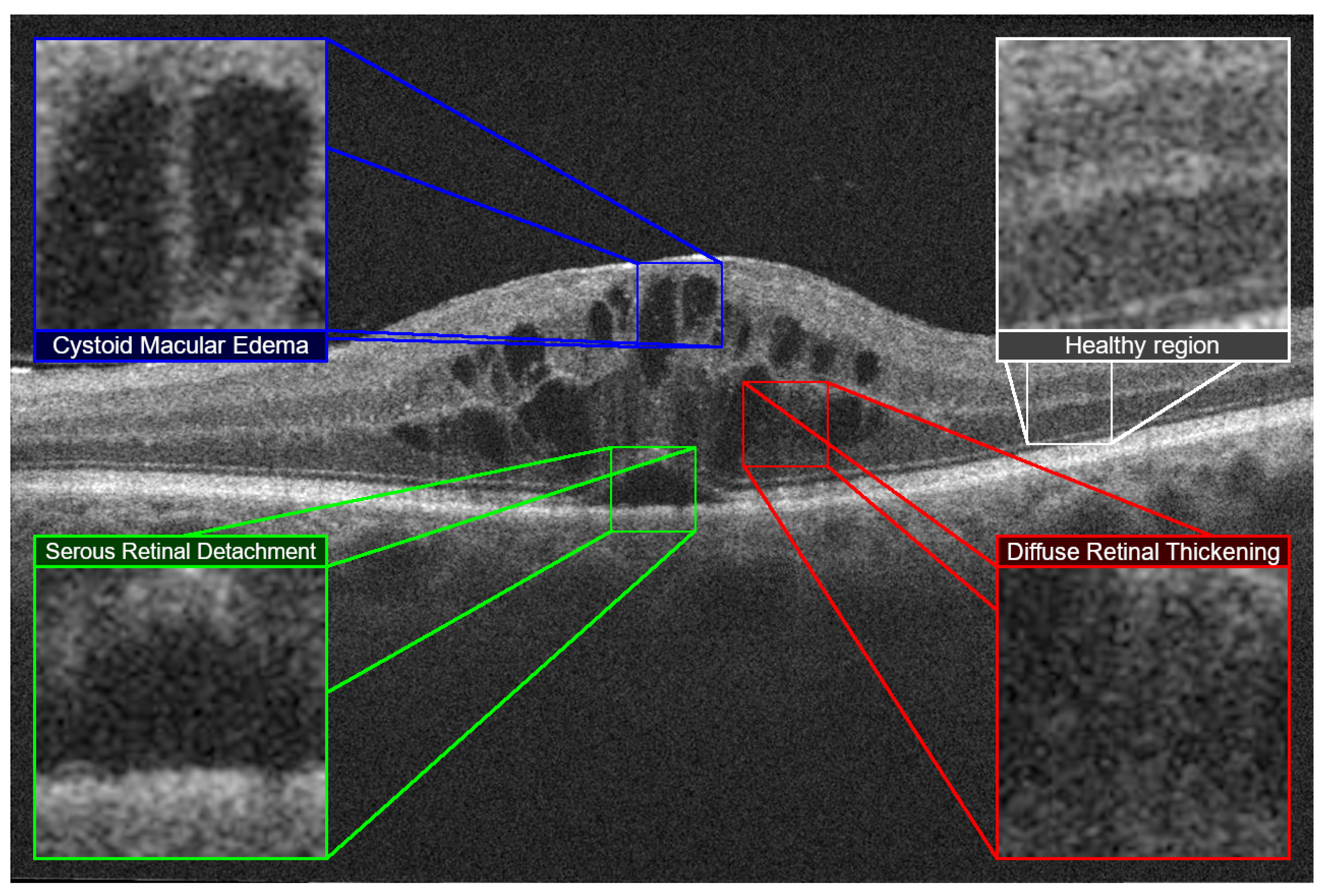
11] established three different types of DME. An accumulation of fluid with cystic properties, an accumulation in the external layers of the retina causing a detachment of the photoreceptor layer and a sponge-like retinal thickening. This DME classification became the reference in the clinical field after being expanded by Panozzo et al. [
12] defining these fluid accumulations as Diffuse Retinal Thickening (DRT), Cystoid Macular Edema (CME) and Serous Retinal Detachment (SRD) with more precise limits, subcategories and including an study on the status of the innermost retinal layers (also called
epiretinal traction). These three main retinal fluid accumulations are represented in
Figure 1:
DRT is characterized by a sponge-like texture, formed by the accumulation and mixture of normal retinal tissues and the retinal fluid leakages. These leakages cause a sensible swelling in the retina, but without noticeable fluid accumulations and reduced visual capabilities of the patient. On the other hand, CME consists of a sensible fluid accumulation causing cystoid spaces that are formed in the retinal layers. These cysts may have defined cell barriers and, depending on its location and size, may cause minor to severe visual impairment. Finally, SRD consists in a subretinal dome-like fluid accumulation [
13]. The more these fluid accumulations are closer to the fovea, the more severe the vision impairment (as this type of accumulation directly affects the photoreceptor layer of the retina).
The characterization of the different types of edema previously described is a crucial task during the diagnosis. Each type represents a different stage and complication of the pathology, as different types of fluid accumulations can affect different regions of the field of view with different levels of intensity. In addition, different types of edemas also require different types of treatment; ranging from monitoring and changing the habits of the patient, pharmacological treatment or, in more severe or refractory cases, surgery. While DRT can be easily reabsorbed, the other types of fluid accumulations represent a greater difficulty depending on the extent of the accumulated damage and possibility of scar tissue formation.
Given the complexity and differences among each of the DME types and the relevance of the DME disease, several automated methodologies have arisen trying to offer a quantitative analysis of these pathological fluid accumulations. Generally, the state of the art has mostly considered the segmentation of CME fluid accumulations and, in a lesser extent, SRD ones (as DRT accumulations do not present a defined border to segment). Most of the works mainly extract specific segmentations of the fluid regions using techniques such as thresholding and candidate filtering based on size, intensity or shape constraints (Wilkins et al. [
14], Roychowdhury et al. [
15]), clustering algorithms (Girish et al. [
16]) and even transformations to alternate domains/graph search-cut strategies (Rashno et al. [
17,
18]). To the best of our knowledge, only two works tried to generate a segmentation on the three clinical types we present, the work of Samagaio et al. [
19] which used texture, intensity and clinical-based features to localize CME, DRT and SRD fluid accumulations in OCT images and the work of de Moura et al. [
20], which uses these detections to further generate a segmentation of the different fluid accumulations. Regarding deep learning techniques, recently, works like the ones proposed by Lee et al. [
21], Venhuizen et al. [
22], Roy et al. [
23] or Chen et al. [
24] also aimed for a precise segmentation, most of these deep approaches deriving from the U-Net architecture of Ronnenberger et al. [
25] which has been used too to characterize some of the segmented fluid accumulations (Lu et al. [
26]).
Despite the variety of proposals, these approaches imply, as main limitation, the necessity of visible defined contours for the analyzed fluid regions. To improve the detections in these diffuse regions, de Moura et al. [
27,
28] proposed a complementary strategy based on a regional identification. Afterwards, in Vidal et al. [
29] this strategy was used to analyze the OCT images and generate an intuitive visualization of the detected region. In this work, and considering satisfactory preliminary results [
30], we propose to expand this strategy to be able to simultaneously detect and characterize the aforementioned types of DME. Using this identifications and characterizations, we also designed a graphical intuitive representation of the defined types of DME to facilitate the inspection and diagnosis by the expert clinicians. Our methodology was trained and tested using a dataset composed by images obtained in clinical practice where experts selected the images with the intraretinal fluid categories of interest, and made sure to include the maximum heterogeneity in both severity and variety within each type of fluid accumulation.
The work hereby presented is divided in three main sections.
Section 2: Materials and methods describes the tools, libraries and dataset used; as well as the methodology followed.
Section 3: Results presents the performed experiments and a brief comment on them. Afterwards, these results are analyzed in
Section 4: Discussion. Finally,
Section 5: Conclusions presents a summary of what was achieved in this work as well as possible future lines of work.
2. Materials and Methods
2.1. Dataset and Software Resources
The dataset used for this work was composed by 177 OCT images coming from a CIRRUSTM HD-OCT 500 Carl Zeiss Meditec and 179 OCT images from a modular HRA + OCT SPECTRALIS® from Heidelberg Engineering, Inc. The OCT images were captured from both left and right eyes with different device configurations and ranging in resolution from pixels to pixels in the Spectralis dataset and pixels to pixels in the Cirrus dataset.
The protocol to obtain these images from clinical practice and posterior study were conducted in accordance with the Declaration of Helsinki, approved by the Ethics Committee of Investigation from A Coruña/Ferrol (2014/437) the 24th of november, 2014.
Two experts have labelled the different regions as belonging to a specific class only those regions where they have a high degree of confidence in the assigned class. Any region that is uncertain (either because it belongs to multiple classes, is part of one of the vague limits or simply because of the idiosyncrasy of the pathology itself) will be indicated as an special separate class. Thus, as shown in
Figure 2, regions with CME have been colored blue, regions with DRT have been colored red and regions with SRD have been colored green. Additionally, the region with tissue not belonging to the considered pathologies is shown in white, and two different cases are marked in black: if they are outside the ROI, the region is ignored because it belongs to a non-ROI region. On the other hand, if the color black is located within the ROI, it is considered a doubtful region and is not assigned explicitly to a class. Regarding software resources, for the development of this project we have used the Pattern Recognition Tools library, Version 5.3.1 [
31] in MATLAB 9.4 R2018a [
32].
2.2. Diabetic Macular Edema Characterization
The proposed methodology can be divided into three main steps. An image analysis step, a ME classification step where we trained models to classify each type of ME, and a map generation step where we generated the maps and final visualizations for the experts using those trained models. These steps can be seen in
Figure 3.
2.3. Image Analysis
In this first phase of image analysis, we have the stages aimed at preparing and extracting the samples from the images of the dataset. These phases include both the automatic determination of the ROI, exhaustive sampling of the OCT images and the generation of the vectors that are used later in the methodology.
2.3.1. Layer Extraction
In this first step, we delimited the ROI to the region of the OCT image where the studied fluid accumulations may appear. The retinal layers that separate the ROI and the rest of the image are the innermost Inner Limiting Membrane (ILM) and the outermost Retinal Pigment Epithelium (RPE). These two layers separate the retina from the choroid (the vascular layer of the eye, which nourishes the rest of the eye structures) and the vitreous humor (a gelatinous transparent fluid inside the eyeballs). All these structures are shown in
Figure 4.
The retinal layers were extracted using the algorithm proposed by Chiu et al. [
33,
34], which obtains the minimum weighted path that connects the horizontal image edges to find an initial layer, and does this iteratively by establishing a minimum distance from the previously detected ones. This method is able to obtain seven different retinal layers, but in this work we only consider the two aforementioned ones: the ILM and the RPE.
2.3.2. Image Sampling
Four independent datasets were generated: one for each type of DME and one for the tissue that is not associated with any fluid accumulation (not necessarily healthy) as were the labels defined by the experts. To sample each image, it is divided into overlapping windows. These samples are, then, assigned with a label based on the ground truth established by the experts. To assign the label, it is first evaluated whether the sample is centered within a defined label region. If this is the case (i.e., the center pixel of a sample has a label that can be non-fluid, CME, DRT or SRD), then that sample is directly considered as belonging to that class.
On the other hand, if a sample does not have a defined category in the central pixel (as may be the case when it falls into one of the previously defined black doubtful regions), it is assigned to the class with the largest area within the sample. An example of this case can be seen in
Figure 5, where the sample would have been assigned to the healthy class, as it is the one with the most surface within the candidate sample limits. This way, we ensure that the detections are adjusted to the samples, while avoiding any confusion in the system if the pathological fluid accumulations have a minimal amount of non-pathological tissue inside. This strategy represents a trade-off balance between reducing the amount of patterns of non-ROI regions and directly losing the ROI border patterns by not considering these samples.
Once all the samples have been labeled, 30 random samples were selected for each class from the total available in the image (giving a theoretical sum of 120 samples per image). In case this quota of 30 samples per class/image was not covered in an OCT image, the debt is accumulated progressively in successive images, trying to extract from them a greater number of random samples until the sample debt for that class was satisfied and the base extraction quota was enough.
Finally, as samples extracted in borderline retinal regions could contain undesired patterns from any of the external regions and it is important to preserve the spatial relationship of the pixels, the biggest rectangle from each sample that only contains pixels inside the ROI was extracted. This way, three balanced datasets were generated using only the set of 284 training images. The CME dataset was composed of 16,980 samples, the DRT dataset was composed of 16,980 samples and the SRD dataset, being less prominent in our dataset and with a smaller surface area when present, the methodology has only been able to extract 16,620 samples.
2.3.3. Feature Extraction
In this work, we analyzed the textures present in the samples to determine the type of fluid accumulation (or absence of it). For this, we needed a library of intensity and texture descriptors. These descriptors have been chosen (and expanded within each category) as they have already proved their usefulness in the field of medical imaging [
35] as well as in the detection of intraretinal fluid [
29,
36]. The complete library of extracted features is shown in
Table 1.
Thus, for each of the samples, we extracted a vector that defines its texture and intensity properties from different points of view using the 357 markers. Then, we used a feature selector to reduce the dimensionality of the vector and keep the most relevant features for each type of DME. To prevent a bias we divided the dataset into two subsets containing the 80% and 20%. This way, that 20% of samples will remain unbiased for further statistical analyses.
As representative feature selector, we used the Sequential Forward Selection (SFS) strategy to extract the top 100 most relevant features [
37]. This selector looks for the features that best group the samples of the same class in the space and at the same time are as different as possible from the rest of the classes. Specifically, this selector evaluates each feature independently when considering it as a possible candidate. This allowed us to later incrementally examine the contribution of each feature to the detection and characterization of DME types.
2.4. ME Classification
Now that we had samples extracted for each of the types of DME, their features and the most relevant features selected for each type of DME; we were ready to train a model for each of the DME types (as we used an one-vs-all approach for each class).
For the training of the models, we had to reduce the sampling rate of each image from 30 samples to 5 per image by randomly resampling them on the previously generated sets. This has been done to allow the testing of more complex models during the expensive training in terms of resources. Additionally, to take advantage of the iterative SFS feature selection, we defined an incremental training strategy. This strategy evaluated subsets of features independently, always incorporating in each iteration the next feature that the SFS selector considered as the next most relevant. This way, we started with the most relevant feature. Then, the performance of the feature is evaluated and finally the new feature was added to the subset, repeating the process until all the features in the ranking are evaluated.
To evaluate each of the subsets of features, we used a modified 10-fold cross-validation strategy adapted to our problem with four different tested machine learning paradigms. Therefore, the division of candidates for training and testing was not done at the sample level, but at the image level. With this strategy, we intended to guarantee that the test was not biased because it contains samples of the same image used in the training. For this image separation calculation, only the images that have relevant samples for each category were taken into account, ensuring that no images that do not contain a valid sample for a particular class were assigned to the training or test sets.
In addition, each of the sub-datasets created for the cross-validation of each feature subset were balanced, ensuring that they contain samples from all the classes but maintaining a 50-50 ratio between the positive and negative labels. To achieve this, for each type of pathology, samples were chosen at random from each of the datasets that provided the samples for the negative class, N being the number of samples of the positive class. For example, when generating an hypothetical CME dataset (thus CME would be the positive class), if the CME dataset had 100 samples, 33 samples would be taken from the DRT, SRD and dataset with samples not belonging to fluid regions. For the training, we have chosen four models that represent different paradigms tested in the domain, which have also obtained positive results in previous experiments related to the identification of intraretinal fluid. Thus, the paradigms we used are a k-Nearest Neighbors (kNN) with k = 15, a Linear Discriminant Classifier (LDC), a Random Forest (RF) and a Support Vector Machine (SVM) with a Radial Basis Function (RBF) or Gaussian as kernel.
Finally, to select the optimal number of features to train the final models, we analyzed the results that were obtained by fitting a degree four polynomial curve to the point cloud corresponding to the cross validations made with incremental feature subsets. By doing so, we analyzed the trend of the results from a global behaviour point of view instead of only each independent subset of characteristics (prone to the inherent variability of the designed training strategy). Finally, we do not keep the model that obtained the best result in the adjusted curve (since it may have simply obtained slightly better results than a previous subset by chance or at the cost of generalisation capabilities because it was significantly more complex). Instead, we study the shape of the fitted curve, keeping the simplest classifier (i.e., trained with fewer characteristics) that has reached a positive peak.
2.5. Map Generation
After the candidate models were trained for each type of DME and we had our chosen classifiers, we can now proceed to use them to generate the intuitive visualizations and characterizations of the DME types.
These maps are generated by thoroughly sampling OCT images. The first step, as mentioned, consists in extracting the ROI from the image and completely dividing it into samples of a given size. These samples are extracted all centered inside the ROI, equally spaced and with a previously configured overlap (the higher the overlap, the higher the granularity the maps will have). The same way as in the training process, if any sample falls outside the ROI albeit being centered in it, the biggest rectangular subsample that contains only relevant patterns is extracted from these samples. From each of these samples, then, three feature vectors are generated. Each feature vector corresponds to the descriptors the feature selection technique considered important to classify each different type of DME during their respective training steps.
By doing so, we can obtain a matrix composed by three feature vectors (one for each type of DME and optimal features defined by the SFS algorithm for each class). These classifications are used by two different complementary algorithms, the Confidence map and the Binary map. Finally, when the two types of maps are generated, its information is used to generate an intuitive visualization of the detection and characterization of the different ME types to facilitate their inspection by the specialists. This visualization consists in a 5-window representation of different points of view of the detections with two main representations:
The “CONFIDENCE” window shows a merged representation of all the pathology confidences and the interactions between them. This window allows experts to study the possible overlap between the types of pathology that cannot be achieved through segmentation. In addition, it clearly presents the interactions that cannot be observed in the windows of the other types on their own. With this same idea (displaying the confidence on the detections), the 3 right-sided windows show, independently, each of the confidences given by the confidence map algorithm, but using a color map with a higher degree of granularity (so an expert clinician can see the nuances between in the confidence levels).
The “DETECTIONS” window represents all the detections, without displaying the confidence the system infers in them. This allows the experts to have a different perspective of the detections, showing a more explicit representation of the independent regions that have tested as positive (as well as an interpolation of the surrounding regions that may also be candidates). In doing so, it complements the information shown in the “CONFIDENCE” window.
An example of the results that can be generated by this strategy is represented in
Figure 6. As can be seen in this image, despite the binary map showing a set of false positives, the confidence maps actually highlight the correct regions as important. The same way, specially in the case of DRT, it is interesting to also analyze any detection, as even expert clinicians have issues detecting where or where not the diffuse limits of the DRT end.
Confidence Map
To create the confidence map, we use the windows as ballots to calculate the confidence of the overlapped area of belonging to a given positive class. As an example, in
Figure 7b, a representation of the overlapping windows in a given image with multiple pathological positive samples is shown. Each sample that overlaps a pixel increases in one the number of "ballots", finally generating a map of the number of votes per pixel.
Depending on the morphology of the retina, overlap and certain regions close to the image borders, the weight of each vote over the total is different (that is, in some regions, the number of windows that overlap an area is lesser than in the internal ones; thus increasing the relevance of each ballot for the final confidence value). This effect can be seen in
Figure 7c, where the ROI borders present less intensity than other areas with (in reality) similar confidence levels. To compensate for this difference, the results are normalized dividing by the total number of windows that contained each pixel (
Figure 7c vs.
Figure 7d). Moreover, as the system supports images of any size, some rounding errors may appear depending on the width and height of the image when sampling. This normalization also solves this issue, removing the grid-like pattern derived from this rounding slight positional deviations. This way, formally, a pixel
x will have a detection confidence level equal to:
Being
N the number of valid windows (inside the area of interest) to which the pixel belongs,
the
ith window feature vector and
M the binary classifier function for a given type of DME. In
Figure 7d, the result of this normalization is shown, where the grid pattern has almost completely disappeared and the borders present a confidence level comparable to other regions with a higher number of overlapped windows.
After the maps are generated, four different color maps are applied. When designing a visualization that could represent all the three types of ME in the same image, we faced an issue: no representation could contain, unambiguously, information about the three types of ME if they overlapped at some point. There is no possibility that three differentiated color palettes that, when two are combined, at some point, generate a color that is already present in the last palette. In
Figure 7e, the final independent colormaps (for the sake of a better understanding of the results) are shown.
For this reason, we divided the results into two representations. The main combined representation uses the basic RGB colors, but varying their value in the HSV color domain to show the confidence values obtained to the clinician. This representation offers a clean view of the interactions between the different ME types in a non-ambiguous way, albeit losing some granularity when representing the confidence levels.
To compensate for this information loss, three different representations (one for each type of DME) are also generated. These representations use a color scale that represents, moving in the hue space, the confidence given by system. These representations offer an interface more readable for a human being with a higher level of granularity (albeit losing information about the interactions between the types of DME). This way, generating a representation with both the combined map and the triple independent representation, both interactions between the DME types and their precise confidence levels are clearly shown.
2.6. Binary Map
To generate the binary maps, once the classification has been made, we perform a nearest-neighbor interpolation. This allows us to obtain a regional identification without an in-depth sampling of the image. In
Figure 8, each one of the steps is graphically represented. In this case, the maps were generated with a low-overlap configuration to better illustrate the comparison between the two complementary visualizations. As it is shown in this image, the nearest pixels are assigned to the class that the selection is closer to. Moreover, as we are only using three independent binary classifiers, this process is done in parallel and without interference. As shown, despite the low overlap that is used when sampling the image, the three different ME regions are clearly represented. Additionally, in this figure we can see certain false positives. However, thanks to the complementary visualization we propose, these false positives would be shown in the complementary confidence maps with a low degree of relevance to the expert. Additionally, we will further discuss strategies to palliate these spurious detections in the discussion section of this work.
This way, while the confidence map aids the clinician to quickly assess the most relevant cases that are present in the image, the binary map represents all the possible candidates, favoring a quick but informed diagnostic.
3. Results
In this section we will proceed to present and explain different performance metrics obtained during the development of this work. Below we will show the results for both the feature selection stage (
Section 3.1) and the training/validation of the models (
Section 3.2).
3.1. Feature Selection
To properly evaluate the meaning and significance of the feature selection, we have divided these results into two different points of view. These two points of view are presented in
Figure 9.
Figure 9a shows the percentage of occurrence of each category over the total of the Top 100 selected features by the SFS strategy. However, this metric is not able to fully illustrate the relevance of each class, since not all the feature categories that were considered have the same number of markers. For this reason,
Figure 9b shows this same ranking, but weighted by the total number of available markers in each category.
We can see how the HOG descriptor is heavily chosen for all the DME types. This detector, based on the analysis of the gradient orientations, is extremely useful in all the three cases: CME with orientations in all the directions, SRD with fusiform shapes and, on the other hand, DRT characterized by the direct absence of defined edges, presenting diffuse patterns (therefore, in the case of DRT, HOG acts as a negative discriminant, marking the absence of edges an indicative of the presence of DRT).
On the other hand, we can observe that DRT also has less selected basic characteristics (GLDS) than for CME and SRD. This is due to the fact that DRT tends to be intermixed with healthy tissue and, specially in images that were taken with the Cirrus capture device, to present value ranges and distributions very similar to healthy tissue. Therefore, it is easier to distinguish DRT cases based on texture patterns rather than simply raw intensity data (again, specially with OCT images generated with the Cirrus OCT device).
This same fact has also influenced the total lack of selection of any of Gabor features. This descriptor, which as can be seen in
Figure 9a, stands out for the net amount of potential markers, being usually selected at the same ratio as HOG features since both offer good results when dealing with oriented gradients. However, Gabor is based on the decomposition of the image into its components in terms of the orientations and frequencies that make up the texture (similar to a Fourier transform). This strategy is not very useful in the case of DRT, since it does not present any regular texture pattern and, as we mentioned, the gray levels without spatial information are not very useful either when identifying this type of ME (which is why, as mentioned, the GLDS characteristics have not been selected either). This same case applies to the LAWS features, as they share similarities with Gabor and HOG descriptors. In the end, as the other aspects that Gabor and LAWS provided in addition to the information already stablished by the HOG features are not useful to detect the DRT type of ME, they were not selected in favor of simply using HOG.
We can observe in
Figure 9b the significant similarity between the characteristics that were selected for CME and SRD. This is due to the similarities in properties of both DME types, although with slight differences that manifest themselves in some of the selection proportions of various descriptors. For example, we see a higher rate of eigenvalues-based markers, which is appropriate due to the self-similarity present in the samples with SRD. There is also a lower rate of features from GLCM, since this type of fluid accumulation presents a simple pattern, always with hyperreflective to hyporeflective gradients as it is located around the retinal photoreceptor layer. Finally, we can also see a higher rate of selection of GLRLIS features compared to the CME ranking, which study the presence of strands of pixels with similar grey levels in different orientations. These are specially useful in this class because SRD-type fluid accumulations are usually either small in size or thin and elongated. In both cases, this descriptor allows to find those very homogeneous, hyporeflective and short strips commonly present in small fluid accumulations or long, hyporeflective and horizontal patterns present in elongated fluid accumulations.
Figure 10 shows in a crude way the results of the indices of the selected features in the top 100. What is particularly noticeable is the sequentiality of the features for the DRT after a certain initial number of selected features. This is mainly because, due to the difficulty of this pathology, the system was not able to select individual markers as relevant and directly began to add entire families of features sequentially.
3.2. Training of the Models
In this section we present the results of the training steps divided into the three different types of DME. In
Figure 11 we can see the results of the CME training process. On the other hand, in
Figure 12 we present the results for the training of the model destined to classify DRT samples. Finally, in
Figure 13, the results for the training of the model to detect SRD samples.
To increase the training speed and the model selection, the increments were done in steps of three features instead of one. As such, in these figures, the real mean test accuracy presented is the one of the closest feature subset to the estimated best candidate (hence the "Closest mean test accuracy" tag).
The final selected model for each type of DME can be seen in
Table 2. In particular, we decided to use the RF instead of the SVM-RBF for the DRT class as, despite the slightly higher closer mean test accuracy, we opted to use a simpler model with 44 features instead of a much more complex and only slightly better model with 99 features. Additionally, if we observe the fitted curves, we can see that the slope (that is, improvement provided by each new feature) is negligible in this ME class.
Looking at the Top 100 features generated by the SFS algorithm, we can see the reason behind this behavior. As shown in
Figure 10, for the DRT class, the selector was only able to find a few features at the beginning that were strictly better than the rest. Afterwards, the selector has chosen to directly include whole families of features instead of individual markers since there is not enough difference in relevance between them to decide on a particular one. For this reason, the training results start at an already high accuracy for this class and then remain almost unchanged (despite being nearly monotonic).
On the other hand, we see a similar behaviour in the case of the SRD, but for very different reasons. In this case, it is due to the fact that, since it is an easier DME type to identify than the other cases, a smaller number of features is enough for its detection and classification (despite the fact that all of them contribute sensibly to the variance of the dataset).
4. Discussion
The trained models returned satisfactory results, returning a mean test accuracy of 84.04% for CME, 78.44% for DRT and 95.40% for SRD, respectively. Additionally, the models tended to favor, in case of doubt, the positive class (a desirable behavior when a system is designed to be used in a medical environment). Moreover, given the robust nature of the generated visual representations, these false positives tend to be subverted in favor of the real, more clustered, detections thanks to the strategy designed to train the classifiers and sample the expert-labeled images.
Compared to other works based on a segmentation, pure precision metrics seem to be inferior. However, this work cannot be compared with other works of the state of the art as it offers a completely innovative way of analyzing these regions (precisely by being able to operate even in the fuzzy regions that these other methodologies cannot really return a proper output). Furthermore, our methodology is based on a classification of a high density sampling and voting paradigm, so even these metrics based on pure hit percentage statistics are in practice significantly enhanced by this exhaustive sampling by each region of the OCT images.
Figure 14 and
Figure 15 show representative examples from the test set with a healthy image and another one with different types of pathological fluid generated with the Spectralis and Cirrus devices, respectively. In addition, the labels of the expert are included to compare with the results. As can be seen, in both cases, the system is capable of satisfactorily detecting the pathological fluid accumulations whenever they are present, although we can see spurious detections in the cases of both
Figure 14a and
Figure 15a. However, these detections can be easily corrected with post-processing strategies as mentioned before, but we decided to include them for a better understanding of the behavior of the system.
In these visualizations, we can observe two major phenomena. The first one consists in the tendency to spurious detections in the images generated by the Spectralis of the SRD-type edema. This may be due to the higher contrast of the images generated by this device, favouring the appearance of common patterns of the SRD-type edema not present in the images that are generated with Cirrus. On the other hand, we can see a clear tendency in the Cirrus images to generate small detections of DRT-type edema. This is due to what was already mentioned in the section where we analyzed the selection of features: the DRT pattern of the images that are generated with the Cirrus device is very similar to the normal tissue in comparison with the "wet" and coarse texture of the DRT fluid accumulations in images that are generated with the Spectralis device.
In the images that were used for testing, we have found an example of each of these extreme cases, represented in
Figure 16. In this figure, we see the particular case of an Spectralis image with dark regions detected as SRD (
Figure 16a) and a particular case of a Cirrus image with slightly thickened layers that was detected as DRT (
Figure 16b).
As a final note, currently, the main limitation of the methodology lies in the dependence on a layer extraction algorithm. Since the vitreous humor has fluid patterns similar to those found in the different classes of DME, a small failure of the layer detector is very likely to lead to false positives (as shown in
Figure 16,
Figure 15 particularly around the DRT detections of the binary visualization or in the example with lower overlap between samples in
Figure 8). Nonetheless, these spurious detections can be compensated by adding, in the case of SRD, positional information to the feature library. A relative horizontal position descriptor would aid the trained model to better discriminate when the sample is not centered, a defining feature of this type of fluid accumulation. Additionally, in the rest of fluid accumulations and given the smaller nature of these misclassifications compared to the true detections, it can easily be fixed by increasing the overlap (which would make most of them disappear and shrink the persistent ones) and performing a morphological aperture with an small kernel (or by directly removing connected components with lesser than a given number of pixels). Also, the use of a predefined library of features slightly limits the sensitivity of the system to these particular cases (although it is compensated by the sampling algorithm overlap that enforces the same region to be studied multiple times). This is specially noticeable in the Cirrus device, as the device itself performs a post-processing to smooth the OCT images that slightly deteriorates the textures present in the image compared to its Spectralis counterpart (specially with DRT, confusing it sometimes with darker non-fluid tissues).
5. Conclusions
In this work, we used an alternative paradigm to the classical segmentation to find and characterize these three types of fluid accumulations, as well as including an extensive analysis of texture and intensity features to its detection. This paradigm, instead of aiming for a precise segmentation, performs a regional analysis. That is, classifies hundreds of samples of the image and infers the final result from the distribution of the detections. With these detections, five complementary visualizations are generated to aid the clinicians with the diagnosis: four complementary confidence maps and a binary one.
In the tests, we show how the main confidence map offer a visualization on the general confidence that the system has on the detections and the relationships between them (as the detections may or may not overlap). On the other hand, the three different confidence maps, one for each type of DME with a different color mapping, allow for a more precise analysis on the detections, but lacking the information about how the different fluid accumulation types may appear together. Finally, the binary map offers a direct interpolation of the results without any information on the confidence the system confers to them. This visualization allows the clinicians to see even the detections that the methodology deemed as false positives (low confidence detections) to allow for a more informed diagnosis in particularly complicated cases or clinical pictures. This way, the methodology is able to offer a representation of all the three types of ME in an intuitive way, robust to diffuse boundaries between regions and independent on the possibility of having a defined segmentation of the fluid accumulations.
Given these positive and satisfactory preliminary results, we plan to further expand our dataset; allowing us to improve our methodology with deep learning techniques and expand it to the 3D domain. Additionally, given the satisfactory viability and reliability the system has reported in the presented tests, in collaboration with clinicians, we will study its performance in a live clinical environment as well as the benefit derived from its application.