Virtual UV Fluorescence Microscopy from Hematoxylin and Eosin Staining of Liver Images Using Deep Learning Convolutional Neural Network
Abstract
Featured Application
Abstract
1. Introduction
1.1. Contribution of the Paper and Related Works
- Possibility to use a digital bright field microscope in place of a fluorescence microscope,
- Limiting the influence of photobleaching and photo damage in the slide microstructure,
- Analysis of the possibility of using deep learning convolutional neural networks to implement this type of conversion.
1.2. Content of the Paper
2. Materials and Methods
2.1. Liver Microscopic Images
2.2. Data and Acquisition System
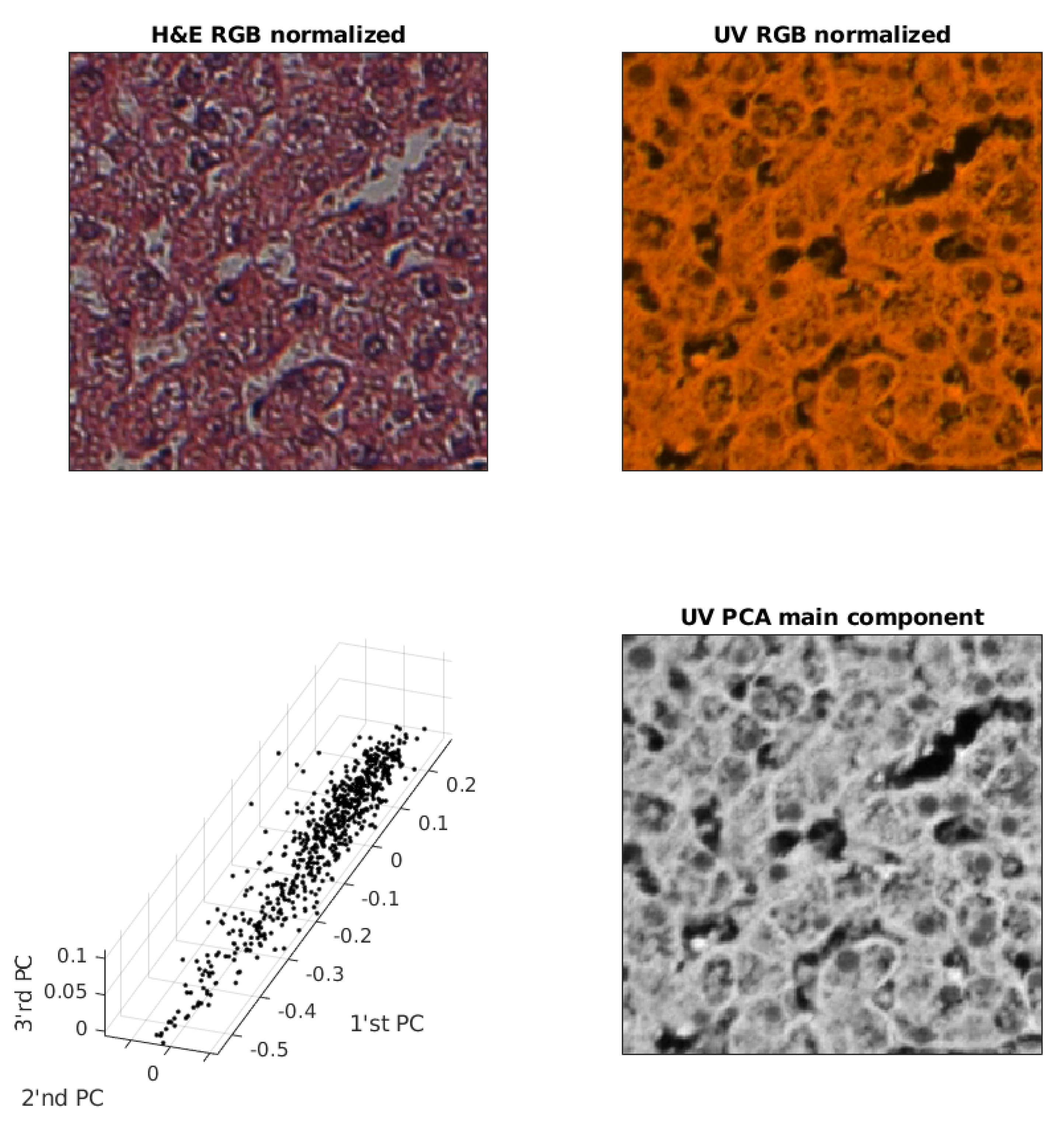
2.3. UV (RGB) Image Redundancy
2.4. Spatial Alignment of Pairs of Images
2.5. Contrast Normalization
2.6. Deep Learning Convolutional Neural Network (ConvNN)
2.7. Evaluation of Results
3. Results
3.1. Mechanical Shifts
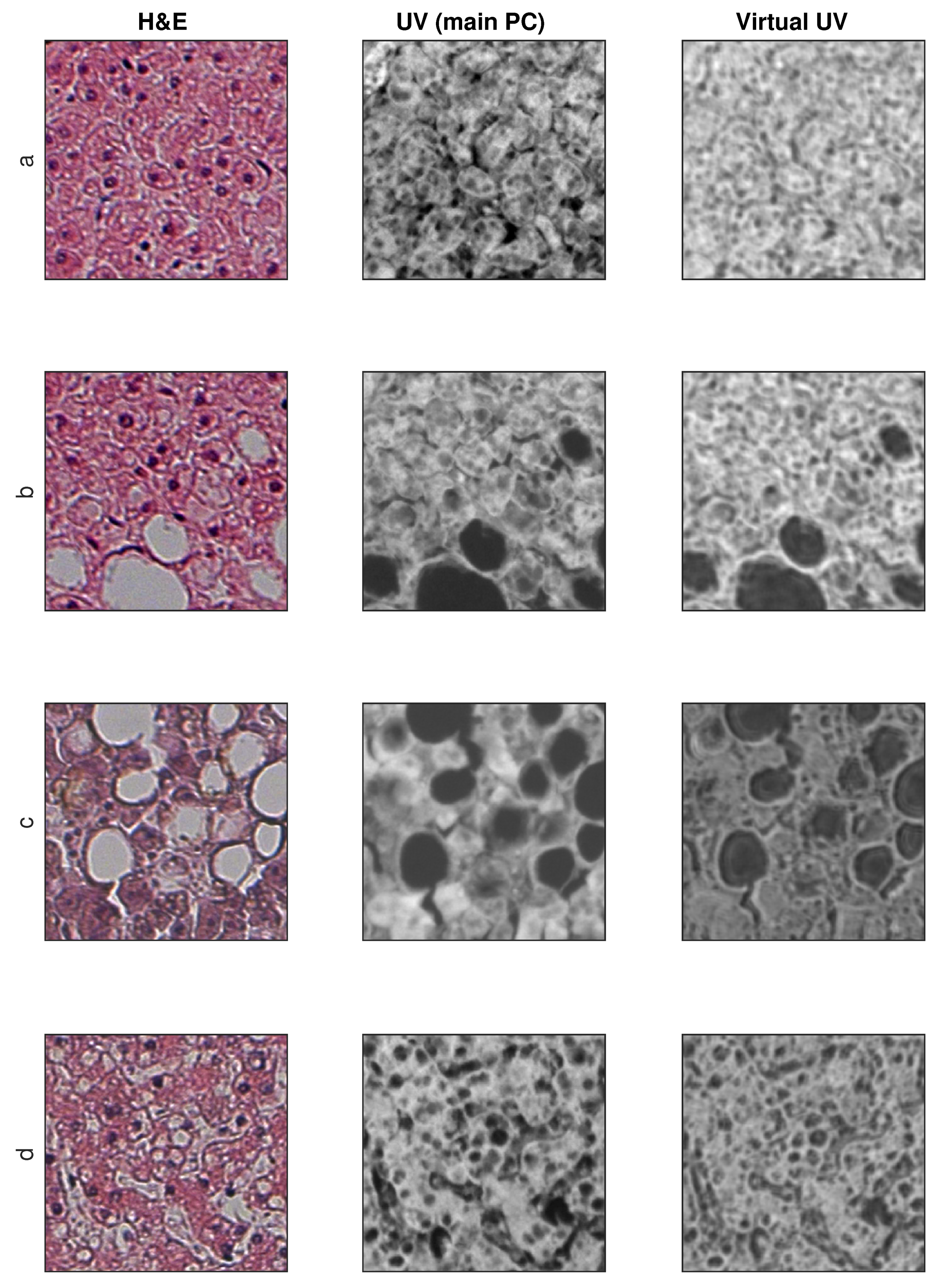
3.2. Exemplary Results
3.3. SSIM and SSIM (Structure Only) Metrics
4. Discussion
4.1. Discussion of Results
4.2. Discussion Related to Other Works
5. Conclusions and Further Work
Author Contributions
Funding
Conflicts of Interest
References
- Heintzmann, R. Introduction to Optics and Photophysics. In Fluorescence Microscopy; Wiley–VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2013; Chapter 1; pp. 1–31. [Google Scholar]
- Kubitscheck, U. Principles of Light Microscopy. In Fluorescence Microscopy; Wiley–VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2013; Chapter 2; pp. 33–95. [Google Scholar]
- Kiernan, J. Histological and Histochemical Methods: Theory and Practice; Cold Spring Harbor Laboratory Press: Banbury, UK, 2015; Volume 6. [Google Scholar]
- Ramsundar, B.; Eastman, P.; Walters, P.; Pande, V. Deep Learning for the Life Sciences; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2019. [Google Scholar]
- Doi, K. Computer-Aided Diagnosis in Medical Imaging: Historical Review, Current Status and Future Potential. Comput. Med. Imaging Graph. 2007, 31, 198–211. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Nishikawa, R.M. (Eds.) Computer-Aided Detection and Diagnosis in Medical Imaging; CRC Press: Boca Raton, FL, USA, 2015. [Google Scholar]
- Murphy, D.B.; Davidson, M.W. Fundamentals of Light Microscopy and Electronic Imaging, 2nd ed.; Wiley-Blackwell: New Jersey, NJ, USA, 2013. [Google Scholar]
- Hawkes, P.; Spence, J.C. Springer Handbook of Microscopy; Springer International Publishing: Cham, Switzerland, 2019. [Google Scholar]
- Dobrucki, J.W. Fluorescence Microscopy. In Fluorescence Microscopy; Wiley–VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2013; Chapter 3; pp. 97–142. [Google Scholar]
- Rodrigues, I.; Sanches, J. Photoblinking/photobleaching differential equation model for intensity decay of fluorescence microscopy images. In Proceedings of the 2010 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Rotterdam, The Netherlands, 14–17 April 2010; pp. 1265–1268. [Google Scholar]
- Ishikawa-Ankerhold, H.C.; Ankerhold, R.; Drummen, G.P. Advanced Fluorescence Microscopy Techniques–FRAP, FLIP, FLAP, FRET and FLIM. Molecules 2012, 17, 4047–4132. [Google Scholar] [CrossRef] [PubMed]
- Tosheva, K.L.; Yuan, Y.; Pereira, P.M.; Culley, S.; Henriques, R. Between life and death: Strategies to reduce phototoxicity in super-resolution microscopy. J. Phys. D Appl. Phys. 2020, 53, 163001. [Google Scholar] [CrossRef]
- Titford, M. Progress in the Development of Microscopical Techniques for Diagnostic Pathology. J. Histotechnol. 2009, 32, 9–19. [Google Scholar] [CrossRef]
- Bayramoglu, N.; Kaakinen, M.; Eklund, L.; Heikkilä, J. Towards Virtual H E Staining of Hyperspectral Lung Histology Images Using Conditional Generative Adversarial Networks. In Proceedings of the 2017 IEEE International Conference on Computer Vision Workshops (ICCVW), Venice, Italy, 22–27 October 2017; pp. 64–71. [Google Scholar]
- Rivenson, Y.; Wang, H.; Wei, Z.; de Haan, K.; Zhang, Y.; Wu, Y.; Günaydın, H.; Zuckerman, J.E.; Chong, T.; Sisk, A.E.; et al. Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning. Nat. Biomed. Eng. 2019, 3, 466–477. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Hui, H.; Zhang, Y.; Tong, W.; Tian, F.; Yang, X.; Liu, J.; Chen, Y.; Tian, J. Deep Learning for Virtual Histological Staining of Bright-Field Microscopic Images of Unlabeled Carotid Artery Tissue. Mol. Imaging Biol. 2020, 22, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; de Haan, K.; Rivenson, Y.; Li, J.; Delis, A.; Ozcan, A. Digital synthesis of histological stains using micro-structured and multiplexed virtual staining of label-free tissue. Light Sci. Appl. 2020, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Cooke, C.L.; Kong, F.; Chaware, A.; Zhou, K.C.; Kim, K.; Xu, R.; Ando, D.M.; Yang, S.J.; Konda, P.C.; Horstmeyer, R. Physics-enhanced machine learning for virtual fluorescence microscopy. arXiv 2020, arXiv:2004.04306. [Google Scholar]
- Vahadane, A.; Kumar, N.; Sethi, A. Learning based super-resolution of histological images. In Proceedings of the 13th IEEE International Symposium on Biomedical Imaging, ISBI 2016, Prague, Czech Republic, 13–16 April 2016; pp. 816–819. [Google Scholar]
- Umehara, K.; Ota, J.; Ishida, T. Application of Super-Resolution Convolutional Neural Network for Enhancing Image Resolution in Chest CT. J. Digit. Imaging 2018, 31, 441–450. [Google Scholar] [CrossRef]
- Huang, Y.; Chung, A.C.S. Improving High Resolution Histology Image Classification with Deep Spatial Fusion Network. In Computational Pathology and Ophthalmic Medical Image Analysis; Stoyanov, D., Taylor, Z., Ciompi, F., Xu, Y., Martel, A., Maier-Hein, L., Rajpoot, N., van der Laak, J., Veta, M., McKenna, S., et al., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 19–26. [Google Scholar]
- Mukherjee, L.; Bui, H.D.; Keikhosravi, A.; Loeffler, A.; Eliceiri, K.W. Super-resolution recurrent convolutional neural networks for learning with multi-resolution whole slide images. J. Biomed. Opt. 2019, 24, 1–15. [Google Scholar] [CrossRef]
- LeCun, Y.; Kavukcuoglu, K.; Farabet, C. Convolutional networks and applications in vision. In Proceedings of the 2010 IEEE International Symposium on Circuits and Systems, Paris, France, 30 May–2 June 2010; pp. 253–256. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning (Adaptive Computation and Machine Learning); MIT Press: Cambridge, MA, USA; London, UK, 2016. [Google Scholar]
- Kumar, V.; Abbas, A.; Aster, J. (Eds.) Robbins Basic Pathology, 10th ed.; Elsevier: Philadelphia, PA, USA, 2017. [Google Scholar]
- Chosia, M.; Domagala, W.; Urasinska, E. Atlas Histopatologii. Atlas of Histopathology; PZWL Wydawnictwo Lekarskie: Warszawa, Poland, 2006. [Google Scholar]
- Mescher, A.L. Junqueira’s Basic Histology: Text and Atlas, 15th ed.; McGraw-Hill Education: New York, NY, USA, 2018. [Google Scholar]
- Li, X.; Gunturk, B.; Zhang, L. Image demosaicing: A systematic survey. In Visual Communications and Image Processing 2008; Pearlman, W.A., Woods, J.W., Lu, L., Eds.; International Society for Optics and Photonics, SPIE: San Jose, CA, USA, 2008; Volume 6822, pp. 489–503. [Google Scholar]
- Linkert, M.; Rueden, C.T.; Allan, C.; Burel, J.M.; Moore, W.; Patterson, A.; Loranger, B.; Moore, J.; Neves, C.; Macdonald, D.; et al. Metadata matters: Access to image data in the real world. J. Cell Biol. 2010, 189, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Krzanowski, W.J. Principles of Multivariate Analysis: A User’s Perspective; Oxford University Press, Inc.: New York, NY, USA, 1988. [Google Scholar]
- Gavrilovic, M.; Azar, J.C.; Lindblad, J.; Wählby, C.; Bengtsson, E.; Busch, C.; Carlbom, I.B. Blind Color Decomposition of Histological Images. IEEE Trans. Med Imaging 2013, 32, 983–994. [Google Scholar] [CrossRef] [PubMed]
- Forczmański, P. On the Dimensionality of PCA Method and Color Space in Face Recognition. In Image Processing and Communications Challenges 4; Choraś, R.S., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 55–63. [Google Scholar]
- Kather, J.; Weis, C.A.; Marx, A.; Schuster, A.; Schad, L.; Zöllner, F. New Colors for Histology: Optimized Bivariate Color Maps Increase Perceptual Contrast in Histological Images. PLoS ONE 2015, 10, e0145572. [Google Scholar] [CrossRef]
- Reddy, B.S.; Chatterji, B.N. An FFT–Based Technique for Translation, Rotation, and Scale–Invariant Image Registration. Trans. Image Proc. 1996, 5, 1266–1271. [Google Scholar] [CrossRef]
- Sada, A.; Kinoshita, Y.; Shiota, S.; Kiya, H. Histogram-Based Image Pre-processing for Machine Learning. In Proceedings of the 2018 IEEE 7th Global Conference on Consumer Electronics (GCCE), Las Vegas, NV, USA, 9–12 October 2018; pp. 272–275. [Google Scholar]
- Everitt, B.S.; Hand, D.J. Finite Mixture Distributions; Chapman & Hall: London, UK, 1981. [Google Scholar]
- Robbins, H.; Monro, S. A Stochastic Approximation Method. Ann. Math. Stat. 1951, 22, 400–407. [Google Scholar] [CrossRef]
- Mei, S.; Montanari, A.; Nguyen, P.M. A mean field view of the landscape of two-layer neural networks. Proc. Natl. Acad. Sci. USA 2018, 115, E7665–E7671. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Gao, F.; Hou, W.; Hao, L. Objective image quality assessment: A survey. Int. J. Comput. Math. 2013, 91, 2374–2388. [Google Scholar] [CrossRef]
- Okarma, K. Current trends and advances in image quality assessment. Elektron. Elektrotechnika 2019, 25, 77–84. [Google Scholar] [CrossRef]
- Zhai, G.; Min, X. Perceptual image quality assessment: A survey. Sci. China Inf. Sci. 2020, 63, 211301. [Google Scholar] [CrossRef]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef] [PubMed]
- Metropolis, N. Monte Carlo Method. In From Cardinals to Chaos: Reflection on the Life and Legacy of Stanislaw Ulam; Book News, Inc.: Portland, OR, USA, 1989; p. 125. [Google Scholar]
- Kroese, D.P.; Brereton, T.; Taimre, T.; Botev, Z.I. Why the Monte Carlo method is so important today. Wiley Interdiscip. Rev. Comput. Stat. 2014, 6, 386–392. [Google Scholar] [CrossRef]
- Trentacoste, M.; Mantiuk, R.; Heidrich, W.; Dufrot, F. Unsharp Masking, Countershading and Halos: Enhancements or Artifacts? Comput. Graph. Forum 2012, 31, 555–564. [Google Scholar] [CrossRef]
- Jing, Y.; Yang, Y.; Feng, Z.; Ye, J.; Yu, Y.; Song, M. Neural Style Transfer: A Review. IEEE Trans. Vis. Comput. Graph. 2020, 26, 3365–3385. [Google Scholar] [CrossRef] [PubMed]
- Shelhamer, E.; Long, J.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 640–651. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef]
- Kumar, V.; Gu, Y.; Basu, S.; Berglund, A.; Eschrich, S.A.; Schabath, M.B.; Forster, K.; Aerts, H.J.W.L.; Dekker, A.; Fenstermacher, D.; et al. Radiomics: The process and the challenges. Magn. Reson. Imaging 2012, 30, 1234–1248. [Google Scholar] [CrossRef]








| No. | Name | Type | Activations | Learnables |
|---|---|---|---|---|
| 1 | imageinput | Image Input | - | |
| images | ||||
| 2 | conv_1 | Convolution | Weights | |
| 128 ] convolutions | Bias | |||
| with stride [1 1] and padding [0 0 0 0] | ||||
| 3 | relu_1 | ReLU | - | |
| 4 | conv_2 | Convolution | Weights | |
| 256 convolutions | Bias | |||
| with stride [1 1] and padding [0 0 0 0] | ||||
| 5 | relu_2 | ReLU | - | |
| 6 | conv_3 | Convolution | Weights | |
| 256 convolutions | Bias | |||
| with stride [1 1] and padding [0 0 0 0] | ||||
| 7 | relu_3 | ReLU | - | |
| 8 | conv_4 | Convolution | Weights | |
| 1 convolutions | Bias | |||
| with stride [1 1] and padding [0 0 0 0] | ||||
| 9 | regressionoutput | Regression | - | - |
| MSE | Output |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oszutowska-Mazurek, D.; Parafiniuk, M.; Mazurek, P. Virtual UV Fluorescence Microscopy from Hematoxylin and Eosin Staining of Liver Images Using Deep Learning Convolutional Neural Network. Appl. Sci. 2020, 10, 7815. https://doi.org/10.3390/app10217815
Oszutowska-Mazurek D, Parafiniuk M, Mazurek P. Virtual UV Fluorescence Microscopy from Hematoxylin and Eosin Staining of Liver Images Using Deep Learning Convolutional Neural Network. Applied Sciences. 2020; 10(21):7815. https://doi.org/10.3390/app10217815
Chicago/Turabian StyleOszutowska-Mazurek, Dorota, Miroslaw Parafiniuk, and Przemyslaw Mazurek. 2020. "Virtual UV Fluorescence Microscopy from Hematoxylin and Eosin Staining of Liver Images Using Deep Learning Convolutional Neural Network" Applied Sciences 10, no. 21: 7815. https://doi.org/10.3390/app10217815
APA StyleOszutowska-Mazurek, D., Parafiniuk, M., & Mazurek, P. (2020). Virtual UV Fluorescence Microscopy from Hematoxylin and Eosin Staining of Liver Images Using Deep Learning Convolutional Neural Network. Applied Sciences, 10(21), 7815. https://doi.org/10.3390/app10217815






