Abstract
Segmenting brain tumors accurately and reliably is an essential part of cancer diagnosis and treatment planning. Brain tumor segmentation of glioma patients is a challenging task because of the wide variety of tumor sizes, shapes, positions, scanning modalities, and scanner’s acquisition protocols. Many convolutional neural network (CNN) based methods have been proposed to solve the problem of brain tumor segmentation and achieved great success. However, most previous studies do not fully take into account multiscale tumors and often fail to segment small tumors, which may have a significant impact on finding early-stage cancers. This paper deals with the brain tumor segmentation of any sizes, but specially focuses on accurately identifying small tumors, thereby increasing the performance of the brain tumor segmentation of overall sizes. Instead of using heavyweight networks with multi-resolution or multiple kernel sizes, we propose a novel approach for better segmentation of small tumors by dilated convolution and multi-task learning. Dilated convolution is used for multiscale feature extraction, however it does not work well with very small tumor segmentation. For dealing with small-sized tumors, we try multi-task learning, where an auxiliary task of feature reconstruction is used to retain the features of small tumors. The experiment shows the effectiveness of segmenting small tumors with the proposed method. This paper contributes to the detection and segmentation of small tumors, which have seldom been considered before and the new development of hierarchical analysis using multi-task learning.
1. Introduction
Among cancers originally from the brain, glioma is one of the most common frequent [1]. Glioma arises from glial cells and can be separated into low-grade glioma and high-grade glioma. The high-grade gliomas are malignant and they often have a mean survival time of 15 months [2]. Low-grade gliomas may be malignant or benign. They develop slowly and can be progressed to high-grade gliomas. Currently, the most common treatment methods for gliomas are radiotherapy, chemotherapy, and surgery. Of all the brain tumor treatments, it is important for segmenting the tumors and their surrounding tissues accurately because it helps doctors to evaluate disease progression, treatment response, and therapy planning for glioma patients. Computed tomography (CT), Positron emission tomography (PET), and magnetic resonance imaging (MRI) are the most common ways of detecting and monitoring tumors. Among these ways, MRI is the first chosen method because it has a high resolution, superior contrast, and no harm to patients’ health. In the conventional method of clinical diagnosis and treatment, the doctor needs to navigate 3D images search and segment tumor areas manually, which is very boring, time-consuming, and requires high expertise. Due to those reasons, the automatic segmentation of brain tumors is critical in reducing the workload of doctors and has a significant impact on improving the diagnostic process, survival prediction, and treatment planning of patients with gliomas. Unlike some types of brain tumors, such as meningioma, which are easy to localize and segment, other tumors like glioblastomas and gliomas are much more troublesome to delineate. Glioma tumors and neighboring edema are dispersed, low-contrast, and appearing in many forms that make them hard for segmentation. Another essential trouble with locating brain tumors is that they can occur everywhere within brain areas, in nearly any shape and size. In addition, the scale and range of each voxel values in MRI is not uniform like other images obtained from CT scans or X-ray. Several factors affect image voxel values such as the MR machine type to be used and different acquisition protocols. Consequently, the same tumor cells may have significantly different grayscale values when being taken at different hospitals or institutes.
Recently, various methods and networks, particularly ones based on deep learning with convolutional neural network (CNN), have been developed to automatically identify and segment brain tumors in medical images. Most models proposed for brain tumor segmentation focus on increasing overall performance. Several studies have worked on the problem in detection and segmentation of small-sized tumors. The tumor size is a very important factor. If their size is big, the survival rate will be low. As a consequence, it is important to detect brain tumors when they are still small. However, it is very tough to detect small brain tumors with human eyes [3]. Devkota et al. [4] proposed a segmentation model for detecting early-stage of brain tumors. The model was developed to segment small, medium, and large size tumors. However, the model was built based on traditional methods and did not achieve high efficiency. Nasor et al. [3] proposed a network to segment brain tumors at their very early-stages. The network utilized k-means clustering, object counting, patch-based technique, and tumor evaluation. The network can segment the tumor area with various sizes, especially for very small-sized tumors. However, the method was evaluated and implemented on a small amount of data with only twenty brain MRI scans.
Most recent brain tumor segmentation methods using deep learning are based on U-Net [5]. However, U-Net architecture uses of the down-sampling layer to reduce feature resolution. The resolution plays a very important role in detecting and segmenting small objects. Although the U-Net uses the skip connection to restore the higher resolution features from the encoder part to decoder part, it often fails to keep the detailed information of the input or to retain the features of small objects [6]. Consequently, U-Net based models often miss out on small objects. Several deep learning methods have been proposed for dealing with the small objects problem. They are often based on multiresolution [7], multiple kernel sizes [8], or using dilated convolution [6] for capturing multiscale features. The multiresolution and multiple kernel sizes work well on 2D but they become very heavy and complex when dealing with 3D MRI data. The dilated convolution is good for capturing multiscale features without adding many weights. However, they are effective on rather large objects, not small objects, because of an enlarged receptive field. The dilated convolution is used in our network for multiscale tumor segmentation, however, we need an additional method for small tumors. In this paper, we try a new approach using multi-task learning with feature reconstruction, which is very effective for retaining the important features for small tumors [9].
Multi-task learning is a technique where the network optimizes two or more tasks at the same time. In addition to the main task, which provides training for the original purpose of the network, auxiliary tasks are used. The aim of the auxiliary task is to improve the model’s convergence or optimization for the benefit of the main task. Those tasks are usually related and share common features. Multi-task learning has been used very successfully in many applications of deep learning such as image classification [10], video captioning [11], voice enhancement [12], or medical image segmentation [13,14]. Based on the idea of an autoencoder, Zhang et al. [9] proposed U-module as another task for better initialization of the parameters of CNN for medical image classification. Zhang used CNN with U-module embedded in the early stage of the network, instead of putting it to the end of the network, for classification [9]. The U-module is added as an auxiliary task for feature reconstruction. It helps to better initialize parameters of the model and at the same time it retains the detailed features as much as possible. In this paper, we use multi-task learning with the auxiliary task of feature reconstruction to preserve detailed features for small tumors, which are often lost in down-sampled layers. We define small tumors as the group of tumors of size about less than 10% of the median. In the case of enhancing tumors, which is the main target of experiments, the median is about the size of 20,000 voxels, therefore the small tumors are defined to be of size less than 2000 voxels. The experiments are performed in the tumors of size less than 1000 voxels and less than 2000 voxels each. Specifically, we devised a new multi-task learning network based on U-Net, where U-module is embedded in the encoding stage of the network. Our network is simple and easy to run in medium hardware specification and has been successful in dealing with small tumors. To the best of our knowledge, this is the first time multi-task learning is used to detect and segment small objects. Even though there have been approaches [13,14] using multi-task learning for medical image segmentation, they have not been targeted for hierarchical analysis. The contribution of the paper can be summarized as follows:
- Most of the previous studies seldom considered the small-sized tumors, which have a significant impact on finding early-stage cancers. This paper focuses on solving the problem of accurately identifying multiscale tumors, especially very small-sized tumors in the brain.
- Instead of using heavyweight networks with multiresolution or multiple kernel sizes, we propose a novel approach for better segmentation of small tumors by dilated convolution and multi-task learning. Using multi-task learning for hierarchical analysis is relatively new and has never been tried before small-tumor detection or segmentation.
2. Related Work
Medical image analysis is the process of analyzing the images to solve practical clinical issues. Medical imaging analysis has helped radiologists and doctors to make diagnostic and treatment planning process more accurate [15]. Some computer-aided diagnosis (CADx) system relied on practical medical analysis as a vital factor and can directly affect the operation of clinical examination and treatment [16,17]. The purpose of medical imaging analysis is to extract useful features, which are helpful for improving clinical diagnosis. In CADx, machine learning has been playing a very important role in popular applications such as tumor segmentation, classification, nodule detection, image-guided therapies, medical image retrieval, and annotation [18,19,20]. In the past, traditional handcrafted features were used for machine learning. The task of selecting certain features and estimating them is complicated. Deep learning is commonly used for the purposes of medical image processing and analysis among the machine learning techniques because useful information and features are automatically learned by the neural network.
Segmentation in the medical image is a requirement for medical application such as brain segmentation [21,22], body organ segmentation [23,24,25], and cardiac ventricle segmentation [26]. The accurate segmentation results are useful in the diagnosis and treatment planning. The relevant information extracted in medical images using the segmentation methods includes shape, length, volume, relative organ location, and abnormalities [27]. An iterative 3D multiscale Otsu threshold method is introduced in [28] for the segmentation of the medical image. The impacts of noise and weak edges are reduced by multilevel image representation. A hybrid methodology for the automatic segmentation of ultrasound images is proposed in [29]. The proposed approach combines knowledge from Fuzzy clustering of spatial constraint-based kernels with the edge-based features of the regularized level set. An experiment was applied to test the method on both real images and synthetically generated images.
Brain tumor segmentation has attracted much attention and many methods have been proposed. In the early works, brain tumor segmentation methods were often based on thresholds, edges, regions, atlases, classifications, and clustering methods [30]. These methods usually do not consume a lot of computational resources, but they do not achieve high performance and require user interaction. The multimodal brain tumor segmentation challenge (BRATS) was introduced for the first time in 2012. Many methods have been proposed for the task of segmenting brain tumors. Prastawa et al. [31] proposed a method that compared the differences between the atlas of the normal healthy brain with the patient scan for brain tumor segmentation. One drawback of the method is that it excludes the tumor-induced mass effect caused by the deformation of adjacent healthy structures, which may cause incorrect registration. Some researchers propose a method, which uses tumor growth models to modify the healthy atlas and performs both registration and segmentation to a new brain atlas [32,33]. These approaches can get the benefit of taking tumor characteristics into account. However, tumor growth models are followed by further complexities. Cordier et al. [34] have proposed a multi-atlas approach that relies on a searching algorithm for a similar patch of images. Numerous other active-contour approaches were proposed [35,36]. All of these methods rely on the symmetry and/or alignment features of the left-right brain. Since it is a challenging task to align a brain with a large tumor onto a template, some methods complete the registration task and the tumor segmentation task at the same time. Some voxel-wise classifiers based on segmentation models such as random forests [37,38] or SVM [39,40] obtained promising results. For example, Geremia et al. [41] suggested a method that used information from neighboring voxel and far-off region, including the symmetrical section from the brain for classifying each voxel in magnetic resonance (MR) images.
Recently, many researchers have applied convolution neural networks (CNNs) to segmenting brain tumors with MRI scans and have obtained very good results. The directions of the study can be divided into three categories. The first group used the 2D convolution in deep learning to make training faster but failed to learn 3D structures, which are one of the most important features in the medical image. The second group used 3D convolution to exploit the 3D features and they often got higher segmentation performance compared with 2D approaches but they require a high computational cost. The third group combined the 2D together with 3D to take advantage of the compactness of the 2D model and the volumetric information in 3D models. Ronneberger et al. [5] proposed U-Net, a fully convolution network with the encoder–decoder architecture. The U-Net is designed to train in an end-to-end manner to create a full resolution segmented mask. Despite this 2D-based approach method ignores the most important 3D features in the medical images, they have accomplished remarkable performance in brain tumor segmentation. Cicek et al. [42] upgraded the original U-Net to 3D U-Net by leveraging 3D operations such as 3D convolution layers and 3D max-pooling layers. Chen et al. [43] proposed a network exploring the group convolution and dilated convolution that helps to not only reduce computing costs but also have good performance for segmentation of brain tumors.
In brain tumors, the tumor substructures have a stratified structure. In particular, the enhanced tumor is found inside the tumor core and the tumor core is included in the whole tumor area. Many methods have utilized this property to increase the efficiency of brain tumor segmentation. Malmi et al. [44] and Pereira et al. [45] introduced a two-stages cascaded approach for brain tumor segmentation. At the first stage, the whole tumor was segmented and the tumor core and enhancing tumor were segmented in the second stage. Wang et al. [46] presented a cascade of three CNN networks to segment whole tumor, tumor core and enhancing tumor, respectively. Although the above method achieved high efficiency, they had to build three almost identical models. It causes the redundancy of the model, complexity in training and ignoring the correlation among these models. In addition, training several models separately makes the quality of the latter models significantly affected by the former models. Liu et al. [47] proposed an end-to-end network, called CU-Net, which exploited the hierarchical structure from brain tumors. However, the network was built based on 2D CNN and processed input MR images on every slice. It leads to the loss of three-dimensional space information, which is very important in medical imaging. Chen et al. [43] utilized an existing parcellation to bring the brain’s location information into patch-based neural networks that improve networks’ brain tumor segmentation performance. Brugger et al. [48] proposed a partially reversible U-Net architecture. The reversible architecture can recover each layer’s outputs from the subsequent layer’s ones, eliminating the need to store activations for backpropagation. Fabian et al. [49] introduced an instantiation of the 3D U-Net [42] with minor modifications. They used patches of size 128 × 128 × 128 with a batch size of two for training, reducing the number of filters, reduced the number of filters right before upsampling, and used instance normalization instead of batch normalization for having more consistent results. Nuechterlein et al. proposed 3D-ESPNet [50], which is extended from ESPNet, a fast and efficient network based on point-wise convolution for 2D semantic segmentation to 3D medical image data. Kao et al. [51] introduced a methodology to integrate human brain connectomics and parcellation for brain tumor segmentation and survival prediction. Wei et al. [52] designed a new separable 3D convolution by dividing each 3D convolution into three branches in a parallel fashion, each with a different orthogonal view, namely axial, sagittal and coronal. They also proposed a separable 3D block that takes advantage of the state-of-the art residual inception architecture. However, these networks are not for the hierarchical analysis nor targeted for small tumors.
Multi-task learning has been successful in many applications of deep learning including the brain tumor segmentation. Mlynarski et al. [53] exploited multi-task learning for brain tumor segmentation with an auxiliary task of classification. Their network proved the effectiveness of the multi-task learning on the brain tumor segmentation. Myronenko [54] proposed a U-Net based model with an additional variational autoencoder branch for reconstructing the input images. The image reconstruction task is jointly trained with the segmentation task. Although this architecture worked well for segmentation in general, their model requires a large memory and too heavy computation cost.
U-Net is a very successful network in biomedical image segmentation. It is the first choice when dealing with medical imaging segmentation. However, U-Net gradually reduces the feature resolution through down-sampling layers. This process leads to the poor-segmentation on small-sized objects. There are several methods that have been proposed to address small object problems. Kampffmeyer et al. [55] applied a patch-based and pixel-to-pixel based method for improving the segmentation performance on small objects in urban remote sensing images. Shareef et al. [8] proposed a small tumor-aware network with multiple kernel sizes for accurate segmentation of small tumors in breast ultrasound images. These methods work well on 2D images but may not operate efficiently on MR images, where the data is in the 3D form.
In this paper, we propose a network that focuses on small-sized tumors in the brain tumor segmentation problem. Our network uses multi-task learning for retaining important features of small tumors, which is often lost during the downsampling process of original U-Net. The experimental results show that the auxiliary task in our model is helpful for the small tumor segmentation task.
3. Materials and Method
In this part, we will first present the concepts of the baseline model and U-Module [9]. Secondary, we will show the architecture of our network. Finally, the strategy to train our network is demonstrated.
3.1. Baseline Network
Our model is based on a variant of U-Net called the dilated multifiber network [43] (DMFNet). We used DMFNet because it consumes a small amount of computation cost and memory, suitable for most GPUs while maintaining good performance. The DMFNet is based on the U-Net with the encoder–decoder network architecture. Firstly, a residual unit with two regular convolution layers is called a fiber [43]. The multifiber design consists of multiple separated residual units, or fibers. The multifiber (MF) unit takes advantage of a multiplexer for information routing and the dilated multifiber (DMF) unit with an adaptive weighting scheme for different dilation rates.
The network exploring the 3D multifiber unit and 3D dilated convolution that helps to reduce computing costs while still having good performance for segmentation of brain tumors. The DMFNet use MF and dilated DMF unit as the basic blocks. In the encoder part, six DMF units extract the multiscale features by using the different kernel sizes in the dilated convolution layers. In the decoder part, the extracted features are upsampled and concatenated with the high-resolution feature map from the encoder part. The Rectified Linear Unit (RELU) functions and batch normalization are conducted prior to every convolution operation of DMF and MF units. Channel grouping is applied to separate the convolution channel into many groups of convolutions. The technique significantly reduces the model parameters between the feature maps.
3.2. U-Module as an Auxiliary Task
Our model is inspired by autoencoder architecture and the U-module [9]. Autoencoder is one type of neural network where the output has the same dimensionality as the input. It is often used to compress data into a latent space presentation or dimension reduction using unsupervised learning methods. In the autoencoder, the encoder learns efficient data codings and preserves as much of the relevant information as possible. The decoder takes the encoding and reconstructs it into a full image. The U-module is based on an autoencoder with the purpose of better parameter initialization of CNNs for medical image classification. The U-module is proved to retain the feature in the next layers of CNNs. In our paper, besides the main task of segmentation, we add U-module to our model as an auxiliary task, which helps to force the model to preserve as much of the relevant and important information as possible.
3.3. Proposed Network Architecture
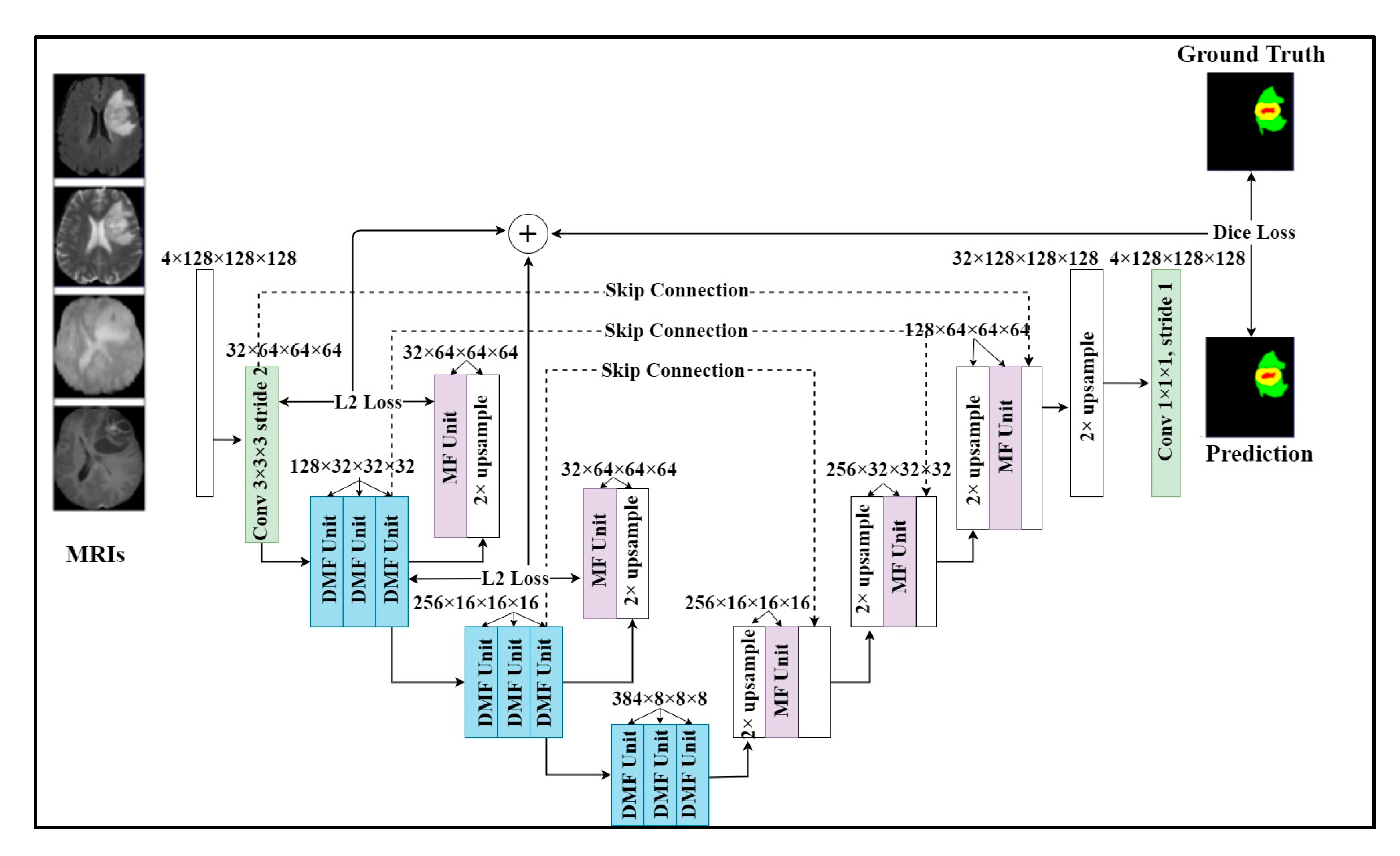
Our proposed method is based on two observations. Firstly, the size of brain tumors is various. Secondly, the U-Net cannot detect the tumor with a small size, especially the enhancing tumors, which have the smallest size. Some of the previous methods, they just focus on improving the performance.
There are still some unsolved issues including missing tumors and poor detection/segmentation performance on small-sized tumors. For small object segmentation, context is one of the most important elements [6]. In a typical U-Net, the down-sampling layers are useful for feature extraction and extending receptive fields. However, they lose an important element, which is the resolution. Through downsampling layers, the feature resolution is gradually reduced. As a result, the features of small objects can be lost. Once those features are lost, they are difficult to recover in spite of the effort of skip connection [5,6]. Therefore, we need a method to be able to retain the features of small objects, even if the resolution is reduced. The architecture of our model is shown in Figure 1. Our model employed a multi-task learning with two tasks. The main task was brain tumor segmentation and additional feature reconstruction was used for an auxiliary task. The model inputted four MRI modalities and outputted a segmented mask of regions including whole tumor, tumor core, and enhancing tumor. In particular, we added an upsampling layer and an MF unit for reconstructing features after encoder layers. By applying this technique, important features were retained in the following layers and reduced the side effects of downsampling layers on small tumors. As shown in Figure 1, U-module was formed by 3 DMF units, one MR unit, and an upsampling layer. The first DMF unit will reduce the size of the feature map. After two DMF units, the feature map resolution was the same and we added an upsampling layer and an MF unit to upsample the feature map to create a reconstructed feature map with the same size as the previous feature map. In detail, the operation of the U-module is described as the following formula:
where the and transitions present the encoder and decoder of the U-module. The encoders compress the original feature map F into a smaller feature map Z. The decoder using the upsampling layer and MF unit to recover the original feature map. The difference between the reconstructed map and the features map is minimized so the small feature map can represent the large feature map. This way, the most important and relevant features of the previous layer will be retained in the next layer. In our model, we used two U-modules at the first and second encoder layers because those layers had the largest feature resolution to be kept. In addition to helping our model retain the properties of small tumors, the U-module can also improve parameter initialization as its original purpose.
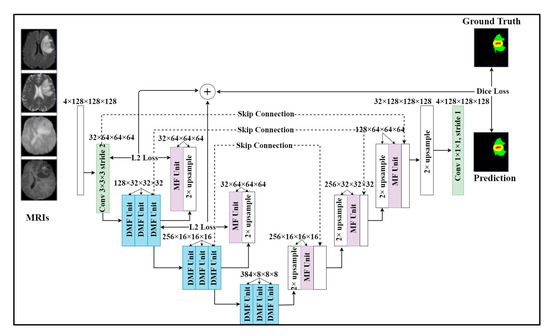
Figure 1.
Our proposed multi-task learning network with two tasks. One task is brain tumor segmentation. We add the additional feature reconstruction task to our model as an auxiliary task to help retain important features when downsampling. The model takes a sequence of MRI modalities as input and generates the brain tumor mask.
3.4. Training Strategy
In this paper, we used two loss functions for the segmentation task and feature reconstruction task separately. The first loss function is the generalized dice loss (GDL) [56]. Most of the previous methods use dice loss to calculate the difference between the predicted mask and ground truth. However, the distribution of the number of voxels in the tumor regions varied greatly in the brain tumor structure. Therefore, we take advantage of the GDL to solve the imbalanced class problem in the labels of subtumors. The GDL is calculated by the following formula:
where the and are the ground truth tumor mask and the predicted tumor mask of class c, respectively. The class weight was used to provide invariance to the different label set properties.
We used mean squared error to measure the difference between the reconstructed feature map and the input feature map of each U-module. The loss for the feature reconstruction task was calculated by the following formula:
where, the and are the reconstruction losses of the first and second U-module block. is the input feature of U-module and is the reconstructed feature map.
There were two tasks in our model. The first one is the tumor segmentation task and the second one is feature reconstruction. Therefore, we had two loss functions for optimization. We need to combine two loss functions together. The combined loss function is the sum of the loss functions of the main task along with the auxiliary task as shown in the following:
where the is the loss function for our main task (tumor segmentation) and the is the loss function for our auxiliary task (feature reconstruction). The and are the weight parameters. Since each loss function has its optimization direction and the range value of each loss function is different, we need to set up a training strategy for getting the best optimization. At the first 20 epochs, we set the and equal 0.5 for better parameter initialization. After, we set the main task weight the = 1 and auxiliary task weight = 0.05 so the network can focus on the main task.
The reconstruction task was used to retain important features and help the network converge better in the training phase. During the inference phase, the reconstruction task was removed. We only used the main task to generate segmented tumor masks.
4. Experimental Results
In this session, we presented the experimental details of our work. Firstly, we will show the dataset that we used in our paper. Second, we will mention data preprocessing, implementation details, and evaluation metrics. Finally, we will present our results.
4.1. Dataset
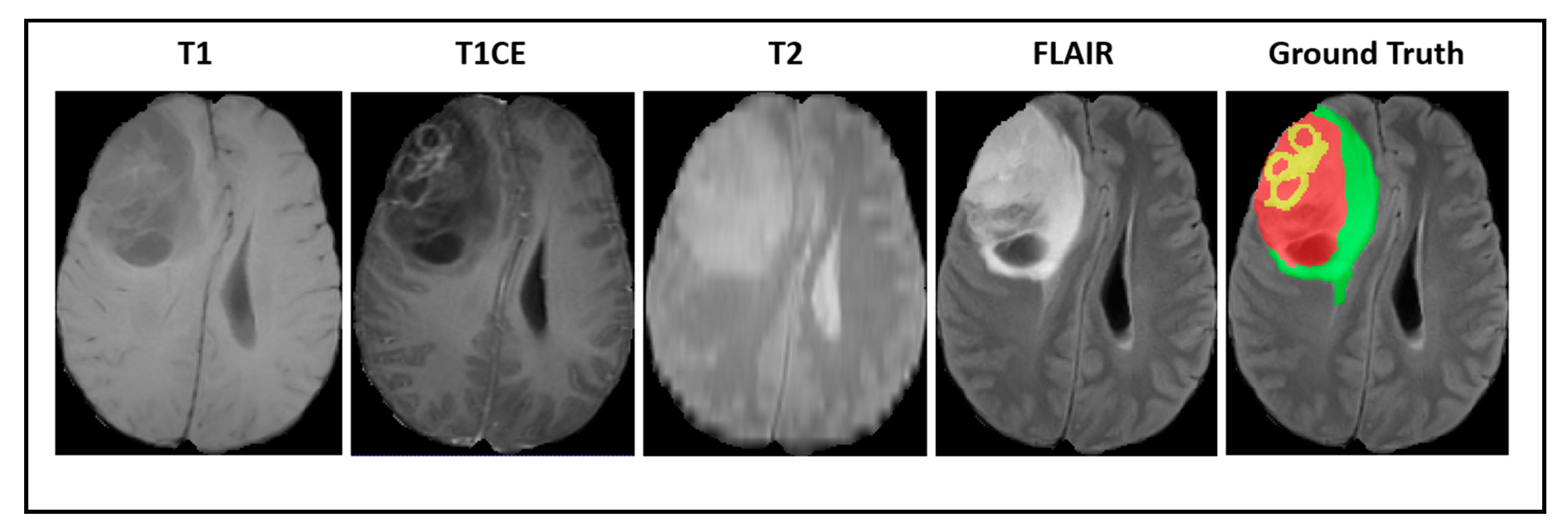
Our experiments were implemented based on the imaging data provided by the Brain Tumor Segmentation Challenge (BraTS2018) [57,58,59]. All of the medical images were provided in 3D volumes with four kinds of MR sequences called native T1weighted (T1), post-contrast T1-weighted (T1CE), T2-weighted (T2), and fluid attenuated inversion recovery (FLAIR). Multimodal scans were acquired from 19 institutions with various clinical protocols and different scanners. Figure 2 shows a visualization of BraTS2018 datasets with tumor masks on each MRI modality.
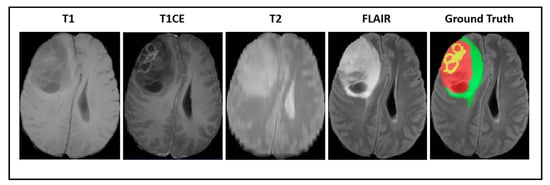
Figure 2.
A visualization of Brain Tumor Segmentation Challenge (BraTS) datasets with a tumor mask where green illustrates the edema, yellow illustrates the enhancing tumor, and red illustrates the necrotic area.
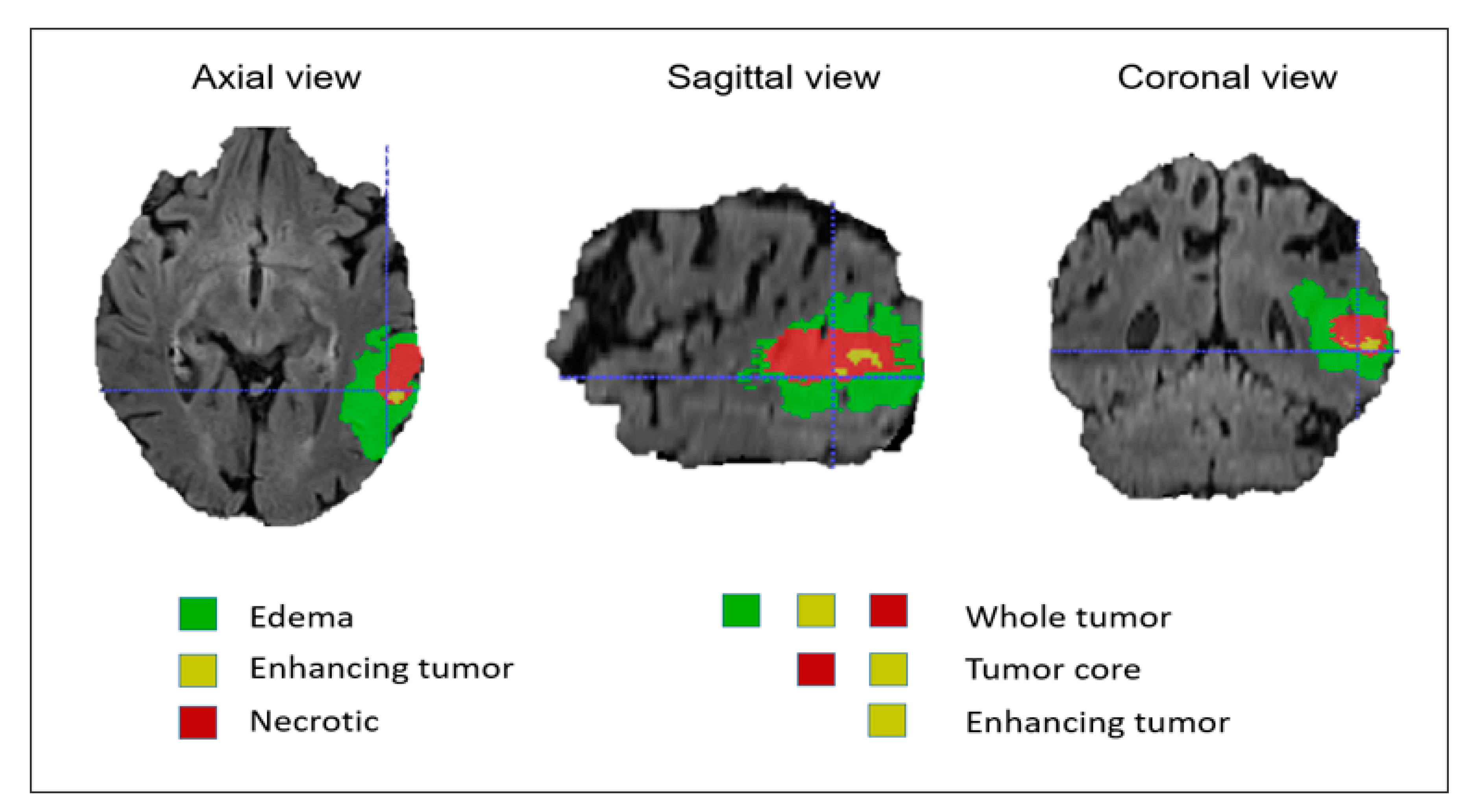
In this dataset, all modalities were coregistered with the same anatomical template and sampled with a dimension of 1 mm × 1 mm × 1 mm. The MR images of all patients were manually segmented by four experts and verified and approved by experienced neuroradiologists. These labels were annotated based on the intratumoral structures in gliomas. The segmentation results were evaluated based on the tumor subregions, including the whole tumor, the tumor core, and the enhancing tumor as shown in Figure 3. The evaluation was done by uploading the segmented files to the online evaluation system CBICA Image Processing Portal (https://ipp.cbica.upenn.edu). The dataset included 285 patients for the training process and 66 patients for validation. The training datasets can be downloaded from the contest’s official website (http://braintumorsegmentation.org).
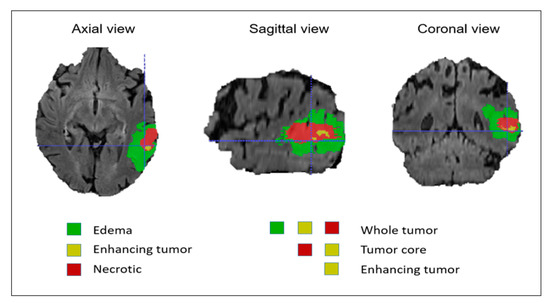
Figure 3.
Brain tumor mask based on intratumoral structures and tumor subregions.
4.2. Data Preprocessing
The preprocessing is very important for the computer vision. In this paper, we used several preprocessing methods including cropping, normalization, and augmentation. The details of data processing technique are described as follows:
Crop images. All the brain MR images are randomly sampled with the size of 128 × 128 × 128 to fit with the CPU memory. This technique still keeps most of the image content within the crop area and does not affect the datasets, but it will reduce the image size and the computational complexity.
Data normalization. In medical images, MRI scans are usually obtained from different scanners and different acquisition protocols. Therefore, MRI intensity value normalization is crucial for compensating heterogeneity between images. All input images are normalized to have a zero mean and unit variance. We also take the threshold of 5–95% intensity of each voxel in the MR image.
Data augmentation. In deep learning, data augmentation techniques are very commonly used techniques for increasing the amount of sample without adding new data. They help deep neural networks enhance generalization. First, they are capable of generating more data from a limited set of data. This is useful for data sets where for the collection of new data, labeling is very hard and time-consuming as for medical imaging. The second is that they help avoid overfitting. For deep learning networks, one problem that can be encountered is that the network can remember large amounts of data, resulting in overfitting. Instead of just learning the key concepts of data, the model memorizes all the inputs, which causes decreasing model efficiency. In this paper, we used some data augmentation methods such as rotation and flipping and scaling with the range of 0–10 degrees, 0–0.5, and 0.9–1.1, respectively.
4.3. Implementation Details
In our proposed network, the size of the training input was 128 × 128 × 128 volume, while the original image size was 240 × 240 × 155. It would be an omission if we only randomly sampled a portion of the original data during the testing process. In addition, we zero pad the original MRI scans with a size of 240 × 240 × 155 to transform the input to the size 240 × 240 × 160 because the depth needs to be dividable by 16. We developed the proposed model on the Pytorch [60] platform with python programming language. We used NVIDIA GeForce RTX 2080Ti GPU with 11 GB of memory.
4.4. Evaluation Metrics
The model segmentation results were evaluated by the dice similarity coefficient (DSC). The DSC is one of the commonly used evaluation metrics in segmentation models. They have a value between 0 and 1. The metric indicates the overlap level between two objects. A value of 0 means there is no overlap and a value of 1 means the two objects completely overlap each other. The formula for calculating DSC is expressed in the equation:
In which, and are the predicted segmentation and ground truth, respectively. denotes the number of true positive, denotes the number of false positive, and denotes the number of false negative segmentation.
4.5. Results and Analysis
4.5.1. Small Tumor Segmentation Performance
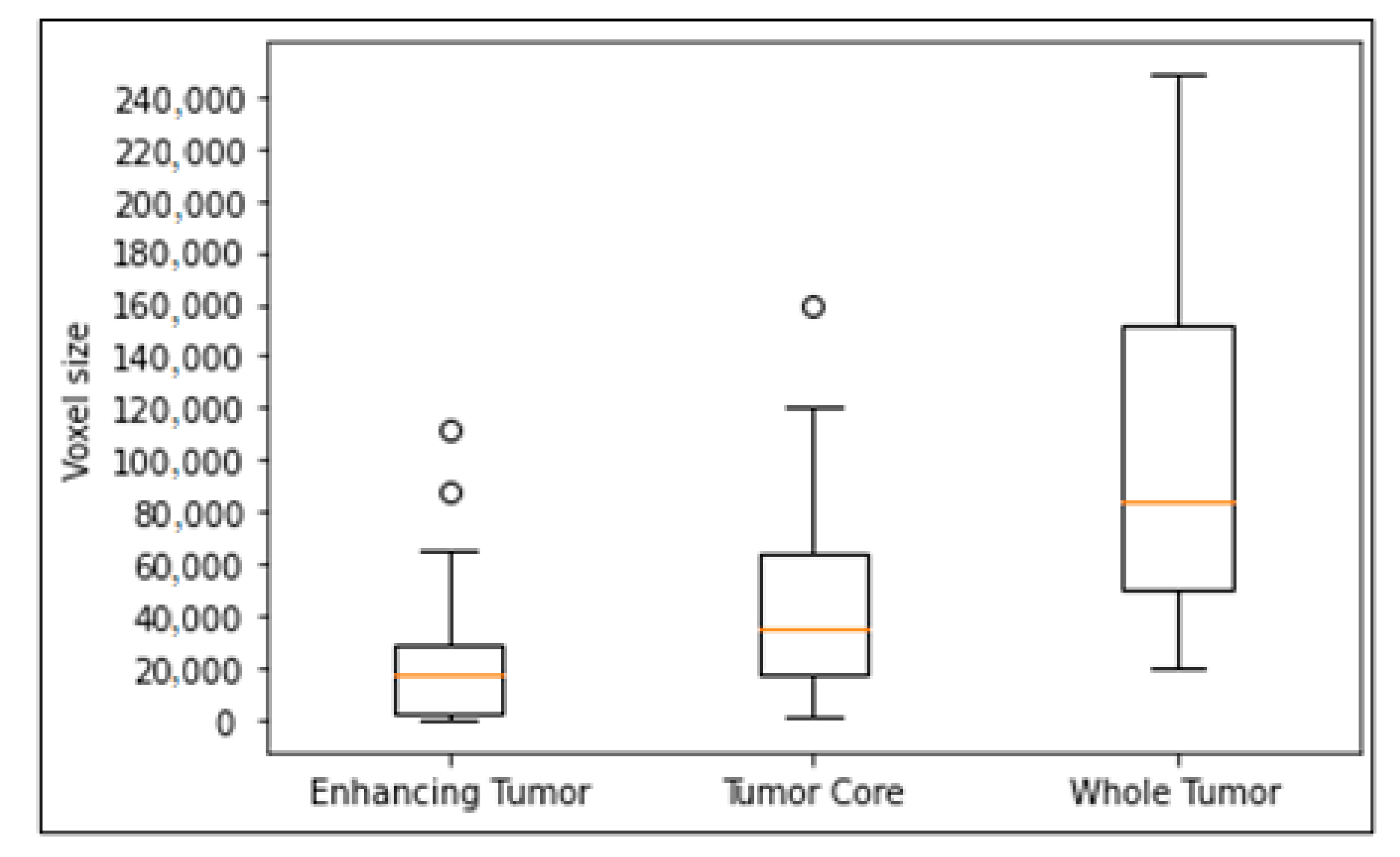
We evaluated the effectiveness of the proposed model with other models on small-sized tumors. We randomly split training into 80% for training and 20% for evaluation. We trained all models on the training set and get the best results of each model on the evaluation set. As shown in Figure 4, the enhancing tumor is a tumor type with the smallest size in brain. Therefore, we proved our effectiveness on small-sized tumors by comparing the segmentation results of our models with other models on small enhancing tumors. The criteria for selecting small-sized tumors are the volume size of tumors in voxels, and thresholds of the volume are selected as 2000 voxels and 1000 voxels respectively.
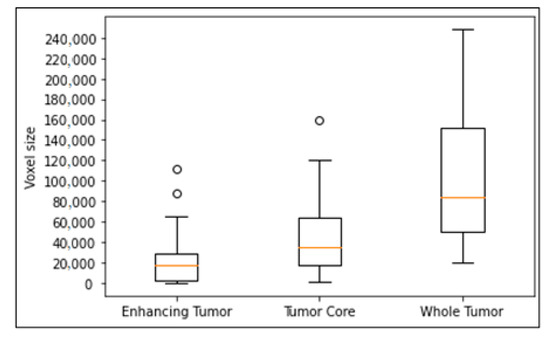
Figure 4.
Box plot of tumor volume with different tumor subregions. Outliers in the circle, the box with 25–75% with the orange line of median, and dash bars indicating 0–100% except outliers.
As shown in Table 1 and Table 2, our model performed better with enhancing tumors smaller than 2000 voxels. Other models such as DMFNet [43], ReversibleUnet [48], and No New-Net [49] often overlook tumors of a very small size. However, our method can detect and segment them.

Table 1.
The performance comparison of small tumors (tumor volume <2000 voxels).

Table 2.
The performance comparison of small tumors (tumor volume <1000 voxels).
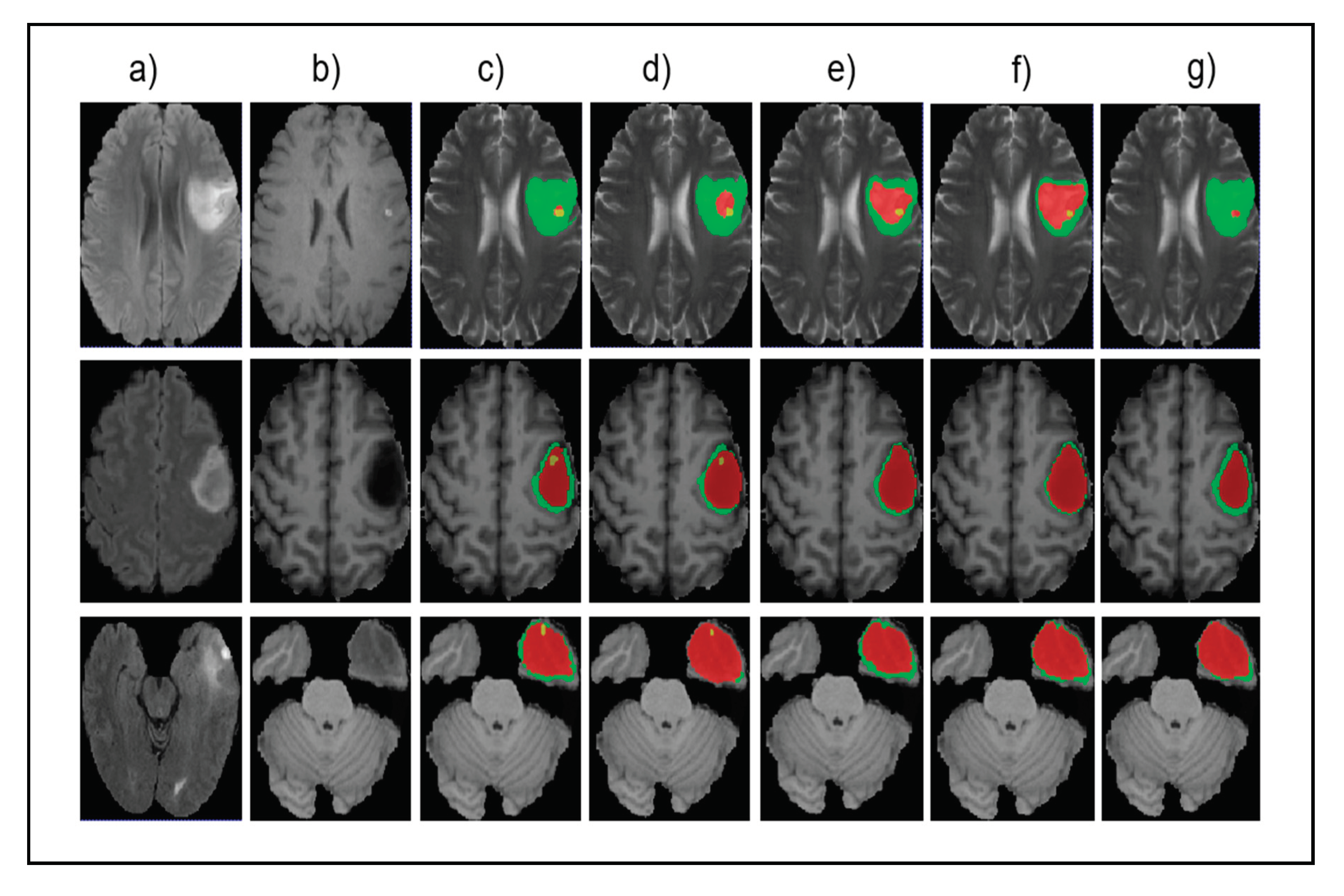
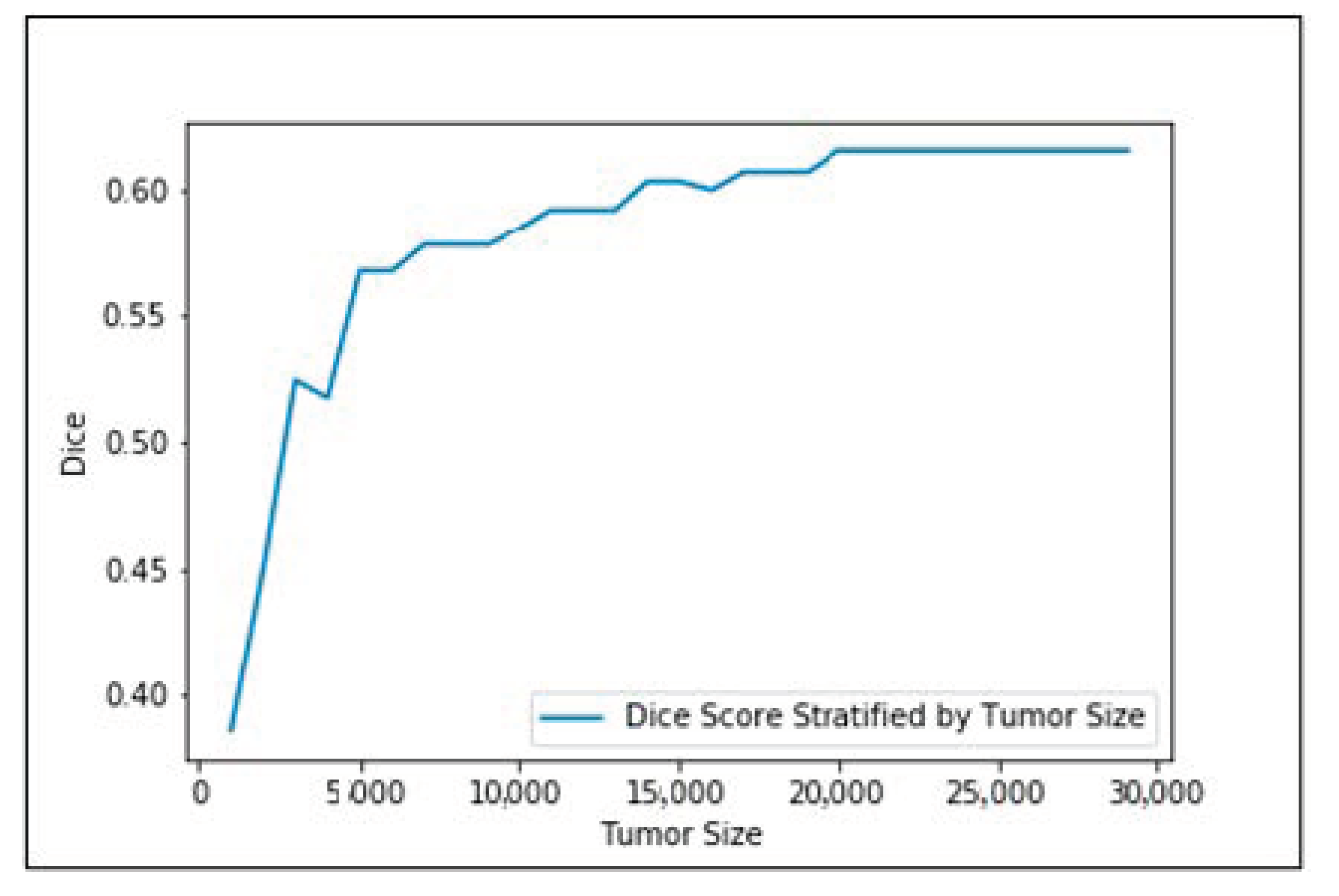
Figure 5 shows the input images, ground truth, and segmentation results of the enhancing tumor predicted by four models including our model, ReversibleUnet [48], No New-Net [49], and DMFNet [43]. As shown in the first row of the figure, ReversibleUnet and No New-Net predicted high false negatives on the enhancing tumor (yellow color), DMFNet failed to segment the enhancing tumor area while our model can accurately segment them. In the second and third rows of Figure 5, we can see that our model could segment the area of small enhancing tumor while other models failed to segment them. Figure 6 shows the dice similarity coefficient (DSC) stratified by tumor size.
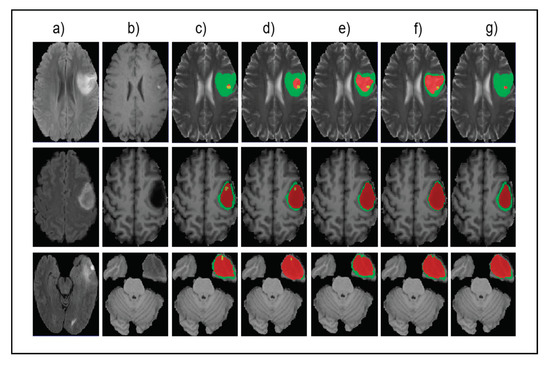
Figure 5.
Qualitative comparison of different models for small tumor segmentation. (a) Input fluid attenuated inversion recovery (FLAIR) image. (b) Input T1 image and (c) ground truth images. (d) Results of our method. (e) Results of ReversibleUnet [48]. (f) Results of No New-Net [49]. (g) Results of DMFNet [43].
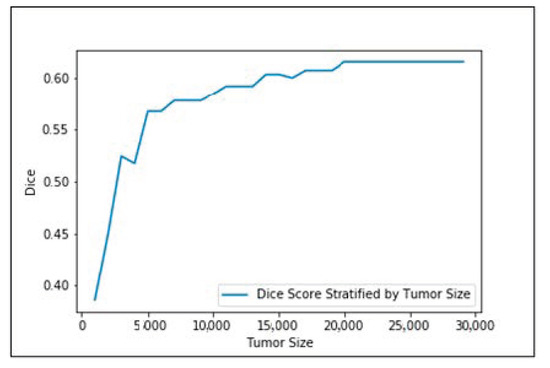
Figure 6.
Dice similarity coefficient (DSC) stratified by tumor size.
4.5.2. Overall Segmentation Performance
We performed the training stage on the BraTS2018 training set and then evaluated the segmentation results on the validation set. The segmentation performances of our proposed model were 81.82%, 89.75%, and 84.05% for the enhancing tumor, tumor core, and whole tumor, respectively. We also set up a table comparing our performance with other segmentation methods such as 3D U-Net [42], 3D-ESPNet [50], Kao et al. [51], DMFNet [43], Reversible U-Net [48], or comparing with winners of the competition such as No New-Net [49] and NVDLMED [54] (single model). As shown in Table 3, our method achieved the best performance on enhancing tumor segmentation compared to other models. Enhanced tumors are directly related to the early development of brain cancer and they are the most important one in early diagnosis of cancers. However, in most cases, the number of small tumors in the dataset could be relatively small. Therefore, the performance improvement of small tumors may not contribute much to the overall performance in some cases. For the whole tumor and the tumor core, our model achieved comparable results. Note that the No New-Net ranked second in the BraTS2018 challenge and its performance on the whole tumor is better than others. However, it is worth knowing that they trained their models with external datasets from other institutes. The NVDLMED [54] method has a very complex model, which takes up almost 32 GB.

Table 3.
The performance comparison of the dice score of different models on the BraTS2018 validation set.
5. Conclusions
In this paper, we built a 3D model for multi-task brain tumor segmentation in MR images. In our model, we leveraged the multi-task learning with U-module to make the tumor segmentation model better for small tumor size. The U-module helped retain features of small-sized tumors, which are easily overlooked because of decreasing feature resolution after each encoder layer of U-Net based models. We proved the efficiency of our proposed brain tumor segmentation algorithm on BraTS2018 dataset. During training, we did not use any external dataset. Our model outperformed other models in terms of small-sized tumor segmentation. The model also had comparable results with other state-of-the-art models on overall performance.
Author Contributions
Conceptualization, G.-S.L. and D.-K.N.; Methodology, D.-K.N.; Writing—review and editing, D.-K.N., M.-T.T. and G.-S.L.; Supervision, G.-S.L., S.-H.K. and H.-J.Y.; Project administration, G.-S.L., S.-H.K. and H.-J.Y.; Funding acquisition, G.-S.L., S.-H.K. and H.-J.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Bio and Medical Technology Development Program of the National Research Foundation (NRF) and funded by the Korean government (MSIT) (NRF-2019M3E5D1A02067961) and a grant (HCRI 19 001-1*H2019-0295) Chonnam National University Hwasun Hospital Institute for Biomedical Science and National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (NRF-2020R1A4A1019191). The corresponding author is Guee-Sang Lee.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bauer, S.; Wiest, R.; Nolte, L.-P.; Reyes, M. A Survey of MRI- Based Medical Image Analysis for Brain Tumor Studies. Phys. Med. Biol. 2013, 58, R97–R129. [Google Scholar] [CrossRef] [PubMed]
- Linda, D.; Neil, K.A.; Jan, J.H.; Martin, J.B.T. Health-Related Quality of Life in High-Grade Glioma Patients. Chin. J. Cancer 2014, 33, 40–45. [Google Scholar]
- Nasor, M.; Obaid, W. Detection and Localization of Early-Stage Multiple Brain Tumors Using Ahybrid Technique of Patch-Based Processing, K-Means Clustering and Object Counting. Int. J. Biomed. Imaging 2020, 2020, 9035096. [Google Scholar] [CrossRef]
- Pries, T.P.; Jahan, R.; Suman, P. Review of Brain Tumor Segmentation, Detection and Classification Algorithms in fMRI Images. In Proceedings of the 2018 International Conference on Computational and Characterization Techniques in Engineering & Sciences (CCTES), Lucknow, India, 14–15 September 2018; pp. 300–303. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for Biomedical Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 234–241. [Google Scholar]
- Hamaguchi, R.; Fujita, A.; Nemoto, K.; Imaizumi, T.; Hikosaka, S. Effective Use of Dilated Convolutions for Segmenting Small Object Instances in Remote Sensing Imagery. In Proceedings of the 2018 IEEE Winter Conference on Applications of Computer Vision (WACV), Lake Tahoe, NV, USA, 12–15 March 2018; pp. 1442–1450. [Google Scholar]
- Li, Y.; Qi, H.; Dai, J.; Ji, X.; Wei, Y. Fully Convolutional Instance-Aware Semantic Segmentation. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2359–2367. [Google Scholar]
- Shareef, B.; Xian, M.; Vakanski, A. Stan: Small Tumor-Aware Network for Breast Ultrasound Image Segmentation. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging (ISBI), Iowa City, IA, USA, 3–7 April 2020; pp. 1–5. [Google Scholar]
- Zhang, K.; Zhou, X.; Wu, J. U-Module: Better Parameters Initialization of Convolutional Neural Network for Medical Image Classification. In Proceedings of the 2019 IEEE International Conference on Image Processing (ICIP), Taipei, Taiwan, 22–25 September 2019; pp. 799–803. [Google Scholar]
- Kuang, Z.; Li, Z.; Zhao, T.; Fan, J. Deep Multi-Task Learning for Large-Scale Image Classification. In Proceedings of the 2017 IEEE Third International Conference on Multimedia Big Data (BigMM), Laguna Hills, CA, USA, 19–21 April 2017; pp. 310–317. [Google Scholar]
- Li, L.; Gong, B. End-to-End Video Captioning with Multitask Reinforcement Learning. In Proceedings of the 2019 IEEE Winter Conference on Applications of Computer Vision (WACV), Waikoloa Village, HI, USA, 7–11 January 2019; pp. 339–348. [Google Scholar]
- Geon, W.L.; Hong, K.K. Multi-Task Learning U-Net for Single-Channel Speech Enhancement and Mask-Based Voice Activity Detection. Appl. Sci. 2020, 10, 3230. [Google Scholar] [CrossRef]
- He, T.; Hu, J.; Song, Y.; Guo, J.; Yi, Z. Multi-Task Learning for the Segmentation of Organs at Risk with Label Dependence. Med. Image Anal. 2020, 61, 101666. [Google Scholar] [CrossRef]
- Abdullah, A.Z.I.; Demetri, T. Semi-supervised Multi-task Learning with Chest X-Ray Images. In Proceedings of the International Workshop on Machine Learning in Medical Imaging, Shenzhen, China, 13 October 2019; pp. 151–159. [Google Scholar]
- Shen, D.; Wu, G.; Suk, H.-I. Deep Learning in Medical Image Analysis. Annu. Rev. Biomed. Eng. 2017, 19, 221–248. [Google Scholar] [CrossRef]
- Liu, Y.; Cheng, H.D.; Huang, J.H.; Zhang, Y.T.; Tang, X.L.; Tian, J.W.; Wang, Y. Computer Aided Diagnosis System for Breast Cancer Based on Color Doppler Flow Imaging. J. Med. Syst. 2012, 36, 3975–3982. [Google Scholar] [CrossRef] [PubMed]
- Wang, G. A Perspective on Deep Imaging. IEEE Access 2016, 4, 8914–8924. [Google Scholar] [CrossRef]
- Saez, A.; Sánchez-Monedero, J.; Gutiérrez, P.A.; Hervas-Martinez, C. Machine Learning Methods for Binary and Multiclass Classification of Melanoma Thickness from Dermoscopic Images. IEEE Trans. Med. Imaging 2015, 35, 1036–1045. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhan, Y.; Shen, D. Incremental Learning with Selective Memory (ILSM): Towards Fast Prostate Localization for Image Guided Radiotherapy. IEEE Trans. Med. Imaging 2013, 16, 378–386. [Google Scholar] [CrossRef]
- Tao, Y.; Peng, Z.; Krishnan, A.; Zhou, X.S. Robust Learning-Based Parsing and Annotation of Medical Radiographs. IEEE Trans. Med. Imaging 2010, 30, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Akkus, Z.; Galimzianova, A.; Hoogi, A.; Rubin, D.L.; Erickson, B.J. Deep Learning for Brain MRI Segmentation: State of the Art and Future Directions. J. Digit. Imaging 2017, 30, 449–459. [Google Scholar] [CrossRef]
- Khan, G.; Khan, N.M. Brain MRI Segmentation using efficient 3D Fully Convolutional Neural Networks. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, 3–6 December 2018; pp. 2351–2356. [Google Scholar]
- Lu, X.; Xie, Q.; Zha, Y.; Wang, D. Fully Automatic Liver Segmentation Combining Multi-Dimensional Graph Cut with Shape Information in 3D CT Images. Sci. Rep. 2018, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Valindria, V.V.; Lavdas, I.; Cerrolaza, J.; Aboagye, E.O.; Rockall, A.G.; Rueckert, D.; Glocker, B. Small Organ Segmentation in Whole-Body MRI Using a Two-Stage FCN and Weighting Schemes. In Proceedings of the International Workshop on Machine Learning in Medical Imaging, Granada, Spain, 16 September 2018; pp. 346–354. [Google Scholar]
- Hu, S.; Hoffman, E.; Reinhardt, J. Automatic Lung Segmentation for Accurate Quantitation of Volumetric X-Ray CT Images. IEEE Trans. Med. Imaging 2001, 20, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Grosgeorge, D.; Petitjean, C.; Caudron, J.; Fares, J.; Dacher, J.-N. Automatic Cardiac Ventricle Segmentation in MR Images: A Validation Study. Int. J. Comput. Assist. Radiol. Surg. 2010, 6, 573–581. [Google Scholar] [CrossRef]
- Vishnuvarthanan, G.; Rajasekaran, M.P.; Subbaraj, P.; Vishnuvarthanan, A. An Unsupervised Learning Method with a Clustering Approach for Tumor Identification and Tissue Segmentation in Magnetic Resonance Brain Images. Appl. Soft Comput. 2016, 38, 190–212. [Google Scholar] [CrossRef]
- Feng, Y.; Zhao, H.; Li, X.; Zhang, X.; Li, H. A Multi-Scale 3D Otsu Thresholding Algorithm for Medical Image Segmentation. Digit. Signal Process. 2017, 60, 186–199. [Google Scholar] [CrossRef]
- Gupta, D.; Anand, R. A Hybrid Edge-Based Segmentation Approach for Ultrasound Medical Images. Biomed. Signal Process. Control 2017, 31, 116–126. [Google Scholar] [CrossRef]
- Dupont, C.; Betrouni, N.; Reyns, N.; Vermandel, M. On Image Segmentation Methods Applied to Glioblastoma: State of Art and New Trends. IRBM 2016, 37, 131–143. [Google Scholar] [CrossRef]
- Prastawa, M. A Brain Tumor Segmentation Framework Based on Outlier Detection. Med. Image Anal. 2004, 8, 275–283. [Google Scholar] [CrossRef]
- Gooya, A.; Pohl, K.M.; Bilello, M.; Cirillo, L.; Biros, G.; Melhem, E.R.; Davatzikos, C. GLISTR: Glioma Image Segmentation and Registration. IEEE Trans. Med. Imaging 2012, 31, 1941–1954. [Google Scholar] [CrossRef]
- Kwon, D.; Shinohara, R.T.; Akbari, H.; Davatzikos, C. Combining Generative Models for Multifocal Glioma Segmentation and Registration. In Proceedings of the Multimodal Brain Image Analysis, Boston, MA, USA, 14–18 September 2014; pp. 763–770. [Google Scholar]
- Cordier, N.; Delingette, H.; Ayache, N. A Patch-Based Approach for the Segmentation of Pathologies: Application to Glioma Labelling. IEEE Trans. Med. Imaging 2015, 35, 1066–1076. [Google Scholar] [CrossRef] [PubMed]
- Cobzas, D.; Birkbeck, N.; Schmidt, M.; Jagersand, M.; Murtha, A. 3D Variational Brain Tumor Segmentation using a High Dimensional Feature Set. In Proceedings of the 2007 IEEE 11th International Conference on Computer Vision, Rio de Janeiro, Brazil, 14–20 October 2007; pp. 1–8. [Google Scholar]
- Popuri, K.; Cobzas, D.; Jagersand, M.; Shah, S.L.; Murtha, A. 3D Variational Brain Tumor Segmentation on a Clustered Feature Set. SPIE Med. Imaging 2009, 7259, 72591. [Google Scholar] [CrossRef]
- Tustison, N.J.; Shrinidhi, K.L.; Wintermark, M.; Durst, C.R.; Kandel, B.M.; Gee, J.C.; Grossman, M.C.; Avants, B.B. Optimal Symmetric Multimodal Templates and Concatenated Random Forests for Supervised Brain Tumor Segmentation (Simplified) with ANTsR. Neuroinformatics 2014, 13, 209–225. [Google Scholar] [CrossRef]
- Le Folgoc, L.; Nori, A.V.; Ancha, S.; Criminisi, A. Lifted Auto-Context Forests for Brain Tumour Segmentation. In Proceedings of the International Workshop on Brain Lesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Athens, Greece, 17–21 October 2016; pp. 171–183. [Google Scholar]
- Bauer, S.; Nolte, L.-P.; Reyes, M. Fully Automatic Segmentation of Brain Tumor Images Using Support Vector Machine Classification in Combination with Hierarchical Conditional Random Field Regularization. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Toronto, ON, Canada, 18–22 September 2011; pp. 354–361. [Google Scholar]
- Lee, C.-H.; Schmidt, M.; Murtha, A.; Bistritz, A.; Sander, J.; Greiner, R. Segmenting Brain Tumors with Conditional Random Fields and Support Vector Machines. In Proceedings of the International Workshop on Computer Vision for Biomedical Image Applications, Beijing, China, 21 October 2005; pp. 469–478. [Google Scholar]
- Geremia, E.; Menze, B.H.; Ayache, N. Spatial Decision Forests for Glioma Segmentation in Multi-Channel Mr Images. In Proceedings of the MICCAI Challenge on Multimodal Brain Tumor Segmentation, Nice, France, 1 October 2012. [Google Scholar]
- Ciçek, Ö.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Athens, Greece, 17–21 October 2016; pp. 424–432. [Google Scholar]
- Chen, C.; Liu, X.; Ding, M.; Zheng, J.; Li, J. 3D Dilated Multi-Fiber Network for Real-Timebrain Tumor Segmentation in MRI. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Shenzhen, China, 13–17 October 2019; pp. 184–192. [Google Scholar]
- Malmi, E.; Parambath, S.; Peyrat, J.M.; Abinahed, J.; Chawla, S. Cabs: A Cascaded Brain Tumor Segmentation Approach. In Proceedings of the MICCAI Brain, Tumor Segmentation (BRATS), Munich, Germany, 5–9 October 2015; pp. 42–47. [Google Scholar]
- Pereira, S.; Oliveira, A.; Ealves, V.; Silva, C.A. On Hierarchical Brain Tumor Segmentation in MRI Using Fully Convolutional Neural Networks: A Preliminary Study. In Proceedings of the 2017 IEEE 5th Portuguese Meeting on Bioengineering (ENBENG), Coimbra, Portugal, 16–18 February 2017; pp. 1–4. [Google Scholar]
- Wang, G.; Li, W.; Ourselin, S.; Vercauteren, T. Automatic Brain Tumor Segmentation Using Cascaded Anisotropic Convolutional Neural Networks. In Proceedings of the International MICCAI Brain Lesion Workshop, Quebec City, QC, Canada, 11–13 September 2018; pp. 178–190. [Google Scholar]
- Liu, H.; Shen, X.; Shang, F.; Ge, F.; Wang, F. CU-Net: Cascaded U-Net with Loss Weighted Sampling for Brain Tumor Segmentation. In Proceedings of the International Workshop, MFCA 2019, Held in Conjunction with MICCAI 2019, Shenzhen, China, 17 October 2019; pp. 102–111. [Google Scholar]
- Brügger, R.; Baumgartner, C.F.; Konukoglu, E. A Partially Reversible U-Net for Memory-Efficient Volumetric Image Segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Shenzhen, China, 13–17 October 2019; pp. 429–437. [Google Scholar]
- Isensee, F.; Kickingereder, P.; Wick, W.; Bendszus, M.; Maier-Hein, K.H. No New-Net. In Proceedings of the International MICCAI Brain Lesion Workshop, Granada, Spain, 16 September 2018; pp. 234–244. [Google Scholar]
- Nuechterlein, N.; Mehta, S. 3D-ESPNet with Pyramidal Refinement for Volumetric Brain Tumor Image Segmentation. In Proceedings of the International MICCAI Brain Lesion Workshop, Granada, Spain, 16 September 2018; pp. 245–253. [Google Scholar]
- Kao, P.-Y.; Ngo, T.; Zhang, A.; Chen, J.W.; Manjunath, B.S. Brain Tumor Segmentation and Tractographic Feature Extraction from Structural MR Images for Overall Survival Prediction. In Proceedings of the International MICCAI Brain Lesion Workshop, Granada, Spain, 16 September 2018; pp. 128–141. [Google Scholar]
- Chen, W.; Liu, B.; Peng, S.; Sun, J.; Qiao, X. S3D-UNet: Separable 3D U-Net for Brain Tumor Segmentation. In Proceedings of the International MICCAI Brain Lesion Workshop, Granada, Spain, 16–20 September 2019; pp. 358–368. [Google Scholar]
- Mlynarski, P.; Delingette, H.; Criminisi, A.; Ayache, N. Deep Learning with Mixed Supervision for Brain Tumor Segmentatio. J. Med. Imaging 2019, 6, 034002. [Google Scholar] [CrossRef]
- Myronenko, A. 3D MRI Brain Tumor Segmentation Using Autoencoder Regularization. In Proceedings of the International MICCAI Brain Lesion Workshop, Granada, Spain, 16–20 September 2019; pp. 311–320. [Google Scholar]
- Kampffmeyer, M.; Salberg, A.-B.; Jenssen, R. Semantic Segmentation of Small Objects and Modeling of Uncertainty in Urban Remote Sensing Images Using Deep Convolutional Neural Networks. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Las Vegas, NV, USA, 26 June–1 July 2016; pp. 680–688. [Google Scholar]
- Sudre, C.H.; Li, W.; Vercauteren, T.; Ourselin, S.; Cardoso, M. Generalised Dice Overlap as a Deep Learning Loss Function for Highly Unbalanced Segmentations. In Proceedings of the International MICCAI Brain Lesion Workshop, Quebec City, QC, Canada, 14 September 2017; pp. 240–248. [Google Scholar]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2014, 34, 1993–2024. [Google Scholar] [CrossRef]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing The Cancer Genome Atlas Glioma MRI Collections With Expert Segmentation Labels and Radiomic Features. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef] [PubMed]
- Bakas, S.; Reyes, M.; Jakab, A.; Bauer, S.; Rempfler, M.; Crimi, A.; Shinohara, R.T.; Berger, C.; Ha, S.M.; Rozycki, M.; et al. Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the Brats Challenge. arXiv 2018, arXiv:1811.02629. Available online: https://arxiv.org/abs/1811.02629 (accessed on 5 November 2018).
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; et al. PyTorch: An Imperative Style, High-Performance Deep Learning Library. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2019; pp. 8026–8037. [Google Scholar]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).