Multi-Time-Scale Features for Accurate Respiratory Sound Classification
Abstract
:Featured Application
Abstract
1. Introduction
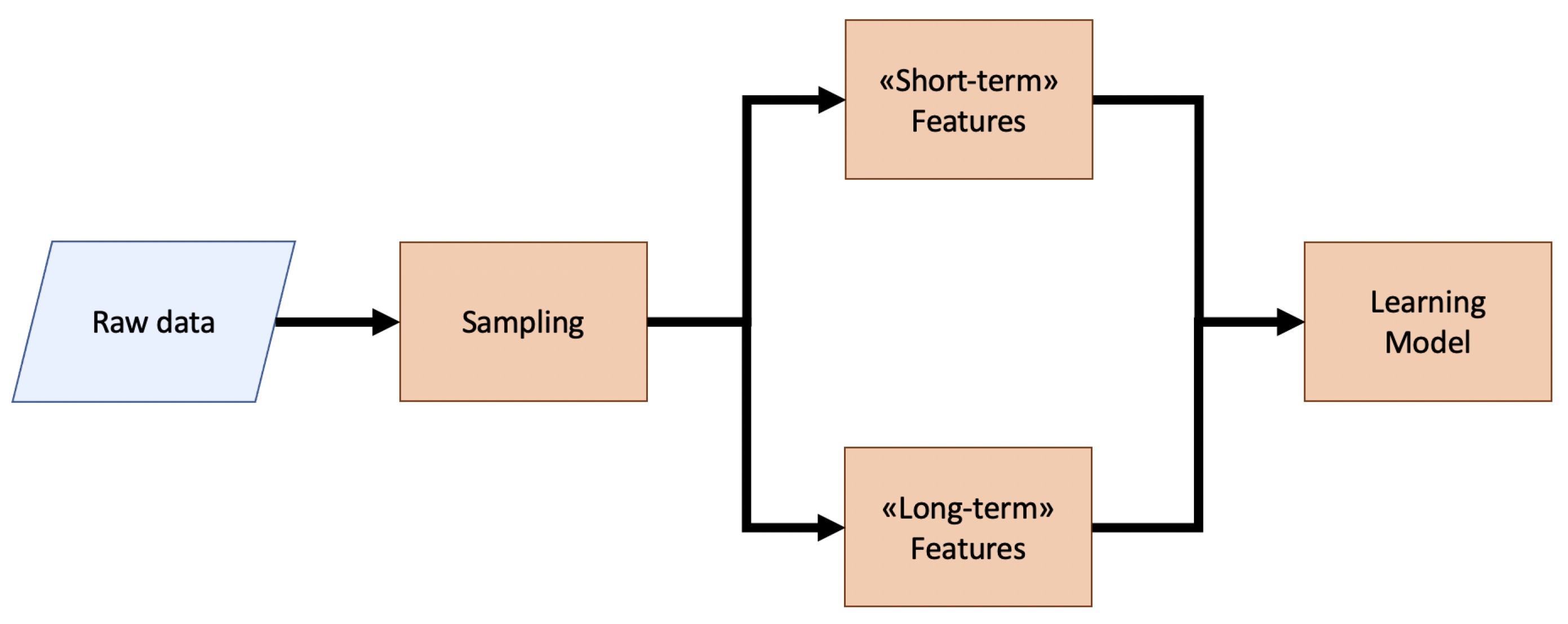
2. Materials and Methods
2.1. The ICBHI Dataset
2.2. Multi-Time-Scale Feature Extraction
2.2.1. Short-Term Features
- Calculate the DFT of the signal in the short-term window;
- Identify M equally spaced frequencies on the Mel scale and build a bank of triangular spectral filters with centered on each corresponding M frequency in Hz;
- Evaluate the spectral output powers of each filter ;
- Estimate MFCCs as
2.2.2. Long-Term Features
2.3. Classification and Performance Assessment
2.3.1. Learning Models
2.3.2. Cross-Validation, Balancing and Performance Metrics
2.3.3. Feature Importance Procedure
3. Results
3.1. Classification Performances
3.2. Feature Importance
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A

References
- European Respiratory Society. The Global Impact of Respiratory Disease. In Forum of International Respiratory Societies, 2nd ed.; European Respiratory Society: Sheffield, UK, 2017. [Google Scholar]
- Williams, S.; Sheikh, A.; Campbell, H.; Fitch, N.; Griffiths, C.; Heyderman, R.S.; Jordan, R.E.; Katikireddi, S.V.; Tsiligianni, I.; Obasi, A. Respiratory research funding is inadequate, inequitable, and a missed opportunity. Lancet Respir. Med. 2020, 8, e67–e68. [Google Scholar] [CrossRef]
- Lai, C.C.; Liu, Y.H.; Wang, C.Y.; Wang, Y.H.; Hsueh, S.C.; Yen, M.Y.; Ko, W.C.; Hsueh, P.R. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARSCoV-2): Facts and myths. J. Microbiol. Immunol. Infect. 2020, 53, 404–412. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.J.; Ni, Z.Y.; Hu, Y.; Liang, W.H.; Ou, C.Q.; He, J.X.; Liu, L.; Shan, H.; Lei, C.L.; Hui, D.S.; et al. Clinical characteristics of 2019 novel coronavirus infection in China. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Yao, L.; Wei, T.; Tian, F.; Jin, D.Y.; Chen, L.; Wang, M. Presumed asymptomatic carrier transmission of COVID-19. JAMA 2020, 323, 1406–1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reichert, S.; Gass, R.; Brandt, C.; Andrès, E. Analysis of respiratory sounds: State of the art. Clin. Med. Circ. Respir. Pulm. Med. 2008, 2, CCRPM-S530. [Google Scholar] [CrossRef] [PubMed]
- Palaniappan, R.; Sundaraj, K.; Sundaraj, S. Artificial intelligence techniques used in respiratory sound analysis–a systematic review. Biomed. Eng./Biomed. Tech. 2014, 59, 7–18. [Google Scholar] [CrossRef]
- Pramono, R.X.A.; Bowyer, S.; Rodriguez-Villegas, E. Automatic adventitious respiratory sound analysis: A systematic review. PLoS ONE 2017, 12, e0177926. [Google Scholar] [CrossRef] [Green Version]
- Mekov, E.; Miravitlles, M.; Petkov, R. Artificial intelligence and machine learning in respiratory medicine. Expert Rev. Respir. Med. 2020, 14, 559–564. [Google Scholar] [CrossRef]
- Sovijarvi, A. Characteristics of breath sounds and adventitious respiratory sounds. Eur. Respir. Rev. 2000, 10, 591–596. [Google Scholar]
- Homs-Corbera, A.; Fiz, J.A.; Morera, J.; Jané, R. Time-frequency detection and analysis of wheezes during forced exhalation. IEEE Trans. Biomed. Eng. 2004, 51, 182–186. [Google Scholar] [CrossRef]
- Kandaswamy, A.; Kumar, C.S.; Ramanathan, R.P.; Jayaraman, S.; Malmurugan, N. Neural classification of lung sounds using wavelet coefficients. Comput. Biol. Med. 2004, 34, 523–537. [Google Scholar] [CrossRef]
- Cnockaert, L.; Migeotte, P.F.; Daubigny, L.; Prisk, G.K.; Grenez, F.; Sá, R.C. A method for the analysis of respiratory sinus arrhythmia using continuous wavelet transforms. IEEE Trans. Biomed. Eng. 2008, 55, 1640–1642. [Google Scholar] [CrossRef] [PubMed]
- Sello, S.; Strambi, S.k.; De Michele, G.; Ambrosino, N. Respiratory sound analysis in healthy and pathological subjects: A wavelet approach. Biomed. Signal Process. Control. 2008, 3, 181–191. [Google Scholar] [CrossRef]
- Jin, F.; Krishnan, S.; Sattar, F. Adventitious sounds identification and extraction using temporal–spectral dominance-based features. IEEE Trans. Biomed. Eng. 2011, 58, 3078–3087. [Google Scholar] [PubMed]
- Sengupta, N.; Sahidullah, M.; Saha, G. Lung sound classification using cepstral-based statistical features. Comput. Biol. Med. 2016, 75, 118–129. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Jin, F.; Krishnan, S.; Sattar, F. Signal feature extraction by multi-scale PCA and its application to respiratory sound classification. Med. Biol. Eng. Comput. 2012, 50, 759–768. [Google Scholar] [CrossRef]
- Charleston-Villalobos, S.; González-Camarena, R.; Chi-Lem, G.; Aljama-Corrales, T. Crackle sounds analysis by empirical mode decomposition. IEEE Eng. Med. Biol. Mag. 2007, 26, 40. [Google Scholar] [CrossRef]
- Lozano, M.; Fiz, J.A.; Jané, R. Estimation of instantaneous frequency from empirical mode decomposition on respiratory sounds analysis. In Proceedings of the 2013 35th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Osaka, Japan, 3–7 July 2013; pp. 981–984. [Google Scholar]
- Lozano, M.; Fiz, J.A.; Jané, R. Automatic differentiation of normal and continuous adventitious respiratory sounds using ensemble empirical mode decomposition and instantaneous frequency. IEEE J. Biomed. Health Inform. 2015, 20, 486–497. [Google Scholar] [CrossRef]
- Perna, D. Convolutional neural networks learning from respiratory data. In Proceedings of the 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Madrid, Spain, 3–6 December 2018; pp. 2109–2113. [Google Scholar]
- Liu, R.; Cai, S.; Zhang, K.; Hu, N. Detection of Adventitious Respiratory Sounds based on Convolutional Neural Network. In Proceedings of the 2019 IEEE International Conference on Intelligent Informatics and Biomedical Sciences (ICIIBMS), Shanghai, China, 21–24 November 2019; pp. 298–303. [Google Scholar]
- Minami, K.; Lu, H.; Kim, H.; Mabu, S.; Hirano, Y.; Kido, S. Automatic classification of large-scale respiratory sound dataset based on convolutional neural network. In Proceedings of the 2019 IEEE 19th International Conference on Control, Automation and Systems (ICCAS), Jeju, Korea, 15–18 October 2019; pp. 804–807. [Google Scholar]
- Acharya, J.; Basu, A. Deep Neural Network for Respiratory Sound Classification in Wearable Devices Enabled by Patient Specific Model Tuning. IEEE Trans. Biomed. Circuits Syst. 2020, 14, 535–544. [Google Scholar] [CrossRef]
- Duda, R.O.; Hart, P.E.; Stork, D.G. Pattern Classification and Scene Analysis; Wiley: New York, NY, USA, 1973; Volume 3. [Google Scholar]
- Rocha, B.M.; Filos, D.; Mendes, L.; Serbes, G.; Ulukaya, S.; Kahya, Y.P.; Jakovljevic, N.; Turukalo, T.L.; Vogiatzis, I.M.; Perantoni, E.; et al. An open access database for the evaluation of respiratory sound classification algorithms. Physiol. Meas. 2019, 40, 035001. [Google Scholar] [CrossRef]
- Pikrakis, A.; Giannakopoulos, T.; Theodoridis, S. A speech/music discriminator of radio recordings based on dynamic programming and bayesian networks. IEEE Trans. Multimed. 2008, 10, 846–857. [Google Scholar] [CrossRef]
- Bachu, R.; Kopparthi, S.; Adapa, B.; Barkana, B. Separation of voiced and unvoiced using zero crossing rate and energy of the speech signal. In Proceedings of the American Society for Engineering Education (ASEE) Zone Conference Proceedings, Pittsburgh, PA, USA, 22–25 June 2008; pp. 1–7. [Google Scholar]
- Rizal, A.; Hidayat, R.; Nugroho, H.A. Entropy measurement as features extraction in automatic lung sound classification. In Proceedings of the 2017 IEEE International Conference on Control, Electronics, Renewable Energy and Communications (ICCREC), Yogyakarta, Indonesia, 26–28 September 2017; pp. 93–97. [Google Scholar]
- Crocker, M.J. Handbook of Acoustics; John Wiley & Sons: Hoboken, NJ, USA, 1998. [Google Scholar]
- Schubert, E.; Wolfe, J.; Tarnopolsky, A. Spectral centroid and timbre in complex, multiple instrumental textures. In Proceedings of the International Conference on Music Perception and Cognition, Evanston, IL, USA, 3–7 August 2004; pp. 112–116. [Google Scholar]
- Lazaro, A.; Sarno, R.; Andre, R.J.; Mahardika, M.N. Music tempo classification using audio spectrum centroid, audio spectrum flatness, and audio spectrum spread based on MPEG-7 audio features. In Proceedings of the 2017 IEEE 3rd International Conference on Science in Information Technology (ICSITech), Bandung, Indonesia, 25–26 October 2017; pp. 41–46. [Google Scholar]
- Misra, H.; Ikbal, S.; Bourlard, H.; Hermansky, H. Spectral entropy based feature for robust ASR. In Proceedings of the 2004 IEEE International Conference on Acoustics, Speech, and Signal Processing, Montreal, QC, Canada, 17–21 May 2004; Volume 1, p. I-193. [Google Scholar]
- Sadjadi, S.O.; Hansen, J.H. Unsupervised speech activity detection using voicing measures and perceptual spectral flux. IEEE Signal Process. Lett. 2013, 20, 197–200. [Google Scholar] [CrossRef]
- Kos, M.; KačIč, Z.; Vlaj, D. Acoustic classification and segmentation using modified spectral roll-off and variance-based features. Digit. Signal Process. 2013, 23, 659–674. [Google Scholar] [CrossRef]
- Logan, B. Mel frequency cepstral coefficients for music modeling. ISMIR 2000, 270, 1–11. [Google Scholar]
- Molau, S.; Pitz, M.; Schluter, R.; Ney, H. Computing mel-frequency cepstral coefficients on the power spectrum. In Proceedings of the 2001 IEEE International Conference on Acoustics, Speech, and Signal Processing. Proceedings (Cat. No. 01CH37221), Salt Lake City, UT, USA, 7–11 May 2001; Volume 1, pp. 73–76. [Google Scholar]
- Müller, M.; Kurth, F.; Clausen, M. Audio Matching via Chroma-Based Statistical Features. ISMIR 2005, 2005, 6. [Google Scholar]
- Heng, R.; Nor, M.J.M. Statistical analysis of sound and vibration signals for monitoring rolling element bearing condition. Appl. Acoust. 1998, 53, 211–226. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Yegnanarayana, B. Artificial Neural Networks; PHI Learning Pvt. Ltd.: New Delhi, India, 2009. [Google Scholar]
- Goodfellow, I.; Bengio, Y.; Courville, A.; Bengio, Y. Deep Learning; MIT Press: Cambridge, UK, 2016; Volume 1. [Google Scholar]
- Han, H.; Guo, X.; Yu, H. Variable selection using mean decrease accuracy and mean decrease gini based on random forest. In Proceedings of the 2016 7th IEEE International Conference on Software Engineering and Service Science (ICSESS), Beijing, China, 26–28 August 2016; pp. 219–224. [Google Scholar]
- Hastie, T.; Tibshirani, R.; Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction; Springer: New York, NY, USA, 2009. [Google Scholar]
- Han, H.; Wang, W.Y.; Mao, B.H. Borderline-SMOTE: A new over-sampling method in imbalanced data sets learning. In International Conference on Intelligent Computing; Springer: Berlin/Heidelberg, Germany, 2005; pp. 878–887. [Google Scholar]
- Maglietta, R.; Amoroso, N.; Boccardi, M.; Bruno, S.; Chincarini, A.; Frisoni, G.B.; Inglese, P.; Redolfi, A.; Tangaro, S.; Tateo, A.; et al. Automated hippocampal segmentation in 3D MRI using random undersampling with boosting algorithm. Pattern Anal. Appl. 2016, 19, 579–591. [Google Scholar] [CrossRef] [Green Version]
- Kruskal, W.H.; Wallis, W.A. Use of Ranks in One-Criterion Variance Analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Pesu, L.; Ademovic, E.; Pesquet, J.C.; Helisto, P. Wavelet packet based respiratory sound classification. In Proceedings of the IEEE Third International Symposium on Time-Frequency and Time-Scale Analysis (TFTS-96), Paris, France, 18–21 June 1996; pp. 377–380. [Google Scholar]
- Güler, E.Ç.; Sankur, B.; Kahya, Y.P.; Raudys, S. Two-stage classification of respiratory sound patterns. Comput. Biol. Med. 2005, 35, 67–83. [Google Scholar] [CrossRef]
- Riella, R.; Nohama, P.; Maia, J. Method for automatic detection of wheezing in lung sounds. Braz. J. Med. Biol. Res. 2009, 42, 674–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayorga, P.; Druzgalski, C.; Morelos, R.; Gonzalez, O.; Vidales, J. Acoustics based assessment of respiratory diseases using GMM classification. In Proceedings of the 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology, Buenos Aires, Argentina, 31 August–4 September 2010; pp. 6312–6316. [Google Scholar]
- Emmanouilidou, D.; Patil, K.; West, J.; Elhilali, M. A multiresolution analysis for detection of abnormal lung sounds. In Proceedings of the 2012 Annual International Conference of the IEEE Engineering in Medicine and Biology Society, San Diego, CA, USA, 28 August–1 September 2012; pp. 3139–3142. [Google Scholar]
- Palaniappan, R.; Sundaraj, K. Respiratory sound classification using cepstral features and support vector machine. In Proceedings of the 2013 IEEE Recent Advances in Intelligent Computational Systems (RAICS), Trivandrum, India, 19–21 December 2013; pp. 132–136. [Google Scholar]
- Sen, I.; Saraclar, M.; Kahya, Y.P. A comparison of SVM and GMM-based classifier configurations for diagnostic classification of pulmonary sounds. IEEE Trans. Biomed. Eng. 2015, 62, 1768–1776. [Google Scholar] [CrossRef] [PubMed]
- Chambres, G.; Hanna, P.; Desainte-Catherine, M. Automatic detection of patient with respiratory diseases using lung sound analysis. In Proceedings of the 2018 IEEE International Conference on Content-Based Multimedia Indexing (CBMI), La Rochelle, France, 4–6 September 2018; pp. 1–6. [Google Scholar]
- Yadav, A.; Dutta, M.K.; Prinosil, J. Machine Learning Based Automatic Classification of Respiratory Signals using Wavelet Transform. In Proceedings of the 2020 IEEE 43rd International Conference on Telecommunications and Signal Processing (TSP), Milan, Italy, 7–9 July 2020; pp. 545–549. [Google Scholar]
- Wu, L.; Li, L. Investigating into segmentation methods for diagnosis of respiratory diseases using adventitious respiratory sounds. In Proceedings of the 2020 IEEE 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montreal, QC, Canada, 20–24 July 2020; pp. 768–771. [Google Scholar]
- Ma, Y.; Xu, X.; Li, Y. LungRN+ NL: An Improved Adventitious Lung Sound Classification Using non-local block ResNet Neural Network with Mixup Data Augmentation. In Proceedings of the Interspeech 2020, Shanghai, China, 25–29 October 2020; pp. 2902–2906. [Google Scholar]
- Yang, Z.; Liu, S.; Song, M.; Parada-Cabaleiro, E.; Schuller12, B.W. Adventitious Respiratory Classification using Attentive Residual Neural Networks. In Proceedings of the Interspeech 2020, Shanghai, China, 25–29 October 2020; pp. 2912–2916. [Google Scholar]
- Pelletier, B.; Hickey, G.M.; Bothi, K.L.; Mude, A. Linking rural livelihood resilience and food security: An international challenge. Food Secur. 2016, 8, 469–476. [Google Scholar] [CrossRef]
- Kristan, M.; Leonardis, A.; Matas, J.; Felsberg, M.; Pflugfelder, R.; Cehovin Zajc, L.; Vojir, T.; Hager, G.; Lukezic, A.; Eldesokey, A.; et al. The visual object tracking vot2017 challenge results. In Proceedings of the IEEE International Conference on Computer Vision Workshops, Venice, Italy, 22–29 October 2017; pp. 1949–1972. [Google Scholar]
- Amoroso, N.; Diacono, D.; Fanizzi, A.; La Rocca, M.; Monaco, A.; Lombardi, A.; Guaragnella, C.; Bellotti, R.; Tangaro, S.; Initiative, A.D.N. Deep learning reveals Alzheimer’s disease onset in MCI subjects: Results from an international challenge. J. Neurosci. Methods 2018, 302, 3–9. [Google Scholar] [CrossRef] [Green Version]
- Choobdar, S.; Ahsen, M.E.; Crawford, J.; Tomasoni, M.; Fang, T.; Lamparter, D.; Lin, J.; Hescott, B.; Hu, X.; Mercer, J.; et al. Assessment of network module identification across complex diseases. Nat. Methods 2019, 16, 843–852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Honma, M.; Kitazawa, A.; Cayley, A.; Williams, R.V.; Barber, C.; Hanser, T.; Saiakhov, R.; Chakravarti, S.; Myatt, G.J.; Cross, K.P.; et al. Improvement of quantitative structure–activity relationship (QSAR) tools for predicting Ames mutagenicity: Outcomes of the Ames/QSAR International Challenge Project. Mutagenesis 2019, 34, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Selesnick, I.W. Wavelet transform with tunable Q-factor. IEEE Trans. Signal Process. 2011, 59, 3560–3575. [Google Scholar] [CrossRef]
- Jakovljević, N.; Lončar-Turukalo, T. Hidden markov model based respiratory sound classification. In International Conference on Biomedical and Health Informatics; Springer: Berlin/Heidelberg, Germany, 2017; pp. 39–43. [Google Scholar]
- Ho, Y.C.; Pepyne, D.L. Simple explanation of the no-free-lunch theorem and its implications. J. Opt. Theory Appl. 2002, 115, 549–570. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Ma, Y.; Xu, X.; Yu, Q.; Zhang, Y.; Li, Y.; Zhao, J.; Wang, G. LungBRN: A Smart Digital Stethoscope for Detecting Respiratory Disease Using bi-ResNet Deep Learning Algorithm. In Proceedings of the 2019 IEEE Biomedical Circuits and Systems Conference (BioCAS), Nara, Japan, 17–19 October 2019; pp. 1–4. [Google Scholar]
- Székely, É.; Henter, G.E.; Gustafson, J. Casting to corpus: Segmenting and selecting spontaneous dialogue for TTS with a CNN-LSTM speaker-dependent breath detecto. In Proceedings of the ICASSP 2019–2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 6925–6929. [Google Scholar]
- Ardakani, A.A.; Kanafi, A.R.; Acharya, U.R.; Khadem, N.; Mohammadi, A. Application of deep learning technique to manage COVID-19 in routine clinical practice using CT images: Results of 10 convolutional neural networks. Comput. Biol. Med. 2020, 121, 103795. [Google Scholar] [CrossRef]
- Mazurowski, M.A.; Habas, P.A.; Zurada, J.M.; Lo, J.Y.; Baker, J.A.; Tourassi, G.D. Training neural network classifiers for medical decision making: The effects of imbalanced datasets on classification performance. Neural Netw. 2008, 21, 427–436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cilli, R.; Monaco, A.; Amoroso, N.; Tateo, A.; Tangaro, S.; Bellotti, R. Machine learning for cloud detection of globally distributed Sentinel-2 images. Remote Sens. 2020, 12, 2355. [Google Scholar] [CrossRef]
- Pikrakis, A.; Giannakopoulos, T.; Theodoridis, S. A Computationally Efficient Speech/Music Discriminator for Radio Recordings; ISMIR: Victoria, BC, Canada, 2006; pp. 107–110. [Google Scholar]
- Hirsch, H.G.; Pearce, D. The Aurora experimental framework for the performance evaluation of speech recognition systems under noisy conditions. In Proceedings of the ASR2000-Automatic Speech Recognition: Challenges for the New Millenium ISCA Tutorial and Research Workshop (ITRW), Paris, France, 18–20 September 2000. [Google Scholar]
Sample Availability: Data used in this work are open access. |






| Learning Models | Accuracy | Precision | Error HC | Error RS |
|---|---|---|---|---|
| Random Forest | ||||
| MLP | ||||
| SVM | ||||
| DNN |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monaco, A.; Amoroso, N.; Bellantuono, L.; Pantaleo, E.; Tangaro, S.; Bellotti, R. Multi-Time-Scale Features for Accurate Respiratory Sound Classification. Appl. Sci. 2020, 10, 8606. https://doi.org/10.3390/app10238606
Monaco A, Amoroso N, Bellantuono L, Pantaleo E, Tangaro S, Bellotti R. Multi-Time-Scale Features for Accurate Respiratory Sound Classification. Applied Sciences. 2020; 10(23):8606. https://doi.org/10.3390/app10238606
Chicago/Turabian StyleMonaco, Alfonso, Nicola Amoroso, Loredana Bellantuono, Ester Pantaleo, Sabina Tangaro, and Roberto Bellotti. 2020. "Multi-Time-Scale Features for Accurate Respiratory Sound Classification" Applied Sciences 10, no. 23: 8606. https://doi.org/10.3390/app10238606
APA StyleMonaco, A., Amoroso, N., Bellantuono, L., Pantaleo, E., Tangaro, S., & Bellotti, R. (2020). Multi-Time-Scale Features for Accurate Respiratory Sound Classification. Applied Sciences, 10(23), 8606. https://doi.org/10.3390/app10238606









