P-Wave Detection Using a Fully Convolutional Neural Network in Electrocardiogram Images
Abstract
1. Introduction
2. Materials and Methods
2.1. Used Databases
2.1.1. Self-Annotated Database (MIT-BIH AF)
2.1.2. Standard Annotated Database (QT Database)
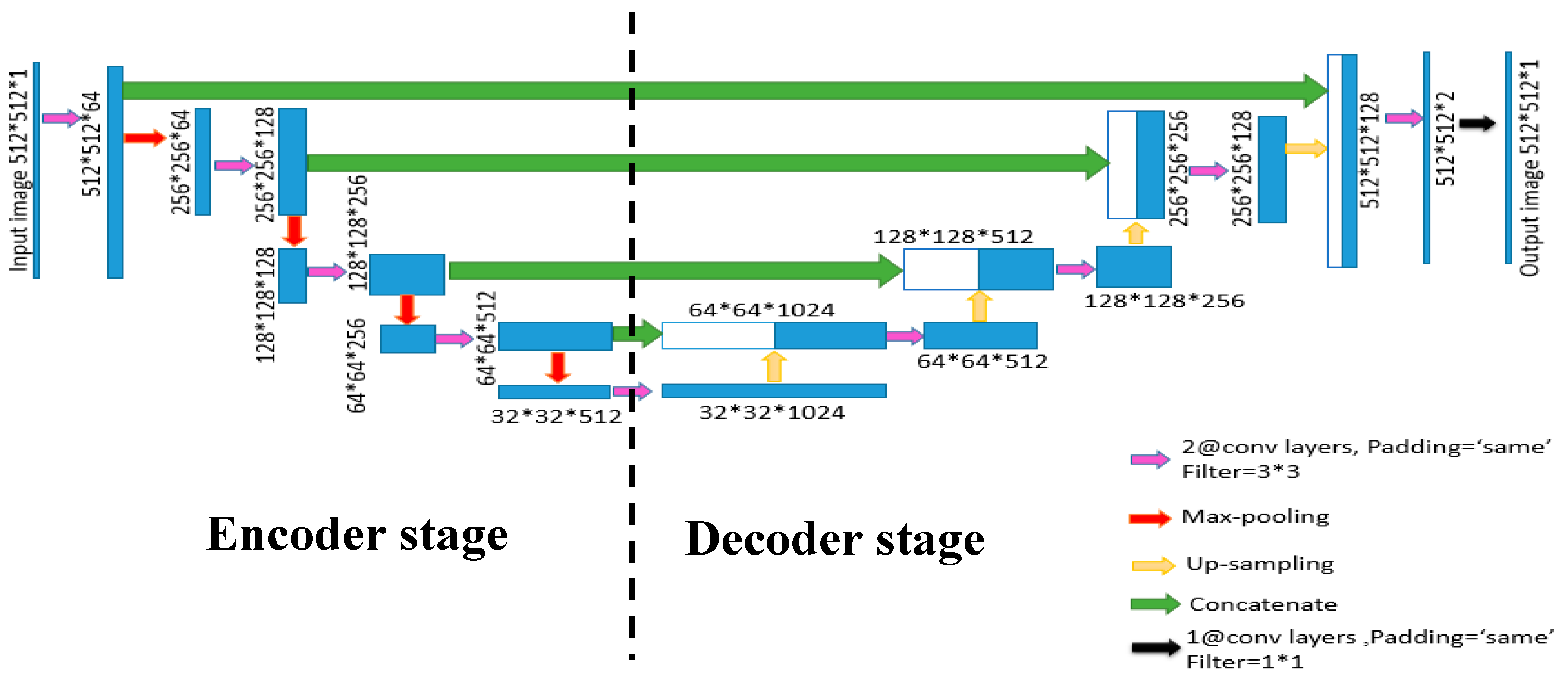
2.2. Proposed P-Wave Image-Based Detection Algorithm
Segmentation via U-Net Architecture
3. Experimental Results
3.1. Experimental Setup
3.2. Evaluation Metrics
3.3. Evaluation Results
4. Performance Validation and Discussion
4.1. Automatically Annotated QT Database Results
4.2. Manually Annotated QT Database
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Goss, J.R.; Australian Institute of Health and Welfare. Projection of Australian Health Care Expenditure by Disease, 2003 to 2033; Australian Institute of Health and Welfare: Canberra, Australia, 2008; ISBN 978-1-74024-862-4.
- Tun, H.M. Analysis on conversion process from paper record ECG to computer based ECG. MOJ Appl. Bionics Biomech. 2017, 1. [Google Scholar] [CrossRef]
- Meek, S. ABC of clinical electrocardiography: Introduction. II—Basic terminology. BMJ 2002, 324, 470–473. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.; Quaresma, R.; Rahman, H. Investigating factors influencing the physicians’ adoption of electronic health record (EHR) in healthcare system of bangladesh: An empirical study. Int. J. Inf. Manag. 2019, 44, 76–87. [Google Scholar] [CrossRef]
- Ravichandran, L.; Harless, C.; Shah, A.J.; Wick, C.A.; Mcclellan, J.H.; Tridandapani, S. Novel Tool for complete digitization of paper electrocardiography data. IEEE J. Transl. Eng. Health Med. 2013, 1, 1800107. [Google Scholar] [CrossRef] [PubMed]
- Waits, G.S.; Soliman, E.Z. Digitizing paper electrocardiograms: Status and challenges. J. Electrocardiol. 2017, 50, 123–130. [Google Scholar] [CrossRef]
- Sadhukhan, D.; Mitra, M. R-peak detection algorithm for ECG using double difference and RR interval processing. Procedia Technol. 2012, 4, 873–877. [Google Scholar] [CrossRef]
- Mitra, R.N.; Pramanik, S.; Mitra, S.; Chaudhuri, B.B. A robust technique for delineation and features extraction of ECG signal from mobile-phone photography. In Proceedings of the 2012 International Conference on Communications, Devices and Intelligent Systems (CODIS), West Bengal, India, 28–29 December 2012; pp. 121–124. [Google Scholar]
- Bonnet, M.H.; Arand, D.L. Heart rate variability in insomniacs and matched normal sleepers. Psychosom. Med. 1998, 60, 610–615. [Google Scholar] [CrossRef]
- Otzenberger, H.; Gronfier, C.; Simon, C.; Charloux, A.; Ehrhart, J.; Piquard, F.; Brandenberger, G. Dynamic heart rate variability: A tool for exploring sympathovagal balance continuously during sleep in men. Am. J. Physiol. Heart Circ. Physiol. 1998, 275, H946–H950. [Google Scholar] [CrossRef]
- Busek, P.; Vanková, J.; Opavský, J.; Salinger, J.; Nevsímalová, S. Spectral analysis of the heart rate variability in sleep. Physiol. Res. 2005, 54, 369–376. [Google Scholar]
- Jurysta, F.; van de Borne, P.; Migeotte, P.-F.; Dumont, M.; Lanquart, J.-P.; Degaute, J.-P.; Linkowski, P. A study of the dynamic interactions between sleep EEG and heart rate variability in healthy young men. Clin. Neurophysiol. 2003, 114, 2146–2155. [Google Scholar] [CrossRef]
- Scholz, U.J.; Bianchi, A.M.; Cerutti, S.; Kubicki, S. Vegetative background of sleep: Spectral analysis of the heart rate variability. Physiol. Behav. 1997, 62, 1037–1043. [Google Scholar] [CrossRef]
- Trinder, J.; Kleiman, J.; Carrington, M.; Smith, S.; Breen, S.; Tan, N.; Kim, Y. Autonomic activity during human sleep as a function of time and sleep stage. J. Sleep Res. 2001, 10, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Tsipouras, M.G.; Fotiadis, D.I.; Sideris, D. Arrhythmia classification using the RR-interval duration signal. In Proceedings of the Computers in Cardiology, Memphis, TN, USA, 22–25 September 2002; pp. 485–488. [Google Scholar]
- Exarchos, T.P.; Tsipouras, M.G.; Exarchos, C.P.; Papaloukas, C.; Fotiadis, D.I.; Michalis, L.K. A methodology for the automated creation of fuzzy expert systems for ischaemic and arrhythmic beat classification based on a set of rules obtained by a decision tree. Artif. Intell. Med. 2007, 40, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Homaeinezhad, M.R.; Atyabi, S.A.; Tavakkoli, E.; Toosi, H.N.; Ghaffari, A.; Ebrahimpour, R. ECG arrhythmia recognition via a neuro-SVM–KNN hybrid classifier with virtual QRS image-based geometrical features. Expert Syst. Appl. 2012, 39, 2047–2058. [Google Scholar] [CrossRef]
- Balasundaram, K.; Masse, S.; Nair, K.; Umapathy, K. A classification scheme for ventricular arrhythmias using wavelets analysis. Med. Biol. Eng. Comput. 2013, 51, 153–164. [Google Scholar] [CrossRef]
- Da Luz, E.J.S.; Schwartz, W.R.; Cámara-Chávez, G.; Menotti, D. ECG-based heartbeat classification for arrhythmia detection: A survey. Comput. Methods Programs Biomed. 2016, 127, 144–164. [Google Scholar] [CrossRef]
- Thong, T.; McNames, J.; Aboy, M.; Goldstein, B. Prediction of paroxysmal atrial fibrillation by analysis of atrial premature complexes. IEEE Trans. Biomed. Eng. 2004, 51, 561–569. [Google Scholar] [CrossRef]
- Bashour, C.A.; Visinescu, M.; Gopakumaran, B.; Wazni, O.; Carangio, F.; Yared, J.-P.; Starr, N. Characterization of premature atrial contraction activity prior to the onset of postoperative atrial fibrillation in cardiac surgery patients. Chest 2004, 126, 831S. [Google Scholar] [CrossRef]
- De Chazal, P.; O’Dwyer, M.; Reilly, R.B. Automatic classification of heartbeats using ecg morphology and heartbeat interval features. IEEE Trans. Biomed. Eng. 2004, 51, 1196–1206. [Google Scholar] [CrossRef]
- Elgendi, M.; Eskofier, B.; Dokos, S.; Abbott, D. Revisiting QRS detection methodologies for portable, wearable, battery-operated, and wireless ECG systems. PLoS ONE 2014, 9, e84018. [Google Scholar] [CrossRef]
- Chen, R.; Huang, Y.; Wu, J. Multi-window detection for P-wave in electrocardiograms based on bilateral accumulative area. Comput. Biol. Med. 2016, 78, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Leutheuser, H.; Gradl, S.; Anneken, L.; Arnold, M.; Lang, N.; Achenbach, S.; Eskofier, B.M. Instantaneous P- and T-wave detection: Assessment of three ECG fiducial points detection algorithms. In Proceedings of the 2016 IEEE 13th International Conference on Wearable and Implantable Body Sensor Networks (BSN), San Francisco, CA, USA, 14–17 June 2016; pp. 329–334. [Google Scholar]
- Laguna, P.; Mark, R.G.; Goldberger, A.; Moody, G.B. A database for evaluation of algorithms for measurement of QT and other waveform intervals in the ECG. In Proceedings of the IEEE Computers in Cardiology, Lund, Sweden, 7–10 September 1997; pp. 673–676. [Google Scholar]
- Moody, G.B.; Mark, R.G. The impact of the MIT-BIH arrhythmia database. IEEE Eng. Med. Biol. Mag. 2001, 20, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Taddei, A.; Distante, G.; Emdin, M.; Pisani, P.; Moody, G.B.; Zeelenberg, C.; Marchesi, C. The European ST-T database: Standard for evaluating systems for the analysis of ST-T changes in ambulatory electrocardiography. Eur. Heart J. 1992, 13, 1164–1172. [Google Scholar] [CrossRef] [PubMed]
- Moody, G.B.; Mark, R.G. MIT-BIH Atrial Fibrillation Database 1992. Available online: https://physionet.org/content/afdb/1.0.0/ (accessed on 20 January 2020).
- Goldberger, A.L.; Amaral, L.A.N.; Glass, L.; Hausdorff, J.M.; Ivanov, P.C.; Mark, R.G.; Mietus, J.E.; Moody, G.B.; Peng, C.-K.; Stanley, H.E. PhysioBank, physiotoolkit, and physioNet: Components of a new research resource for complex physiologic signals. Circulation 2000, 101, e215–e220. [Google Scholar] [CrossRef]
- Cuiwei Li; Chongxun Zheng; Changfeng Tai detection of ECG characteristic points using wavelet transforms. IEEE Trans. Biomed. Eng. 1995, 42, 21–28. [CrossRef]
- Senhadji, L.; Wang, F.; Hernandez, A.I.; Carrault, G. Wavelets extrema representation for QRS-T cancellation and P wave detection. In Proceedings of the Computers in Cardiology, Memphis, TN, USA, 22–25 September 2002; pp. 37–40. [Google Scholar]
- Sovilj, S.; Jeras, M.; Magjarevic, R. Real time P-wave detector based on wavelet analysis. In Proceedings of the 12th IEEE Mediterranean Electrotechnical Conference (IEEE Cat. No.04CH37521), Dubrovnik, Croatia, 12–15 May 2004; pp. 403–406. [Google Scholar]
- Wan, X.; Qin, S.; Liang, X.; Ding, J. A new approach to wavelet-based P-wave detection. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi 2006, 23, 722–725. [Google Scholar]
- Martinez, J.P.; Almeida, R.; Olmos, S.; Rocha, A.P.; Laguna, P. A wavelet-based ECG delineator: Evaluation on standard databases. IEEE Trans. Biomed. Eng. 2004, 51, 570–581. [Google Scholar] [CrossRef]
- Xie, G.; Nie, Z.; Xiang, H.; Zeng, Z. Detection of P wave through wavelet transform and neural network. Sheng Wu Yi Xue Gong Cheng Xue Za Zhi 1999, 16, 320–323. [Google Scholar]
- Lenis, G.; Pilia, N.; Oesterlein, T.; Luik, A.; Schmitt, C.; Dössel, O. P wave detection and delineation in the ECG based on the phase free stationary wavelet transform and using intracardiac atrial electrograms as reference. Biomed. Eng. Biomed. Tech. 2016, 61, 37–56. [Google Scholar] [CrossRef]
- Goutas, A.; Ferdi, Y.; Herbeuval, J.-P.; Boudraa, M.; Boucheham, B. Digital fractional order differentiation-based algorithm for P and T-waves detection and delineation. ITBM RBM 2005, 26, 127–132. [Google Scholar] [CrossRef]
- Laguna, P.; Jané, R.; Caminal, P. Automatic Detection of Wave Boundaries in Multilead ECG Signals: Validation with the CSE Database. Comput. Biomed. Res. 1994, 27, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Panigrahy, D.; Sahu, P.K. P and T wave detection and delineation of ECG signal using differential evolution (DE) optimization strategy. Australas. Phys. Eng. Sci. Med. 2018, 41, 225–241. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.Q. A new method to detect P-wave based on quadratic function. In Advanced Materials Research; Trans Tech Publications: Zurich, Switzerland, 2011; Volume 267, pp. 462–467. [Google Scholar]
- Sbrollini, A.; Mercanti, S.; Agostinelli, A.; Morettini, M.; Di Nardo, F.; Fioretti, S.; Burattini, L. AThrIA: A new adaptive threshold identification algorithm for electrocardiographic P waves. In 2017 Computing in Cardiology (CinC); IEEE: Piscataway, NJ, USA, 2017; pp. 1–4. [Google Scholar]
- de Azevedo Botter, E.; Nascimento, C.L.; Yoneyama, T. A neural network with asymmetric basis functions for feature extraction of ECG P waves. IEEE Trans. Neural. Netw. 2001, 12, 1252–1255. [Google Scholar] [CrossRef] [PubMed]
- Abrishami, H.; Campbell, M.; Han, C.; Czosek, R.; Zhou, X. P-QRS-T localization in ECG using deep learning. In Proceedings of the 2018 IEEE EMBS International Conference on Biomedical & Health Informatics (BHI), Las Vegas, NV, USA, 4–7 March 2018; pp. 210–213. [Google Scholar]
- Peimankar, A.; Puthusserypady, S. An Ensemble of Deep Recurrent Neural Networks for P-wave Detection in Electrocardiogram. In Proceedings of the ICASSP 2019—2019 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Brighton, UK, 12–17 May 2019; pp. 1284–1288. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep con-volutional neural networks. In Proceedings of the Neural Information Processing Systems, Stateline, NV, USA, 3–8 December 2012; pp. 1097–1105. [Google Scholar]
- Girshick, R.; Donahue, J.; Darrell, T.; Malik, J. Rich feature hierarchies for accurate object detection and semantic segmentation. arXiv 2013, arXiv:1311.2524. [Google Scholar]
- Ciresan, D.C.; Gambardella, L.M.; Giusti, A.; Schmidhuber, J. Deep neural networks segment neuronal membranes in electron microscopy images. In Proceedings of the Neural Information Processing Systems, Stateline, NV, USA, 3–8 December 2012; pp. 2843–2851. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. arXiv 2015, arXiv:1505.04597. [Google Scholar]
- Kao, P.-Y.; Chen, J.W.; Manjunath, B.S. Improving 3D U-net for brain tumor segmentation by utilizing lesion prior. arXiv 2019, arXiv:1907.00281. [Google Scholar]
- Dong, H.; Yang, G.; Liu, F.; Mo, Y.; Guo, Y. Automatic brain tumor detection and segmentation using u-net based fully convolutional networks. In Medical Image Understanding and Analysis; Valdés Hernández, M., González-Castro, V., Eds.; Springer International Publishing: Cham, Switzerland, 2017; Volume 723, pp. 506–517. ISBN 978-3-319-60963-8. [Google Scholar]
- Gu, J.; Wang, Z.; Kuen, J.; Ma, L.; Shahroudy, A.; Shuai, B.; Liu, T.; Wang, X.; Wang, G.; Cai, J.; et al. Recent advances in convolutional neural networks. Pattern Recognit. 2018, 77, 354–377. [Google Scholar] [CrossRef]
- Beraza, I.; Romero, I. Comparative study of algorithms for ECG segmentation. Biomed. Signal Process. Control. 2017, 34, 166–173. [Google Scholar] [CrossRef]
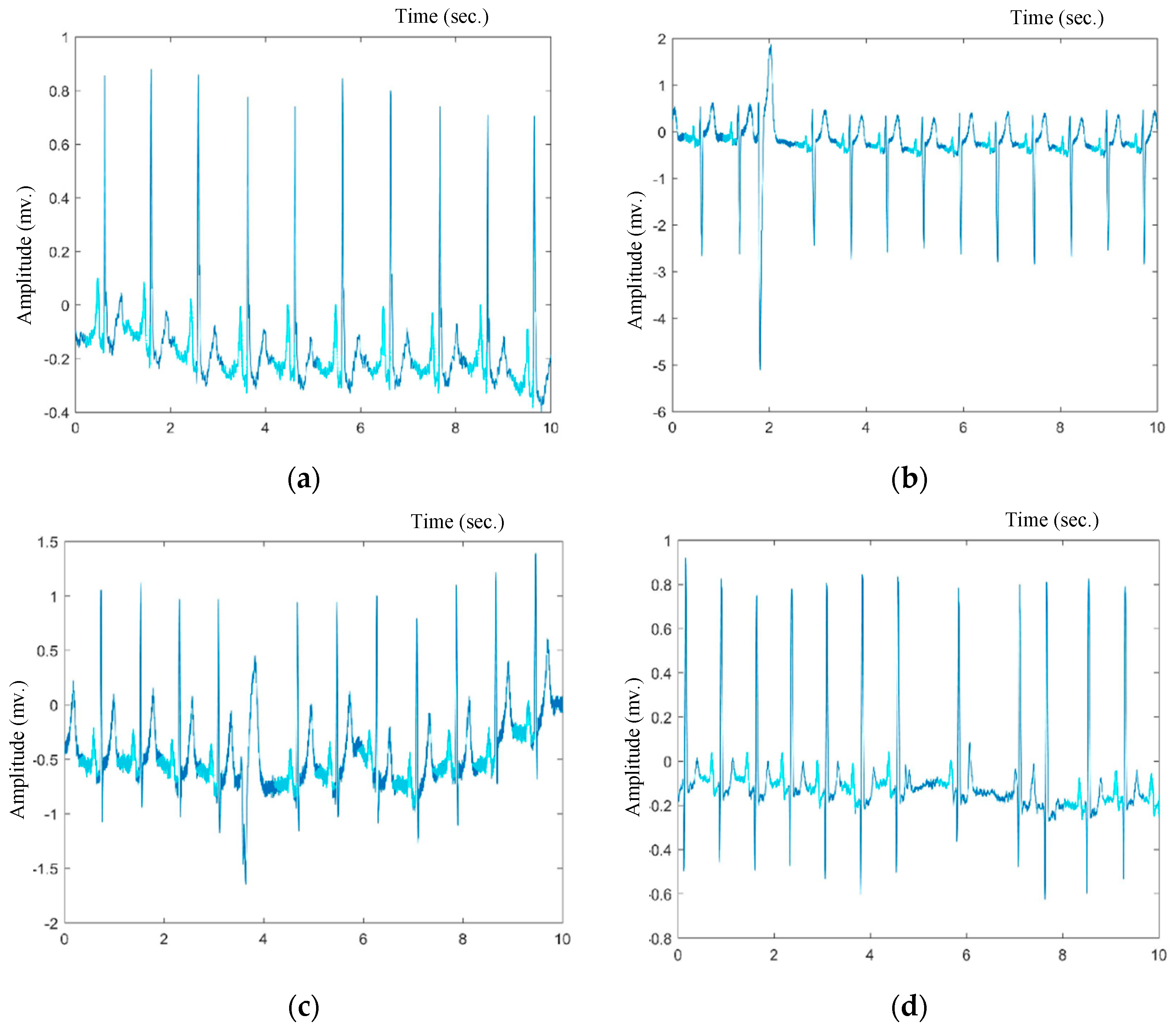



| Database | # Annotated Beats | # Records | Record Duration |
|---|---|---|---|
| MIT-BIH AF [29,30] | 8280 | 23 | 5 min |
| QT Manual Annotation [26] | 3194 | 103 | 15 min |
| QT Automatic Annotation [26] | 11,201 | 105 | 15 min |
| Record | TP | FP | FN | SE (%) | +P (%) |
|---|---|---|---|---|---|
| 04015 | 137 | 7 | 2 | 98.6 | 95.1 |
| 04043 | 175 | 5 | 2 | 98.9 | 97.2 |
| 04048 | 120 | 0 | 0 | 100 | 100 |
| 04126 | 254 | 10 | 4 | 98.4 | 96.2 |
| 04746 | 108 | 1 | 1 | 99.1 | 99.1 |
| 04908 | 165 | 3 | 1 | 99.4 | 98.2 |
| 04936 | 110 | 0 | 0 | 100 | 100 |
| 05091 | 100 | 0 | 0 | 100 | 100 |
| 05121 | 123 | 4 | 0 | 100 | 96.8 |
| 05261 | 127 | 0 | 0 | 100 | 100 |
| 06426 | 118 | 0 | 0 | 100 | 100 |
| 06453 | 134 | 0 | 6 | 100 | 95.7 |
| 06995 | 100 | 0 | 0 | 100 | 100 |
| 07162 | 108 | 6 | 2 | 98.1 | 94.7 |
| 07859 | 144 | 11 | 0 | 100 | 92.9 |
| 07879 | 107 | 1 | 1 | 99.1 | 99.1 |
| 07910 | 91 | 3 | 4 | 95.7 | 96.8 |
| 08215 | 128 | 1 | 2 | 98.4 | 99.2 |
| 08219 | 116 | 2 | 1 | 99.14 | 98.3 |
| 08378 | 129 | 1 | 0 | 100 | 99.23 |
| 08405 | 129 | 1 | 0 | 100 | 99.23 |
| 08434 | 89 | 12 | 11 | 89 | 88.11 |
| 08455 | 108 | 6 | 2 | 98.18 | 94.73 |
| 2920 | 74 | 39 | 98.78 | 97.41 |
| Record | TP | FP | FN | SE (%) | +P (%) |
|---|---|---|---|---|---|
| Sel100 | 374 | 0 | 3 | 99.2 | 100 |
| Sel102 | 45 | 0 | 0 | 100 | 100 |
| Sel103 | 108 | 0 | 0 | 100 | 100 |
| Sel104 | 109 | 1 | 2 | 98.2 | 99.1 |
| Sel114 | 73 | 0 | 0 | 100 | 100 |
| Sel116 | 394 | 1 | 4 | 99.0 | 99.7 |
| Sel117 | 255 | 0 | 1 | 99.6 | 100 |
| Sel123 | 232 | 0 | 0 | 100 | 100 |
| Sel213 | 392 | 1 | 6 | 98.5 | 99.7 |
| Sel221 | 333 | 1 | 20 | 94.3 | 99.7 |
| Sel223 | 363 | 0 | 1 | 99.7 | 100 |
| Sel230 | 356 | 15 | 36 | 90.8 | 96 |
| Sel231 | 242 | 0 | 1 | 99.6 | 100 |
| Sel232 | 267 | 0 | 0 | 100 | 100 |
| Sel233 | 390 | 3 | 24 | 94.2 | 99.2 |
| Sel301 | 434 | 0 | 0 | 100 | 100 |
| Sel302 | 481 | 1 | 1 | 99.8 | 99.8 |
| Sel306 | 277 | 1 | 6 | 97.9 | 99.6 |
| Sel307 | 283 | 1 | 0 | 100 | 99.6 |
| Sel308 | 334 | 3 | 4 | 98.8 | 99.1 |
| Sel310 | 588 | 0 | 0 | 100 | 100 |
| Sel803 | 320 | 1 | 33 | 90.65 | 99.7 |
| Sel808 | 297 | 10 | 13 | 95.8 | 96.7 |
| Sel811 | 233 | 0 | 0 | 100 | 100 |
| Sel820 | 361 | 1 | 1 | 99.7 | 99.7 |
| Sel821 | 323 | 1 | 1 | 99.7 | 99.7 |
| Sel840 | 338 | 1 | 2 | 99.4 | 99.7 |
| Sel847 | 263 | 0 | 0 | 100 | 100 |
| Sel853 | 316 | 2 | 3 | 99 | 99.4 |
| Sel871 | 302 | 0 | 1 | 99.7 | 100 |
| Sel872 | 323 | 0 | 0 | 100 | 100 |
| Sel873 | 286 | 0 | 0 | 100 | 100 |
| Sel883 | 292 | 13 | 16 | 94.8 | 95.8 |
| Sel891 | 352 | 0 | 0 | 100 | 100 |
| Sel16265 | 317 | 3 | 3 | 99.1 | 99.1 |
| Sel16272 | 282 | 0 | 0 | 100 | 100 |
| Sel16273 | 367 | 0 | 0 | 100 | 100 |
| Sel16420 | 353 | 0 | 1 | 99.7 | 100 |
| Sel16483 | 362 | 0 | 0 | 100 | 100 |
| Sel16539 | 306 | 0 | 0 | 100 | 100 |
| Sel16773 | 334 | 17 | 25 | 93 | 95.1 |
| Sel16786 | 307 | 0 | 0 | 100 | 100 |
| Sel16795 | 253 | 0 | 1 | 99.6 | 100 |
| Sel17453 | 346 | 0 | 0 | 100 | 100 |
| Sele0104 | 267 | 0 | 0 | 100 | 100 |
| Sele0106 | 298 | 0 | 0 | 100 | 100 |
| Sele0107 | 267 | 8 | 8 | 97.1 | 97.1 |
| Sele0110 | 289 | 0 | 1 | 99.6 | 100 |
| Sele0111 | 301 | 0 | 0 | 100 | 100 |
| Sele0112 | 178 | 33 | 34 | 84 | 84.4 |
| Sele0114 | 209 | 3 | 3 | 98.6 | 98.6 |
| Sele0116 | 233 | 0 | 0 | 100 | 100 |
| Sele0121 | 476 | 0 | 0 | 100 | 100 |
| Sele0122 | 471 | 0 | 0 | 100 | 100 |
| Sele0124 | 370 | 1 | 2 | 99.5 | 99.7 |
| Sele0126 | 313 | 6 | 28 | 91.8 | 98.1 |
| Sele0129 | 202 | 0 | 0 | 100 | 100 |
| Sele0133 | 128 | 1 | 1 | 99.2 | 99.2 |
| Sele0136 | 242 | 1 | 1 | 99.6 | 99.6 |
| Sele0166 | 271 | 0 | 0 | 100 | 100 |
| Sele0170 | 299 | 0 | 0 | 100 | 100 |
| Sele0203 | 120 | 0 | 0 | 100 | 100 |
| Sele0210 | 354 | 0 | 0 | 100 | 100 |
| Sele0211 | 506 | 1 | 1 | 99.8 | 99.8 |
| Sele0303 | 347 | 0 | 1 | 99.7 | 100 |
| Sele0405 | 399 | 0 | 10 | 97.5 | 100 |
| Sele0406 | 216 | 0 | 0 | 100 | 100 |
| Sele0409 | 408 | 1 | 1 | 99.7 | 99.7 |
| Sele0411 | 383 | 0 | 1 | 99.7 | 100 |
| Sele0509 | 71 | 0 | 0 | 100 | 100 |
| Sele0603 | 278 | 1 | 1 | 99.6 | 99.6 |
| Sele0604 | 343 | 1 | 0 | 99.7 | 99.7 |
| Sele0606 | 458 | 0 | 1 | 99.8 | 100 |
| Sele0607 | 394 | 0 | 0 | 100 | 100 |
| Sele0609 | 373 | 1 | 1 | 99.7 | 99.7 |
| Sele0612 | 250 | 0 | 1 | 99.6 | 100 |
| Sele0704 | 353 | 1 | 12 | 96.7 | 99.7 |
| Sel30 | 336 | 0 | 0 | 100 | 100 |
| Sel31 | 348 | 2 | 1 | 99.7 | 99.4 |
| Sel32 | 360 | 0 | 0 | 100 | 100 |
| Sel33 | 174 | 0 | 0 | 100 | 100 |
| Sel34 | 256 | 1 | 0 | 100 | 99.6 |
| Sel35 | 70 | 0 | 0 | 100 | 100 |
| Sel36 | 141 | 6 | 8 | 94.6 | 95.9 |
| Sel37 | 190 | 1 | 1 | 99.5 | 99.5 |
| Sel38 | 164 | 0 | 0 | 100 | 100 |
| Sel39 | 377 | 0 | 0 | 100 | 100 |
| Sel40 | 300 | 0 | 1 | 99.7 | 100 |
| Sel41 | 301 | 0 | 0 | 100 | 100 |
| Sel42 | 412 | 0 | 0 | 100 | 100 |
| Sel43 | 448 | 1 | 2 | 99.5 | 99.7 |
| Sel44 | 352 | 0 | 0 | 100 | 100 |
| Sel45 | 309 | 1 | 1 | 99.6 | 99.6 |
| Sel46 | 84 | 1 | 0 | 100 | 98.8 |
| Sel47 | 294 | 0 | 0 | 100 | 100 |
| Sel48 | 453 | 1 | 1 | 99.8 | 99.8 |
| Sel49 | 216 | 0 | 0 | 100 | 100 |
| Sel50 | 103 | 0 | 0 | 100 | 100 |
| Sel51 | 249 | 1 | 1 | 99.6 | 99.6 |
| Sel52 | 468 | 1 | 0 | 100 | 99.8 |
| Sel17152 | 469 | 0 | 0 | 100 | 100 |
| Sel14046 | 410 | 0 | 0 | 100 | 100 |
| Sel14157 | 43 | 0 | 0 | 100 | 100 |
| Sel14172 | 217 | 1 | 1 | 99.5 | 99.5 |
| Sel15814 | 337 | 0 | 0 | 100 | 100 |
| 31,511 | 153 | 334 | 98.97 | 99.45 |
| Publication | Method | # Beats | Annotation (File Name) | SE [%] | Accuracy [%] | +P [%] |
|---|---|---|---|---|---|---|
| Proposed Algorithm | U-Net | 111,201 | Automatic.(pu) | 98.4 | 98.74 | 98.74 |
| Peimankar, Puthusserypady [45] | LSTM | 111,201 | Automatic.(pu) | 97.22 | 98.48 | N/R |
| Record | TP | FP | FN | SE (%) | +P (%) |
|---|---|---|---|---|---|
| Sel100 | 10 | 0 | 0 | 100 | 100 |
| Sel103 | 10 | 0 | 0 | 100 | 100 |
| Sel104 | 5 | 1 | 0 | 100 | 83 |
| Sel114 | 16 | 0 | 1 | 94.1 | 100 |
| Sel116 | 17 | 2 | 0 | 100 | 89.5 |
| Sel117 | 10 | 0 | 1 | 91 | 100 |
| Sel123 | 10 | 0 | 0 | 100 | 100 |
| Sel213 | 23 | 1 | 2 | 92 | 95.8 |
| Sel223 | 10 | 0 | 0 | 100 | 100 |
| Sel230 | 16 | 0 | 0 | 100 | 100 |
| Sel231 | 17 | 0 | 0 | 100 | 100 |
| Sel233 | 10 | 1 | 1 | 91 | 91 |
| Sel301 | 10 | 0 | 0 | 100 | 100 |
| Sel302 | 10 | 0 | 0 | 100 | 100 |
| Sel306 | 12 | 0 | 0 | 100 | 100 |
| Sel307 | 9 | 1 | 0 | 100 | 90 |
| Sel308 | 16 | 2 | 1 | 94.1 | 88.9 |
| Sel803 | 10 | 0 | 0 | 100 | 100 |
| Sel808 | 10 | 0 | 0 | 100 | 100 |
| Sel811 | 10 | 0 | 0 | 100 | 100 |
| Sel820 | 9 | 1 | 1 | 90 | 90 |
| Sel821 | 9 | 0 | 1 | 90 | 100 |
| Sel840 | 15 | 1 | 2 | 88.2 | 93.7 |
| Sel847 | 11 | 0 | 0 | 100 | 100 |
| Sel853 | 10 | 0 | 0 | 100 | 100 |
| Sel871 | 22 | 1 | 1 | 95.6 | 95.6 |
| Sel872 | 10 | 0 | 0 | 100 | 100 |
| Sel873 | 11 | 0 | 0 | 100 | 100 |
| Sel883 | 10 | 0 | 0 | 100 | 100 |
| Sel891 | 18 | 0 | 1 | 94.7 | 100 |
| Sel16265 | 10 | 0 | 0 | 100 | 100 |
| Sel16272 | 10 | 1 | 1 | 91 | 91 |
| Sel16273 | 10 | 0 | 0 | 100 | 100 |
| Sel16420 | 10 | 0 | 1 | 91 | 100 |
| Sel16483 | 10 | 0 | 0 | 100 | 100 |
| Sel16539 | 10 | 0 | 0 | 100 | 100 |
| Sel16773 | 8 | 2 | 1 | 89 | 80 |
| Sel16786 | 10 | 0 | 0 | 100 | 100 |
| Sel16795 | 10 | 0 | 0 | 100 | 100 |
| Sel17453 | 10 | 0 | 0 | 100 | 100 |
| Sele0104 | 10 | 0 | 0 | 100 | 100 |
| Sele0106 | 10 | 0 | 0 | 100 | 100 |
| Sele0107 | 9 | 2 | 3 | 75 | 82 |
| Sele0110 | 10 | 0 | 1 | 91 | 100 |
| Sele0111 | 10 | 0 | 0 | 100 | 100 |
| Sele0112 | 14 | 3 | 2 | 87.5 | 82.3 |
| Sele0114 | 10 | 0 | 0 | 100 | 100 |
| Sele0116 | 10 | 0 | 0 | 100 | 100 |
| Sele0121 | 10 | 0 | 0 | 100 | 100 |
| Sele0122 | 10 | 0 | 0 | 100 | 100 |
| Sele0124 | 17 | 1 | 0 | 100 | 94.4 |
| Sele0126 | 7 | 2 | 1 | 87.5 | 77.7 |
| Sele0129 | 10 | 0 | 0 | 100 | 100 |
| Sele0133 | 10 | 0 | 0 | 100 | 100 |
| Sele0136 | 7 | 1 | 1 | 87.5 | 87.5 |
| Sele0166 | 12 | 0 | 0 | 100 | 100 |
| Sele0170 | 10 | 0 | 0 | 100 | 100 |
| Sele0203 | 10 | 0 | 0 | 100 | 100 |
| Sele0210 | 10 | 0 | 0 | 100 | 100 |
| Sele0211 | 9 | 1 | 1 | 90 | 90 |
| Sele0303 | 10 | 0 | 1 | 91 | 100 |
| Sele0405 | 10 | 0 | 1 | 91 | 100 |
| Sele0406 | 9 | 1 | 0 | 100 | 90 |
| Sele0409 | 10 | 0 | 0 | 100 | 100 |
| Sele0411 | 10 | 0 | 0 | 100 | 100 |
| Sele0509 | 10 | 0 | 0 | 100 | 100 |
| Sele0603 | 9 | 1 | 1 | 91 | 91 |
| Sele0604 | 10 | 0 | 0 | 100 | 100 |
| Sele0606 | 6 | 3 | 1 | 85.7 | 66.6 |
| Sele0607 | 10 | 0 | 0 | 100 | 100 |
| Sele0609 | 8 | 1 | 1 | 88.8 | 88.8 |
| Sele0612 | 10 | 0 | 0 | 100 | 100 |
| Sele0704 | 10 | 0 | 0 | 100 | 100 |
| Sel30 | 10 | 0 | 0 | 100 | 100 |
| Sel31 | 10 | 0 | 0 | 100 | 100 |
| Sel32 | 10 | 0 | 0 | 100 | 100 |
| Sel33 | 10 | 0 | 0 | 100 | 100 |
| Sel34 | 9 | 0 | 0 | 100 | 100 |
| Sel36 | 1 | 0 | 0 | 100 | 100 |
| Sel38 | 10 | 0 | 0 | 100 | 100 |
| Sel39 | 10 | 0 | 0 | 100 | 100 |
| Sel40 | 9 | 0 | 1 | 91 | 100 |
| Sel41 | 10 | 0 | 0 | 100 | 100 |
| Sel42 | 10 | 0 | 0 | 100 | 100 |
| Sel43 | 8 | 1 | 0 | 100 | 88.8 |
| Sel44 | 8 | 0 | 0 | 100 | 100 |
| Sel45 | 8 | 0 | 0 | 100 | 100 |
| Sel46 | 8 | 0 | 0 | 100 | 100 |
| Sel47 | 10 | 0 | 0 | 100 | 100 |
| Sel48 | 10 | 0 | 0 | 100 | 100 |
| Sel49 | 10 | 0 | 0 | 100 | 100 |
| Sel51 | 9 | 0 | 1 | 90 | 100 |
| Sel52 | 10 | 0 | 0 | 100 | 100 |
| Sel17152 | 10 | 0 | 0 | 100 | 100 |
| Sel14046 | 10 | 0 | 1 | 91 | 100 |
| Sel14157 | 10 | 0 | 0 | 100 | 100 |
| Sel14172 | 16 | 0 | 0 | 100 | 100 |
| Sel15814 | 10 | 0 | 0 | 100 | 100 |
| 1027 | 31 | 32 | 97.24 | 97.22 |
| Publication | Method | # Beats | Annotation (File Name) | SE [%] | Accuracy [%] | +P [%] |
|---|---|---|---|---|---|---|
| Proposed Algorithm | U-Net | 3194 | Manual.(q1c) | 97.24 | 98.9 | 97.22 |
| Martinez et al. [35] | Wavelets | 3194 | Manual.(q1c) | 98.87 | N/R | 91.03 |
| Laguna et al. [39] | Low-pass differentiation | 3194 | Manual.(q1c) | 97.70 | N/R | 91.17 |
| Lenis, G. [37] | Wavelets | 3194 | Manual.(q1c) | 100 | N/R | 88.15 |
| Panigrahy [40] | Differential Evolution | 3194 | Manual.(q1c) | 98.9 | 98.5 | N/R |
| Abrishami, H. [44] | ConvNet | 3194 | Manual.(q1c) | N/R | 96.2 | N/R |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Costandy, R.N.; Gasser, S.M.; El-Mahallawy, M.S.; Fakhr, M.W.; Marzouk, S.Y. P-Wave Detection Using a Fully Convolutional Neural Network in Electrocardiogram Images. Appl. Sci. 2020, 10, 976. https://doi.org/10.3390/app10030976
Costandy RN, Gasser SM, El-Mahallawy MS, Fakhr MW, Marzouk SY. P-Wave Detection Using a Fully Convolutional Neural Network in Electrocardiogram Images. Applied Sciences. 2020; 10(3):976. https://doi.org/10.3390/app10030976
Chicago/Turabian StyleCostandy, Rana N., Safa M. Gasser, Mohamed S. El-Mahallawy, Mohamed W. Fakhr, and Samir Y. Marzouk. 2020. "P-Wave Detection Using a Fully Convolutional Neural Network in Electrocardiogram Images" Applied Sciences 10, no. 3: 976. https://doi.org/10.3390/app10030976
APA StyleCostandy, R. N., Gasser, S. M., El-Mahallawy, M. S., Fakhr, M. W., & Marzouk, S. Y. (2020). P-Wave Detection Using a Fully Convolutional Neural Network in Electrocardiogram Images. Applied Sciences, 10(3), 976. https://doi.org/10.3390/app10030976





