The Impact of Antimicrobial Substances on the Methanogenic Community during Methane Fermentation of Sewage Sludge and Cattle Slurry
Abstract
:1. Introduction
2. Materials and Methods
2.1. Substrates and Inoculum
2.2. Methane Fermentation
2.3. Genomic DNA Isolation from Digestate Samples
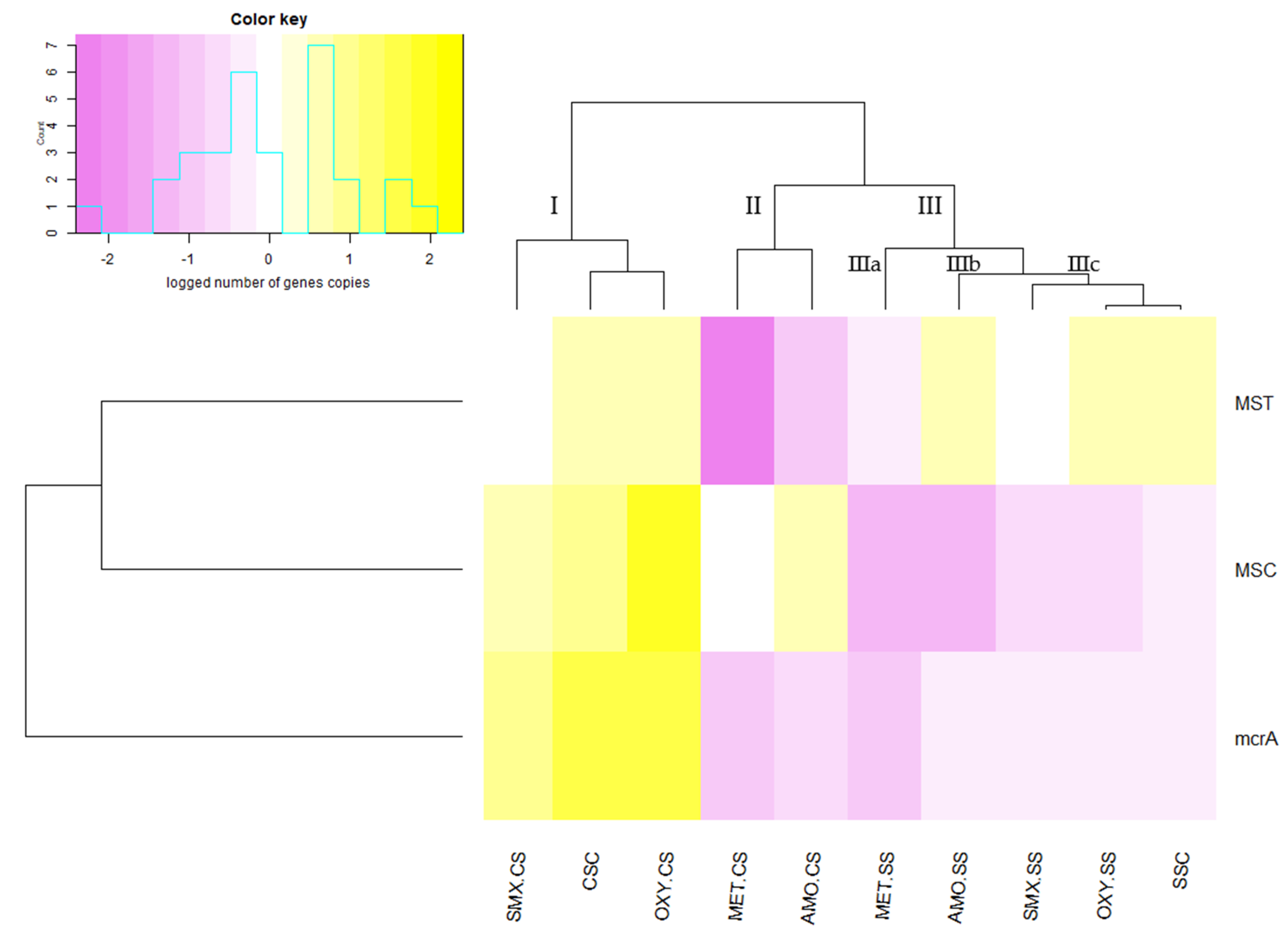
2.4. Analysis of the Genes mcrA, MSC and MSC by Quantitative Real-Time Polymerase Chain Reactions (qPCR)
2.5. Data Analysis
3. Results and Discussion
3.1. The Effect of Antimicrobials on Biogas Yield
3.2. Quantification of mcrA Gene
3.3. The Effects of Antimicrobials on Changes in the Diversity of Methanogenic Microorganisms
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Thompson, T.M.; Young, B.R.; Baroutian, S. Advances in the pretreatment of brown macroalgae for biogas production. Fuel Process. Technol. 2019, 195, 106151. [Google Scholar] [CrossRef]
- Achinas, S.; Achinas, V.; Euverink, G.J.W. A Technological Overview of Biogas Production from Biowaste. Engineering 2017, 3, 299–307. [Google Scholar] [CrossRef]
- Hijazi, O.; Munro, S.; Zerhusen, B.; Effenberger, M. Review of life cycle assessment for biogas production in Europe. Renew. Sustain. Energy Rev. 2016, 54, 1291–1300. [Google Scholar] [CrossRef]
- Gao, P.; Gu, C.; Wei, X.; Li, X.; Chen, H.; Jia, H.; Liu, Z.; Xue, G.; Ma, C. The role of zero valent iron on the fate of tetracycline resistance genes and class 1 integrons during thermophilic anaerobic co-digestion of waste sludge and kitchen waste. Water Res. 2017, 111, 92–99. [Google Scholar] [CrossRef]
- De Vrieze, J.; Saunders, A.M.; He, Y.; Fang, J.; Nielsen, P.H.; Verstraete, W.; Boon, N. Ammonia and temperature determine potential clustering in the anaerobic digestion microbiome. Water Res. 2015, 75, 312–323. [Google Scholar] [CrossRef]
- Czatzkowska, M.; Harnisz, M.; Korzeniewska, E.; Koniuszewska, I. Inhibitors of the methane fermentation process with particular emphasis on the microbiological aspect: A review. Energy Sci. Eng. 2020, 8, 1880–1897. [Google Scholar] [CrossRef] [Green Version]
- Cheng, D.; Feng, Y.; Liu, Y.; Xue, J.; Li, Z. Dynamics of xxytetracycline, sulfamerazine, and ciprofloxacin and related antibiotic resistance genes during swine manure composting. J. Environ. Manag. 2019, 230, 102–109. [Google Scholar] [CrossRef]
- Qiao, M.; Ying, G.; Singer, A.C.; Zhu, Y. Review of antibiotic resistance in China and its environment. Environ. Int. 2019, 111, 160–172. [Google Scholar] [CrossRef] [Green Version]
- Arikan, O.A.; Sikora, L.J.; Mulbry, W.; Khan, S.U.; Rice, C.; Foster, G.D. The fate and effect of oxytetracycline during the anaerobic digestion of manure from therapeutically treated calves. Process Biochem. 2006, 41, 1637–1643. [Google Scholar] [CrossRef]
- Lindsey, M.E.; Meyer, M.; Thurman, E.M. Analysis of trace levels of sulfonamide and tetracycline antimicrobials in groundwater and surface water using solid-phase extraction and liquid chromatography/mass spectrometry. Anal. Chem. 2001, 73, 4640–4646. [Google Scholar] [CrossRef]
- Lee, J.; Park, K.Y. Impact of hydrothermal pretreatment on anaerobic digestion efficiency for lignocellulosic biomass: Influence of pretreatment temperature on the formation of biomass-degrading byproducts. Chemosphere 2020, 256, 127116. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Han, W.; Chen, S.; Dong, W.; Qiao, M.; Hu, C.; Liu, B. Fifteen-Year Application of Manure and Chemical Fertilizers Differently Impacts Soil ARGs and Microbial Community Structure. Front. Microbiol. 2020, 11, 62. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Shi, Y.; Gao, L.; Liu, J.; Cai, Y. Occurrence, distribution and potential affecting factors of antibiotics in sewage sludge of wastewater treatment plants in China. Sci. Total Environ. 2013, 445–446, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Kumar, K.; Gupta, S.C.; Baidoo, S.K.; Chander, Y.; Rosen, C.J. Antibiotic Uptake by Plants from Soil Fertilized with Animal Manure. J. Environ. Qual. 2005, 34, 2082–2085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, E.Y.; Van Boeckel, T.P.; Martinez, E.M.; Pant, S.; Gandra, S.; Levin, S.A.; Goossens, H.; Laxminarayan, R. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc. Natl. Acad. Sci. USA 2018, 115, E3463–E3470. [Google Scholar] [CrossRef] [Green Version]
- Drlica, K.; Malik, M.; Kerns, R.J.; Zhao, X. Quinolone-mediated bacterial death. Antimicrob. Agents Chemother. 2008, 52, 385–392. [Google Scholar] [CrossRef] [Green Version]
- Detman, A.; Mielecki, D.; Pleśniak, Ł.; Bucha, M.; Janiga, M.; Matyasik, I.; Chojnacka, A.; Jȩdrysek, M.O.; Błaszczyk, M.K.; Sikora, A. Methane-yielding microbial communities processing lactate-rich substrates: A piece of the anaerobic digestion puzzle. Biotechnol. Biofuels 2018, 11, 116. [Google Scholar] [CrossRef]
- European Medicines Agency. Sales of Veterinary Antimicrobial Agents in 30 European Countries in 2015. Trends from 2010 to 2015, Seventh ESVAC Report. Available online: https://www.ema.europa.eu/en/documents/report/seventh-esvac-report-sales-veterinary-antimicrobial-agents-30-european-countries-2015_en.pdf (accessed on 10 November 2020).
- Gao, P.; Munir, M.; Xagoraraki, I. Correlation of tetracycline and sulfonamide antibiotics with corresponding resistance genes and resistant bacteria in a conventional municipal wastewater treatment plant. Sci. Total Environ. 2012, 421–422, 173–183. [Google Scholar] [CrossRef]
- Wei, R.C.; Ge, F.; Huang, S.Y.; Chen, M.; Wang, R. Occurrence of veterinary antibiotics in animal wastewater and surface water around farms in Jiangsu Province, China. Chemosphere 2011, 82, 1408–1414. [Google Scholar] [CrossRef]
- Zhao, L.; Dong, Y.H.; Wang, H. Residues of veterinary antibiotics in manures from feedlot livestock in eight provinces of China. Sci. Total Environ. 2010, 408, 1069–1075. [Google Scholar] [CrossRef]
- Martínez-Carballo, E.; González-Barreiro, C.; Scharf, S.; Gans, O. Environmental monitoring study of selected veterinary antibiotics in animal manure and soils in Austria. Environ. Pollut. 2007, 148, 570–579. [Google Scholar] [CrossRef] [PubMed]
- Kohanski, M.A.; Dwyer, D.J.; Collins, J.J. How antibiotics kill bacteria: From targets to networks. Nat. Rev. Microbiol. 2010, 8, 423–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmidt, T.; McCabe, B.K.; Harris, P.W.; Lee, S. Effect of trace element addition and increasing organic loading rates on the anaerobic digestion of cattle slaughterhouse wastewater. Bioresour. Technol. 2018, 264, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Aydin, S.; Ince, B.; Ince, O. Application of real-time PCR to determination of combined effect of antibiotics on Bacteria, Methanogenic Archaea, Archaea in anaerobic sequencing batch reactors. Water Res. 2015, 76, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yu, Y.; Lee, C. Thermo-alkaline pretreatment of waste activated sludge at low-temperatures: Effects on sludge disintegration, methane production, and methanogen community structure. Bioresour. Technol. 2013, 144, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Ji, J.-Y.; Xing, Y.-J.; Ma, Z.-T.; Zhang, M.; Zheng, P. Acute toxicity of pharmaceutical wastewaters containing antibiotics to anaerobic digestion treatment. Chemosphere 2013, 91, 1094–1098. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Contreras, C.; Vidal, G. Methanogenic toxicity evaluation of chlortetracycline hydrochloride. Electron. J. Biotechnol. 2015, 18, 445–450. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, S.M.; Ullman, J.L.; Teel, A.L.; Watts, R.J.; Frear, C. The effects of the antibiotics ampicillin, florfenicol, sulfamethazine, and tylosin on biogas production and their degradation efficiency during anaerobic digestion. Bioresour. Technol. 2013, 149, 244–252. [Google Scholar] [CrossRef]
- Coban, H.; Ertekin, E.; Ince, O.; Turker, G.; Akyol, Ç.; Ince, B. Degradation of oxytetracycline and its impacts on biogas-producing microbial community structure. Bioprocess Biosyst. Eng. 2016, 39, 1051–1060. [Google Scholar] [CrossRef]
- Spielmeyer, A. Occurrence and fate of antibiotics in manure during manure treatments: A short review. Sustain. Chem. Pharm. 2018, 9, 76–86. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, K. Effects of antibiotics on anaerobic digestion resistance genes and archaeal communities in cattle manure. J. Phys. Conf. Ser. 2020, 1549, 032076. [Google Scholar] [CrossRef]
- Bajkacz, S.; Felis, E.; Kycia-Słocka, E.; Harnisz, M.; Korzeniewska, E. Development of a new SLE-SPE-HPLC-MS/MS method for the determination of selected antibiotics and their transformation products in anthropogenically altered solid environmental matrices. Sci. Total Environ. 2020, 726, 138071. [Google Scholar] [CrossRef] [PubMed]
- Rusanowska, P.; Harnisz, M.; Zieliński, M.; Dębowski, M.; Korzeniewska, E.; Kisielewska, M.; Amenda, E. Individual and Synergistic Effects of Metronidazole, Amoxicillin, and Ciprofloxacin on Methane Fermentation with Sewage Sludge. Clean Soil Air Water 2019, 48, 1900281. [Google Scholar] [CrossRef]
- Yu, Y.; Kim, J.; Hwang, S. Use of real-time PCR for group-specific quantification of aceticlastic methanogens in anaerobic processes: Population dynamics and community structures. Biotechnol. Bioeng. 2006, 93, 424–433. [Google Scholar] [CrossRef]
- Denman, S.E.; Tomkins, N.W.; McSweeney, C.S. Quantitation and diversity analysis of ruminal methanogenic populations in response to the antimethanogenic compound bromochloromethane. FEMS Microbiol. Ecol. 2007, 62, 313–322. [Google Scholar] [CrossRef] [Green Version]
- Ward, J.H., Jr. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Rusanowska, P.; Zieliński, M.; Dębowski, M.; Harnisz, M.; Korzeniewska, E.; Amenda, E. Inhibition of Methane Fermentation by Antibiotics Introduced to Municipal Anaerobic Sludge. Proceedings 2018, 2, 1274. [Google Scholar] [CrossRef] [Green Version]
- Boxall, A.B.A. The environmental side effects of medication: How are human and veterinary medicines in soils and water bodies affecting human and environmental health? EMBO Rep. 2004, 5, 1110–1116. [Google Scholar] [CrossRef] [Green Version]
- Bound, J.P.; Voulvoulis, N. Household disposal of pharmaceuticals as a pathway for aquatic contamination in the United Kingdom. Environ. Health Perspect. 2005, 113, 1705–1711. [Google Scholar] [CrossRef] [Green Version]
- Loftin, K.A.; Henny, C.; Adams, C.D.; Surampali, R.; Mormile, M.R. Inhibition of microbial metabolism in anaerobic lagoons by selected sulfonamides, tetracyclines, lincomycin, and tylosin tartrate. Environ. Toxicol. Chem. 2005, 24, 782–788. [Google Scholar] [CrossRef]
- Sarmah, A.K.; Meyer, M.T.; Boxall, A.B.A. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere 2006, 65, 725–759. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Zhen, G.; Liu, Y.; Hojo, T.; Ledezma, A.; Li, Y. Long-term effect of the antibiotic cefalexin on methane production during waste activated sludge anaerobic digestion. Bioresour. Technol. 2014, 169, 644–651. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Du, K.; Yuan, R.; Chen, H.; Wang, F.; Zhou, B. Effects of sulfonamide antibiotics on digestion performance and microbial community during swine manure anaerobic digestion. Environ. Eng. Res. 2021, 26, 190471. [Google Scholar] [CrossRef]
- Sun, J.P.; Zheng, P.; Hu, B.L. Combined effect of antibiotics on anaerobic digestion of piggery wastewater. Huan Jing Ke Xue 2009, 30, 19–24. [Google Scholar]
- Lallai, A.; Mura, G.; Onnis, N. The effects of certain antibiotics on biogas production in the anaerobic digestion of pig waste slurry. Bioresour. Technol. 2002, 82, 205–208. [Google Scholar] [CrossRef]
- Hashemi, H.; Amin, M.; Ebrahimi, A.; Ebrahimi, A. Effects of oxytetracycline, tylosin, and amoxicillin antibiotics on specific methanogenic activity of anaerobic biomass. Int. J. Environ. Health Eng. 2012, 1, 37. [Google Scholar] [CrossRef]
- Awad, Y.M.; Kim, S.C.; Abd El-Azeem, S.A.M.; Kim, K.H.; Kim, K.R.; Kim, K.; Jeon, C.; Lee, S.S.; Ok, Y.S. Veterinary antibiotics contamination in water, sediment, and soil near a swine manure composting facility. Environ. Earth Sci. 2014, 71, 1433–1440. [Google Scholar] [CrossRef]
- Lueders, T.; Chin, K.J.; Conrad, R.; Friedrich, M. Molecular analyses of methyl-coenzyme M reductase α-subunit (mcrA) genes in rice field soil and enrichment cultures reveal the methanogenic phenotype of a novel archaeal lineage. Environ. Microbiol. 2001, 3, 194–204. [Google Scholar] [CrossRef]
- Lwin, K.O.; Matsui, H. Comparative analysis of the methanogen diversity in horse and pony by using mcrA gene and archaeal 16S rRNA Gene clone libraries. Archaea 2014, 83574, 483574. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.; Kim, J.; Hwang, K.; O’Flaherty, V.; Hwang, S. Quantitative analysis of methanogenic community dynamics in three anaerobic batch digesters treating different wastewaters. Water Res. 2009, 43, 157–165. [Google Scholar] [CrossRef]
- Ma, K.; Conrad, R.; Lu, Y. Responses of methanogen mcrA genes and their transcripts to an alternate dry/wet cycle of paddy field soil. Appl. Environ. Microbiol. 2012, 78, 445–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, T.; Kimura, M.; Asakawa, S. Distinct members of a stable methanogenic archaeal community transcribe mcrA genes under flooded and drained conditions in Japanese paddy field soil. Soil Biol. Biochem. 2009, 41, 276–285. [Google Scholar] [CrossRef]
- Alvarado, A.; Montañez-hernández, L.E.; Palacio-molina, S.L.; Oropeza-navarro, R.; Miriam, P. Microbial trophic interactions and mcrA gene expression in monitoring of anaerobic digesters. Front. Microbiol. 2014, 5, 597. [Google Scholar] [CrossRef] [PubMed]
- Stone, J.J.; Clay, S.A.; Zhu, Z.; Wong, K.L.; Porath, L.R.; Spellman, G.M. Effect of antimicrobial compounds tylosin and chlortetracycline during batch anaerobic swine manure digestion. Water Res. 2009, 43, 4740–4750. [Google Scholar] [CrossRef] [PubMed]
- De Vrieze, J.; Hennebel, T.; Van den Brande, J.; Bilad, R.M.; Bruton, T.A.; Vankelecom, I.F.J.; Verstraete, W.; Boon, N. Anaerobic digestion of molasses by means of a vibrating and non-vibrating submerged anaerobic membrane bioreactor. Biomass Bioenergy 2014, 68, 95–105. [Google Scholar] [CrossRef]
- McHugh, S.; O’Reilly, C.; Mahony, T.; Colleran, E.; O’Flaherty, V. Anaerobic granular sludge bioreactor technology. Rev. Environ. Sci. Biotechnol. 2003, 2, 225–245. [Google Scholar] [CrossRef]
- Sawayama, S.; Tsukahara, K.; Yagishita, T. Phylogenetic description of immobilized methanogenic community using real-time PCR in a fixed-bed anaerobic digester. Bioresour. Technol. 2006, 97, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Sundberg, C.; Al-Soud, W.A.; Larsson, M.; Alm, E.; Yekta, S.S.; Svensson, B.H.; Sørensen, S.J.; Karlsson, A. 454 Pyrosequencing Analyses of Bacterial and Archaeal Richness in 21 Full-Scale Biogas Digesters. FEMS Microbiol. Ecol. 2013, 85, 612–626. [Google Scholar] [CrossRef] [Green Version]
- Karakashev, D.; Batstone, D.J.; Angelidaki, I. Influence of Environmental Conditions on Methanogenic Compositions in Anaerobic Biogas Reactors. Appl. Environ. Microbiol. 2005, 71, 331–338. [Google Scholar] [CrossRef] [Green Version]
- Goberna, M.; Gadermaier, M.; García, C.; Wett, B.; Insam, H. Adaptation of Methanogenic Communities to the Cofermentation of Cattle Excreta and Olive Mill Wastes at 37 °C and 55 °C. Appl. Environ. Microbiol. 2010, 76, 6564–6571. [Google Scholar] [CrossRef] [Green Version]
- Wett, B.; Takács, I.; Batstone, D.; Wilson, C.; Murthy, S. Anaerobic model for high-solids or high-temperature digestion—Additional pathway of acetate oxidation. Water Sci. Technol. 2014, 69, 1634–1640. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, B.; Wright, A.D.G. Comparative metagenomic analysis of bacterial populations in three full-scale mesophilic anaerobic manure digesters. Appl. Microbiol. Biotechnol. 2014, 98, 2709–2717. [Google Scholar] [CrossRef] [PubMed]
- Werner, J.J.; Knights, D.; Garcia, M.L.; Scalfone, N.B.; Smith, S.; Yarasheski, K.; Cummings, T.A.; Beers, A.R.; Knight, R.; Angenent, L.T. Bacterial community structures are unique and resilient in full-scale bioenergy systems. Proc. Natl. Acad. Sci. USA 2011, 108, 4158–4163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ike, M.; Inoue, D.; Miyano, T.; Liu, T.T.; Sei, K.; Soda, S.; Kadoshin, S. Microbial population dynamics during startup of a full-scale anaerobic digester treating industrial food waste in Kyoto eco-energy project. Bioresour. Technol. 2010, 101, 3952–3957. [Google Scholar] [CrossRef] [PubMed]
- Demirel, B.; Scherer, P. Production of methane from sugar beet silage without manure addition by a single-stage anaerobic digestion process. Biomass Bioenergy 2008, 32, 203–209. [Google Scholar] [CrossRef]
- Degueurce, A.; Trémier, A.; Peu, P. Dynamic effect of leachate recirculation on batch mode solid state anaerobic digestion: Influence of recirculated volume, leachate to substrate ratio and recirculation periodicity. Bioresour. Technol. 2016, 216, 553–561. [Google Scholar] [CrossRef] [PubMed]
- Osińska, A.; Korzeniewska, E.; Harnisz, M.; Felis, E.; Bajkacz, S.; Jachimowicz, P.; Niestępski, S.; Konopka, I. Small-scale wastewater treatment plants as a source of the dissemination of antibiotic resistance genes in the aquatic environment. J. Hazard. Mater. 2020, 381, 121221. [Google Scholar] [CrossRef]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Okoh, A. Antibiotic use in agriculture and its consequential resistance in environmental sources: Potential public health implications. Molecules 2018, 23, 795. [Google Scholar] [CrossRef] [Green Version]
- An, J.; Chen, H.; Wei, S.; Gu, J. Antibiotic contamination in animal manure, soil, and sewage sludge in Shenyang, northeast China. Environ. Earth Sci. 2015, 74, 5077–5086. [Google Scholar] [CrossRef]
- Sui, Q.; Zhang, J.; Chen, M.; Tong, J.; Wang, R.; Wei, Y. Distribution of antibiotic resistance genes (ARGs) in anaerobic digestion and land application of swine wastewater. Environ. Pollut. 2016, 213, 751–759. [Google Scholar] [CrossRef]
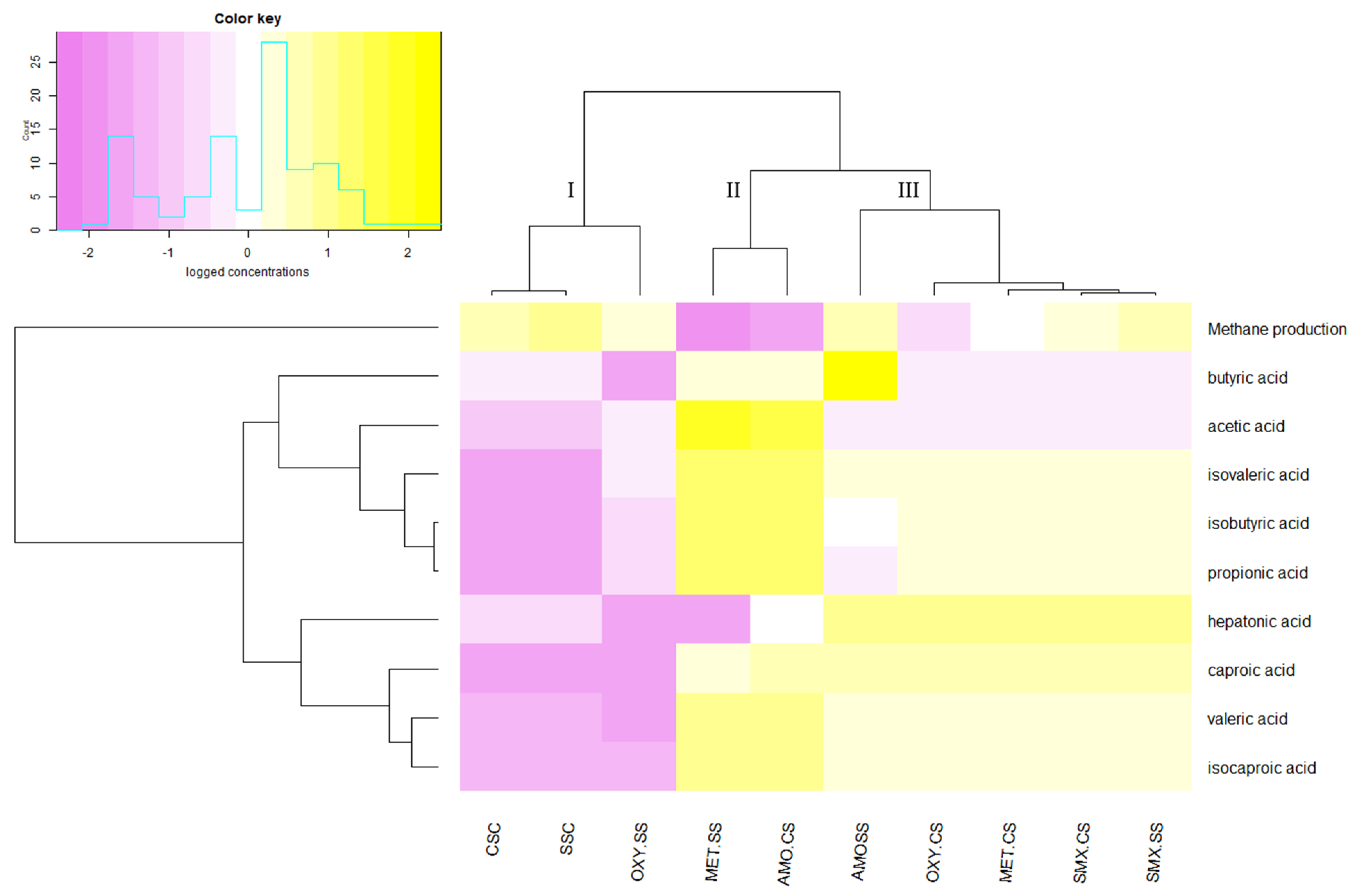


| TS a gD−1 b (mg) | VS c gD−1 (mg) | pH | TP d gTS−1 (mg) | TN e gTS−1 (mg) | |
|---|---|---|---|---|---|
| SS | 55.7 ± 1.5 | 42.8 ± 2.3 | 8.01 ± 0.4 | 0.6 ± 0.2 | 2.1 ± 0.4 |
| CS | 150.0 ± 10.8 | 123.1 ± 14.5 | 8.2 ± 0.5 | 1.6 ± 2.4 | 5.3 ± 2.8 |
| Inoculum | 38.8 ± 5.2 | 25.2 ± 3.8 | 8.1 ± 0.5 | 0.9 ± 0.4 | 5.5 ± 1.9 |
| Antibiotic Concentration (µg mL−1) | Substrate | CH4 * Production (L kg−1 VS) | CH4 Content in Biogas (%) | VFAs Concentration (g L−1) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Acetic Acid | Propionic Acid | Iso-Butyric Acid | Butyric Acid | Iso-Valeric Acid | Valeric Acid | Iso-Caproic Acid | Caproic Acid | Heptanoic Acid | ||||
| MET (512) | SS | 44.3 * ± 3.5 | 12.8 * ± 4.0 | 17.52 * ± 1.85 | 4.96 * ± 0.99 | 5.04 * ± 1.20 | 3.69 ± 0.79 | 5.89 * ± 1.02 | 2.73 * ± 0.84 | 4.36 * ± 0.78 | 0.67 * ± 0.11 | 0.01 ± 0.01 |
| CS | 143.4 * ± 44.0 | 70.7 ± 4.7 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| AMO (1024) | SS | 198.1 * ± 17.3 | 43.9 ± 2.7 | 0.83 ± 0.17 | 0.38 ± 0.13 | 0.44 ± 0.13 | 253.00 * ± 15.89 | 0.00 | 0.00 | 0.49 ± 0.09 | 0.00 | 0.00 |
| CS | 51.2 * ± 27.0 | 61.5 * ± 11.4 | 16.11 * ± 2.09 | 5.96 * ± 0.99 | 6.46 * ± 1.12 | 4.56 * ± 0.89 | 7.75 * ± 1.45 | 2.93 * ± 0.43 | 4.34 * ± 0.85 | 0.89 ± 0.29 | 0.23 ± 0.11 | |
| OXY (1024) | SS | 181.1 * ± 14.3 | 69.8 ± 0.5 | 0.95 ± 0.1 | 0.14 ± 0.09 | 0.12 ± 0.05 | 0.07 ± 0.04 | 0.53 ± 0.21 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.01 ± 0.01 |
| CS | 91.5 * ± 33.1 | 68.2 ± 5.4 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| SMX (512) | SS | 209.4 ± 3.2 | 70.2 ± 1.0 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| CS | 183.1 * ± 54.1 | 71.4 ± 3.9 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | |
| CONTROL | SSC | 272.8 ± 21.1 | 65.5 ± 2.3 | 0.26 ± 0.10 | 0.03 ± 0.03 | 0.03 ± 0.03 | 0.00 | 0.04 ± 0.03 | 0.02 ± 0.02 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.04 ± 0.02 |
| CSC | 201.2 ± 9.7 | 70.8 ± 3.5 | 0.26 ± 0.10 | 0.03 ± 0.03 | 0.03 ± 0.03 | 0.00 | 0.04 ± 0.03 | 0.02 ± 0.02 | 0.01 ± 0.01 | 0.01 ± 0.01 | 0.04 ± 0.02 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koniuszewska, I.; Czatzkowska, M.; Harnisz, M.; Korzeniewska, E. The Impact of Antimicrobial Substances on the Methanogenic Community during Methane Fermentation of Sewage Sludge and Cattle Slurry. Appl. Sci. 2021, 11, 369. https://doi.org/10.3390/app11010369
Koniuszewska I, Czatzkowska M, Harnisz M, Korzeniewska E. The Impact of Antimicrobial Substances on the Methanogenic Community during Methane Fermentation of Sewage Sludge and Cattle Slurry. Applied Sciences. 2021; 11(1):369. https://doi.org/10.3390/app11010369
Chicago/Turabian StyleKoniuszewska, Izabela, Małgorzata Czatzkowska, Monika Harnisz, and Ewa Korzeniewska. 2021. "The Impact of Antimicrobial Substances on the Methanogenic Community during Methane Fermentation of Sewage Sludge and Cattle Slurry" Applied Sciences 11, no. 1: 369. https://doi.org/10.3390/app11010369





