COVID-19 Lesion Segmentation Using Lung CT Scan Images: Comparative Study Based on Active Contour Models
Abstract
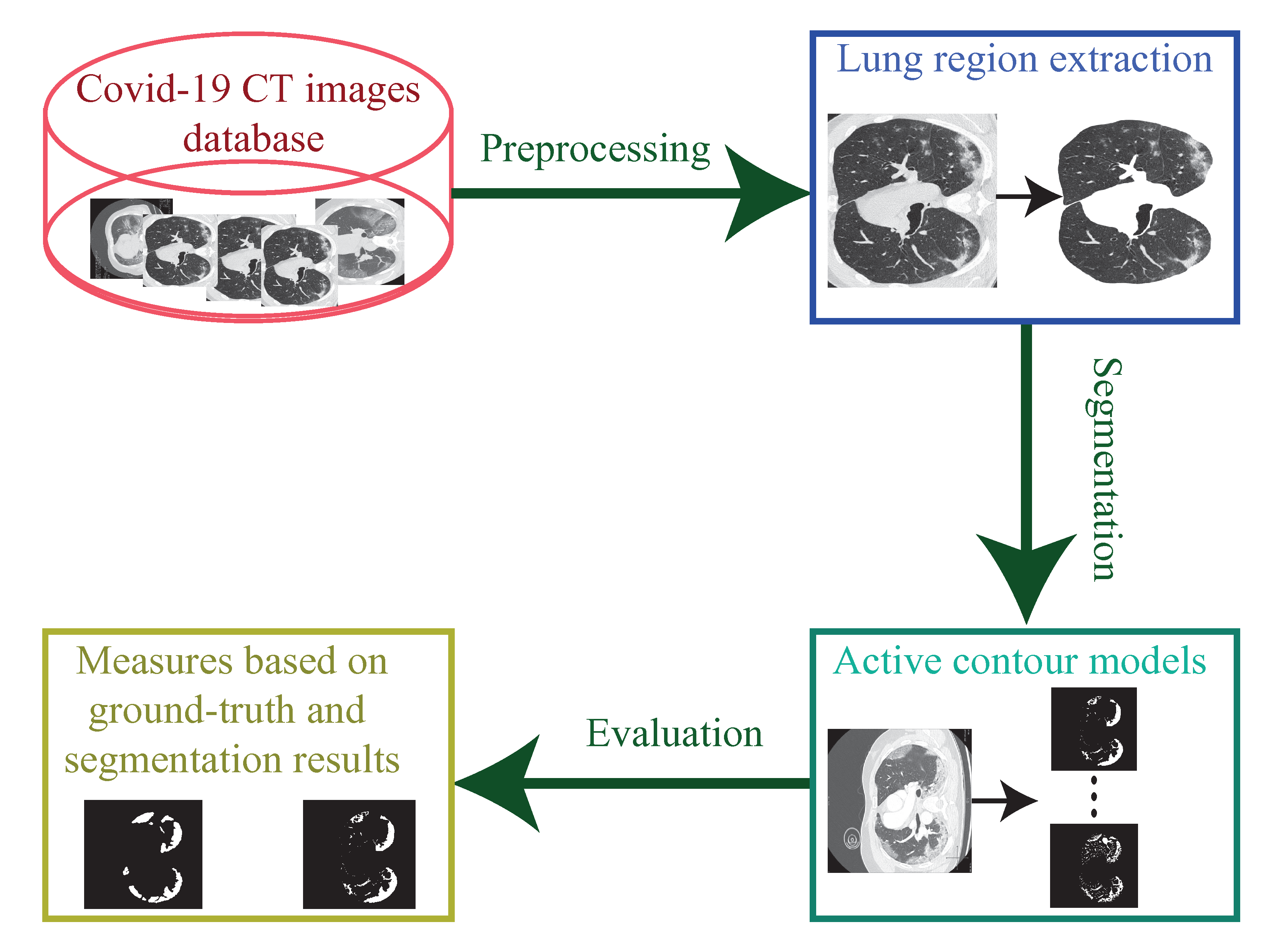
:1. Introduction
- A survey of active contour models: One of the most important aspects of detecting diseases such as pneumonia from medical images is identifying the region of infection. Although deep learning methods are well suited for this goal, if there are not enough images to train deep learning methods, the experimental results of the paper show that active contour methods achieve promising results. Therefore, our main contribution is to verify whether the active contour model-based image processing methods can be useful when only one image is available;
- Study on COVID-19 as a current topic: To the best of the authors’ knowledge, this paper is the first attempt to study active contour models and comparisons on images of the disease;
- Pointing out a line of research for the next researchers: We examine different methods and show which of them are effective for the topic and where the problems lie.
2. Methodology
2.1. Background of ACMS
2.1.1. Traditional ACM
2.1.2. Geometric Active Contours (Gac) Models
- High computational cost due to computation of gradient of the curvature approximation of the current level set at each iteration;
- Since the GAC model is in terms of the curvature and the gradient, only local boundary information is used. This leads to the GAC model being affected by input noise. This problem is shown in Figure 4.
2.1.3. The C-V Model
- Similar to GAC model, C-V model also needs to calculate the curvature approximation , which has a high computational expense;
- Despite power of global segmentation of the C-V model with a proper initial contour, it cannot extract the interior contour without setting the initial contour inside the object and fails to extract all the objects. This problem has been shown in Figure 5.
2.1.4. Deep Learning Approaches
2.2. The State-of-the-Art Methods
2.2.1. Magnetostatic Active Contour (MAC) Model
- Significant improvement in initialization invariance;
- Significant improvement in convergence capability; the contour attracts into deep concave regions;
- Is not affected by stationary point and saddle point problems;
- It is able to capture complex geometries;
- It is able to capture multiple objects with a single initial contour.
2.2.2. Online Region-Based Active Contour Model (Oracm)
- Decreasing efficiency without changing the accuracy of the image segmentation process;
- Accurate segmentation of all object regions in both inside and outside for medical, real, and synthetic images with holes, complex backgrounds, weak edges, and high noise.
2.2.3. Selective Binary and Gaussian Filtering Regularized Level Set (Sbgfrls)
- Robust against noise because the image statistical information is used to stop the curve evolution on the desired boundaries;
- Good performance on images with weak edges or even without edges;
- Initial curve can be defined anywhere to extract the interior boundaries of the objects
- Difficult to use ACM with SBGFRLS on different images because it needs to be tuned according to the image which is why it cannot be used on real-time video images;
- The slowness of the method caused by propagating the SPF function results in the boundary of the level set function only using at the level set. Updating only the boundary of the level set is the main cause of the slowness.
2.2.4. Level Set Active Contour Model (Lsacm)
2.2.5. Region-Scalable Fitting and Optimized Laplacian of Gaussian Energy (Rsfolge)
2.2.6. Adaptive Local-Fitting (Alf) Method
2.2.7. Fuzzy Region-Based Active Contour Model (Frbacm)
2.2.8. Global and Local Signed Energy-Based Pressure Force (Glsepf)
2.3. Database
3. Evaluation
3.1. Lung Region Extraction
3.2. Comparison the Active Contour Methods
3.3. Result
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rodriguez-Morales, A.J.; Cardona-Ospina, J.A.; Gutiérrez-Ocampo, E.; Villamizar-Peña, R.; Holguin-Rivera, Y.; Escalera-Antezana, J.P.; Alvarado-Arnez, L.E.; Bonilla-Aldana, D.K.; Franco-Paredes, C.; Henao-Martinez, A.F.; et al. Clinical, laboratory and imaging features of COVID-19: A systematic review and meta-analysis. Travel Med. Infect. Dis. 2020, 34, 101623. [Google Scholar] [CrossRef]
- Wu, Y.H.; Gao, S.H.; Mei, J.; Xu, J.; Fan, D.P.; Zhao, C.W.; Cheng, M.M. JCS: An Explainable COVID-19 Diagnosis System by Joint Classification and Segmentation. arXiv 2020, arXiv:2004.07054. [Google Scholar]
- Huang, Z.; Zhao, S.; Li, Z.; Chen, W.; Zhao, L.; Deng, L.; Song, B. The battle against coronavirus disease 2019 (COVID-19): Emergency management and infection control in a radiology department. J. Am. Coll. Radiol. 2020, 17, 710–716. [Google Scholar] [CrossRef] [PubMed]
- Ribbens, A.; Hermans, J.; Maes, F.; Vandermeulen, D.; Suetens, P. Unsupervised segmentation, clustering, and groupwise registration of heterogeneous populations of brain MR images. IEEE Trans. Med. Imaging 2013, 33, 201–224. [Google Scholar] [CrossRef] [PubMed]
- Gong, M.; Liang, Y.; Shi, J.; Ma, W.; Ma, J. Fuzzy c-means clustering with local information and kernel metric for image segmentation. IEEE Trans. Image Process. 2012, 22, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Kuo, J.W.; Mamou, J.; Aristizábal, O.; Zhao, X.; Ketterling, J.A.; Wang, Y. Nested graph cut for automatic segmentation of high-frequency ultrasound images of the mouse embryo. IEEE Trans. Med. Imaging 2015, 35, 427–441. [Google Scholar] [CrossRef]
- Li, G.; Chen, X.; Shi, F.; Zhu, W.; Tian, J.; Xiang, D. Automatic liver segmentation based on shape constraints and deformable graph cut in CT images. IEEE Trans. Image Process. 2015, 24, 5315–5329. [Google Scholar] [CrossRef]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: Munich, Germany, 2015; pp. 234–241. [Google Scholar]
- Zhang, K.; Zhang, L.; Lam, K.M.; Zhang, D. A level set approach to image segmentation with intensity inhomogeneity. IEEE Trans. Cybern. 2015, 46, 546–557. [Google Scholar] [CrossRef]
- Ding, K.; Xiao, L.; Weng, G. Active contours driven by region-scalable fitting and optimized Laplacian of Gaussian energy for image segmentation. Signal Process. 2017, 134, 224–233. [Google Scholar] [CrossRef]
- Ding, K.; Xiao, L. A Simple Method to improve Initialization Robustness for Active Contours driven by Local Region Fitting Energy. arXiv 2018, arXiv:1802.10437. [Google Scholar]
- Dong, B.; Weng, G.; Jin, R. Active contour model driven by Self Organizing Maps for image segmentation. Expert Syst. Appl. 2021, 177, 114948. [Google Scholar] [CrossRef]
- Liu, H.; Rashid, T.; Habes, M. Cerebral Microbleed Detection Via Fourier Descriptor with Dual Domain Distribution Modeling. In Proceedings of the 2020 IEEE 17th International Symposium on Biomedical Imaging Workshops (ISBI Workshops), Iowa City, IA, USA, 4 April 2020; pp. 1–4. [Google Scholar]
- Kim, W.; Kim, C. Active contours driven by the salient edge energy model. IEEE Trans. Image Process. 2012, 22, 1667–1673. [Google Scholar]
- Lecellier, F.; Fadili, J.; Jehan-Besson, S.; Aubert, G.; Revenu, M.; Saloux, E. Region-based active contours with exponential family observations. J. Math. Imaging Vis. 2010, 36, 28. [Google Scholar] [CrossRef] [Green Version]
- Zheng, C.; Deng, X.; Fu, Q.; Zhou, Q.; Feng, J.; Ma, H.; Liu, W.; Wang, X. Deep Learning-based Detection for COVID-19 from Chest CT using Weak Label. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Xu, Z.; Feng, J.; Jin, C.; Han, X.; Wu, H.; Shi, H. Longitudinal Assessment of COVID-19 Using a Deep Learning–based Quantitative CT Pipeline: Illustration of Two Cases. Radiol. Cardiothorac. Imaging 2020, 2, e200082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, L.; Han, R.; Ai, T.; Yu, P.; Kang, H.; Tao, Q.; Xia, L. Serial Quantitative Chest CT Assessment of COVID-19: Deep-Learning Approach. Radiol. Cardiothorac. Imaging 2020, 2, e200075. [Google Scholar] [CrossRef] [Green Version]
- Qi, X.; Jiang, Z.; Yu, Q.; Shao, C.; Zhang, H.; Yue, H.; Ma, B.; Wang, Y.; Liu, C.; Meng, X.; et al. Machine learning-based CT radiomics model for predicting hospital stay in patients with pneumonia associated with SARS-CoV-2 infection: A multicenter study. medRxiv 2020. [Google Scholar] [CrossRef]
- Gozes, O.; Frid-Adar, M.; Greenspan, H.; Browning, P.D.; Zhang, H.; Ji, W.; Bernheim, A.; Siegel, E. Rapid ai development cycle for the coronavirus (covid-19) pandemic: Initial results for automated detection & patient monitoring using deep learning ct image analysis. arXiv 2020, arXiv:2003.05037. [Google Scholar]
- Li, L.; Qin, L.; Xu, Z.; Yin, Y.; Wang, X.; Kong, B.; Bai, J.; Lu, Y.; Fang, Z.; Song, Q.; et al. Artificial intelligence distinguishes covid-19 from community acquired pneumonia on chest ct. Radiology 2020, 200905. [Google Scholar] [CrossRef]
- Jin, S.; Wang, B.; Xu, H.; Luo, C.; Wei, L.; Zhao, W.; Hou, X.; Ma, W.; Xu, Z.; Zheng, Z.; et al. AI-assisted CT imaging analysis for COVID-19 screening: Building and deploying a medical AI system in four weeks. medRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Shan, F.; Gao, Y.; Wang, J.; Shi, W.; Shi, N.; Han, M.; Xue, Z.; Shen, D.; Shi, Y. Lung infection quantification of covid-19 in ct images with deep learning. arXiv 2020, arXiv:2003.04655. [Google Scholar]
- Tang, L.; Zhang, X.; Wang, Y.; Zeng, X. Severe COVID-19 pneumonia: Assessing inflammation burden with volume-rendered chest CT. Radiol. Cardiothorac. Imaging 2020, 2, e200044. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.; Yu, N.; Cai, S.; Zhou, J.; Sheng, J.; Liu, K.; Zhou, H.; Guo, Y.; Niu, G. Quantitative computed tomography analysis for stratifying the severity of Coronavirus Disease 2019. J. Pharm. Anal. 2020, 10, 123–129. [Google Scholar] [CrossRef]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Panwar, H.; Gupta, P.; Siddiqui, M.K.; Morales-Menendez, R.; Bhardwaj, P.; Singh, V. A deep learning and grad-CAM based color visualization approach for fast detection of COVID-19 cases using chest X-ray and CT-Scan images. Chaos Solitons Fractals 2020, 140, 110190. [Google Scholar] [CrossRef]
- Sarker, L.; Islam, M.M.; Hannan, T.; Ahmed, Z. COVID-densenet: A deep learning architecture to detect covid-19 from chest radiology images; 2020; Preprints. [Google Scholar]
- Shi, F.; Wang, J.; Shi, J.; Wu, Z.; Wang, Q.; Tang, Z.; He, K.; Shi, Y.; Shen, D. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for covid-19. arXiv 2020, arXiv:2004.02731/. [Google Scholar]
- Islam, M.M.; Karray, F.; Alhajj, R.; Zeng, J. A review on deep learning techniques for the diagnosis of novel coronavirus (COVID-19). IEEE Access 2021, 9, 30551–30572. [Google Scholar] [CrossRef]
- Nayak, J.; Naik, B.; Dinesh, P.; Vakula, K.; Rao, B.K.; Ding, W.; Pelusi, D. Intelligent system for COVID-19 prognosis: A state-of-the-art survey. Appl. Intell. 2021, 51, 2908–2938. [Google Scholar] [CrossRef]
- Soomro, T.A.; Zheng, L.; Afifi, A.J.; Ali, A.; Yin, M.; Gao, J. Artificial intelligence (AI) for medical imaging to combat coronavirus disease (COVID-19): A detailed review with direction for future research. Artif. Intell. Rev. 2021, 1–31. Available online: https://link.springer.com/article/10.1007/s10462-021-09985-z (accessed on 23 August 2021).
- Jenssen, H.B. COVID-19 ct Segmentation Dataset. Available online: http://medicalsegmentation.com/covid19/ (accessed on 4 October 2020).
- Xu, T.; Cheng, I.; Mandal, M. An improved fluid vector flow for cavity segmentation in chest radiographs. In Proceedings of the 2010 20th International Conference on Pattern Recognition, Istanbul, Turkey, 23–26 August 2010; pp. 3376–3379. [Google Scholar]
- Ronfard, R. Region-based strategies for active contour models. Int. J. Comput. Vis. 1994, 13, 229–251. [Google Scholar] [CrossRef]
- Huang, R.; Pavlovic, V.; Metaxas, D.N. A graphical model framework for coupling MRFs and deformable models. In Proceedings of the 2004 IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Washington, DC, USA, 27 June–2 July 2004; Volume 2. [Google Scholar]
- Caselles, V.; Kimmel, R.; Sapiro, G. Geodesic active contours. Int. J. Comput. Vis. 1997, 22, 61–79. [Google Scholar] [CrossRef]
- Xie, X.; Mirmehdi, M. MAC: Magnetostatic active contour model. IEEE Trans. Pattern Anal. Mach. Intell. 2008, 30, 632–646. [Google Scholar] [CrossRef] [Green Version]
- Chan, T.F.; Vese, L.A. Active contours without edges. IEEE Trans. Image Process. 2001, 10, 266–277. [Google Scholar] [CrossRef] [Green Version]
- Talu, M.F. ORACM: Online region-based active contour model. Expert Syst. Appl. 2013, 40, 6233–6240. [Google Scholar] [CrossRef]
- Kass, M.; Witkin, A.; Terzopoulos, D. Snakes: Active contour models. Int. J. Comput. Vis. 1988, 1, 321–331. [Google Scholar] [CrossRef]
- Wang, T.; Cheng, I.; Basu, A. Fluid vector flow and applications in brain tumor segmentation. IEEE Trans. Biomed. Eng. 2009, 56, 781–789. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Cheng, H.D.; Huang, J.; Tian, J.; Tang, X.; Liu, J. Probability density difference-based active contour for ultrasound image segmentation. Pattern Recognit. 2010, 43, 2028–2042. [Google Scholar] [CrossRef]
- Cohen, L.D.; Cohen, I. Finite-element methods for active contour models and balloons for 2-D and 3-D images. IEEE Trans. Pattern Anal. Mach. Intell. 1993, 15, 1131–1147. [Google Scholar] [CrossRef] [Green Version]
- Sethian, J.A.; Sethian, J. Level Set Methods: Evolving Interfaces in Geometry, Fluid Mechanics, Computer Vision, and Materials Science; Cambridge University Press: Cambridge, UK, 1996; Volume 1999. [Google Scholar]
- Zhang, K.; Zhang, L.; Song, H.; Zhou, W. Active contours with selective local or global segmentation: A new formulation and level set method. Image Vis. Comput. 2010, 28, 668–676. [Google Scholar] [CrossRef]
- Chen, X.; Williams, B.M.; Vallabhaneni, S.R.; Czanner, G.; Williams, R.; Zheng, Y. Learning active contour models for medical image segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 15–20 June 2019; pp. 11632–11640. [Google Scholar]
- Marcos, D.; Tuia, D.; Kellenberger, B.; Zhang, L.; Bai, M.; Liao, R.; Urtasun, R. Learning deep structured active contours end-to-end. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 19–21 June 2018; pp. 8877–8885. [Google Scholar]
- Gur, S.; Wolf, L.; Golgher, L.; Blinder, P. Unsupervised microvascular image segmentation using an active contours mimicking neural network. In Proceedings of the IEEE International Conference on Computer Vision, Seoul, Korea, 27 October–2 November 2019; pp. 10722–10731. [Google Scholar]
- Rupprecht, C.; Huaroc, E.; Baust, M.; Navab, N. Deep active contours. arXiv 2016, arXiv:1607.05074. [Google Scholar]
- Ma, D.; Liao, Q.; Chen, Z.; Liao, R.; Ma, H. Adaptive local-fitting-based active contour model for medical image segmentation. Signal Process. Image Commun. 2019, 76, 201–213. [Google Scholar] [CrossRef]
- Fang, J.; Liu, H.; Zhang, L.; Liu, J.; Liu, H. Fuzzy region-based active contours driven by weighting global and local fitting energy. IEEE Access 2019, 7, 184518–184536. [Google Scholar] [CrossRef]
- Liu, H.; Fang, J.; Zhang, Z.; Lin, Y. A novel active contour model guided by global and local signed energy-based pressure force. IEEE Access 2020, 8, 59412–59426. [Google Scholar] [CrossRef]
- Rajinikanth, V.; Kadry, S.; Thanaraj, K.P.; Kamalanand, K.; Seo, S. Firefly-Algorithm Supported Scheme to Detect COVID-19 Lesion in Lung CT Scan Images using Shannon Entropy and Markov-Random-Field. arXiv 2020, arXiv:2004.09239. [Google Scholar]
- Qiu, Y.; Liu, Y.; Li, S.; Xu, J. Miniseg: An extremely minimum network for efficient covid-19 segmentation. arXiv 2020, arXiv:2004.09750. [Google Scholar]
- Howard, A.G.; Zhu, M.; Chen, B.; Kalenichenko, D.; Wang, W.; Weyand, T.; Andreetto, M.; Adam, H. Mobilenets: Efficient convolutional neural networks for mobile vision applications. arXiv 2017, arXiv:1704.04861. [Google Scholar]
- Fan, D.P.; Zhou, T.; Ji, G.P.; Zhou, Y.; Chen, G.; Fu, H.; Shen, J.; Shao, L. Inf-net: Automatic covid-19 lung infection segmentation from ct images. IEEE Trans. Med. Imaging 2020, 39, 2626–2637. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.C.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–12 September 2018; pp. 801–818. [Google Scholar]
- Tan, M.; Le, Q. Efficientnet: Rethinking model scaling for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; PMLR: Long Beach, CA, USA, 2019; pp. 6105–6114. [Google Scholar]
- Lo, S.Y.; Hang, H.M.; Chan, S.W.; Lin, J.J. Efficient dense modules of asymmetric convolution for real-time semantic segmentation. In Proceedings of the ACM Multimedia Asia; 2019; pp. 1–6. Available online: https://scholar.google.com/scholar?q=Efficient+dense+modules+of+asymmetric+convolution+for+real-time+semantic+segmentation&hl=en&as_sdt=0&as_vis=1&oi=scholart (accessed on 23 August 2021).
- Paszke, A.; Chaurasia, A.; Kim, S.; Culurciello, E. Enet: A deep neural network architecture for real-time semantic segmentation. arXiv 2016, arXiv:1606.02147. [Google Scholar]
- Mehta, S.; Rastegari, M.; Shapiro, L.; Hajishirzi, H. Espnetv2: A light-weight, power efficient, and general purpose convolutional neural network. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16-20 June 2019; pp. 9190–9200. [Google Scholar]








| Range of ages of patients | 32–86 (years) |
| No. of total patients (men + women + NA) | 49 (=27 + 11 + 11) |
| Minimum no. of COVID-19 CT images per patient | 1 |
| Maximum no. of COVID-19 CT images per patient | 13 |
| Total no. of COVID-19 CT images | 100 |
| Initial Contour | C-V | SBGFRLS | MAC | ORACM | LSACM | RSFOLGE | ALF | FRAGL | GLSEPF |
|---|---|---|---|---|---|---|---|---|---|
| Inside | Failed | Failed | Done | Done | Failed | Failed | Failed | Failed | Failed |
| Outside | Failed | Failed | Done | Done | Failed | Done | Done | Failed | Failed |
| Cross | Done | Failed | Done | Done | Done | Done | Failed | Done | Done |
| Measure | C-V | MAC | ORACM | LSACM | RSFOLGE | ALF | FRAGL | GLSEPF |
|---|---|---|---|---|---|---|---|---|
| Dice (%) | 93.55 | 95.94 | 96.30 | 95.77 | 89.88 | 92.12 | 96.44 | 95.60 |
| Jaccard (%) | 88.31 | 92.32 | 93.06 | 92.01 | 82.49 | 85.70 | 93.21 | 91.77 |
| Bfscore (%) | 66.82 | 61.40 | 74.13 | 60.46 | 63.50 | 57.05 | 65.55 | 71.96 |
| Precision (%) | 84.44 | 92.37 | 77.73 | 96.89 | 73.03 | 93.33 | 91.33 | 68.24 |
| Recall (%) | 58.15 | 48.25 | 72.41 | 45.67 | 61.44 | 43.34 | 53.18 | 78.81 |
| Iteration | 158 | 8500 | 5 | 200 | 250 | 8 | 10 | 30 |
| Time (s) | 55 | 700 | 1.4 | 12 | 41.40 | 100 | 1.8 | 5.5 |
| MiniSeg | MobileNet | Inf-Net | DeepLabv3+ | EfficientNet | EDANet | ENet | ESPNetv2 | GLSEPF (ACM) |
|---|---|---|---|---|---|---|---|---|
| 84.95 | 81.19 | 76.50 | 79.58 | 80.25 | 82.86 | 81.26 | 77.84 | 78.81 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Akbari, Y.; Hassen, H.; Al-Maadeed, S.; Zughaier, S.M. COVID-19 Lesion Segmentation Using Lung CT Scan Images: Comparative Study Based on Active Contour Models. Appl. Sci. 2021, 11, 8039. https://doi.org/10.3390/app11178039
Akbari Y, Hassen H, Al-Maadeed S, Zughaier SM. COVID-19 Lesion Segmentation Using Lung CT Scan Images: Comparative Study Based on Active Contour Models. Applied Sciences. 2021; 11(17):8039. https://doi.org/10.3390/app11178039
Chicago/Turabian StyleAkbari, Younes, Hanadi Hassen, Somaya Al-Maadeed, and Susu M. Zughaier. 2021. "COVID-19 Lesion Segmentation Using Lung CT Scan Images: Comparative Study Based on Active Contour Models" Applied Sciences 11, no. 17: 8039. https://doi.org/10.3390/app11178039
APA StyleAkbari, Y., Hassen, H., Al-Maadeed, S., & Zughaier, S. M. (2021). COVID-19 Lesion Segmentation Using Lung CT Scan Images: Comparative Study Based on Active Contour Models. Applied Sciences, 11(17), 8039. https://doi.org/10.3390/app11178039







