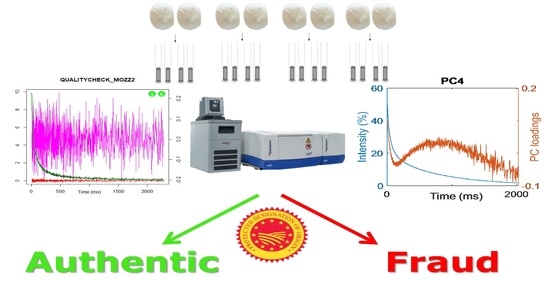
Spotting Frozen Curd in PDO Buffalo Mozzarella Cheese Through Insights on Its Supramolecular Structure Acquired by 1H TD-NMR Relaxation Experiments
Abstract
:Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Mozzarella Cheese Samples
2.2. Nuclear Magnetic Resonance (NMR) Relaxometry
2.3. Chemometric Analysis
3. Results and Discussion
3.1. T2 Relaxation
3.2. Quadratic Discriminant Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reid, L.M.; O’Donnell, C.P.; Downey, G. Recent technological advances for the determination of food authenticity. Trends Food Sci. Technol. 2006, 17, 344–353. [Google Scholar] [CrossRef]
- Danezis, G.P.; Tsagkaris, A.S.; Camin, F.; Brusic, V.; Georgiou, C.A. Food authentication: Techniques, trends & emerging approaches. TrAC Trends Anal. Chem. 2016, 85, 123–132. [Google Scholar] [CrossRef] [Green Version]
- Caira, S.; Pinto, G.; Nicolai, M.A.; Novi, G.; Addeo, F.; Scaloni, A. A non-canonical phosphorylation site in β-casein A from non-Mediterranean water buffalo makes quantifiable the adulteration of Italian milk with foreign material by combined isoelectrofocusing-immunoblotting procedures. Food Chem. 2019, 277, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Caira, S.; Pinto, G.; Nicolai, M.A.; Chianese, L.; Addeo, F. Simultaneously tracing the geographical origin and presence of bovine milk in Italian water buffalo Mozzarella cheese using MALDI-TOF data of casein signature peptides. Anal. Bioanal. Chem. 2016, 408, 5609–5621. [Google Scholar] [CrossRef] [PubMed]
- Di Luccia, A.; Picariello, G.; Trani, A.; Alviti, G.; Loizzo, P.; Faccia, M.; Addeo, F. Occurrence of β-casein fragments in cold-stored and curdled river buffalo (Bubalus bubalis L.) milk. J. Dairy Sci. 2009, 92, 1319–1329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manzo, N.; Biondi, L.; Nava, D.; Capuano, F.; Pizzolongo, F.; Fiore, A.; Romano, R. On the Possibility to Trace Frozen Curd in Buffalo Mozzarella Cheese. J. Food Res. 2017, 6, 104. [Google Scholar] [CrossRef] [Green Version]
- Mezzenga, R.; Schurtenberger, P.; Burbidge, A.; Michel, M. Understanding foods as soft materials. Nat. Mater. 2005, 4, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Kuo, M.-I.; Anderson, M.; Gunasekaran, S. Determining Effects of Freezing on Pasta Filata and Non-Pasta Filata Mozzarella Cheeses by Nuclear Magnetic Resonance Imaging. J. Dairy Sci. 2003, 86, 2525–2536. [Google Scholar] [CrossRef] [Green Version]
- Kuo, M.-I.; Gunasekaran, S.; Johnson, M.; Chen, C. Nuclear Magnetic Resonance Study of Water Mobility in Pasta Filata and Non-Pasta Filata Mozzarella. J. Dairy Sci. 2001, 84, 1950–1958. [Google Scholar] [CrossRef]
- Italian Ministry of Agriculure & Forestry. Disciplinary of Production of Mozzarella di Bufala Campana DOP, 11 February 2008, According to CE Regulation n. 1107. 12 June 1996. Available online: https://www.mozzarelladop.it/wp-content/uploads/2019/03/disciplinare_mozzarella_2008-2.pdf (accessed on 15 December 2020).
- Hills, B.; Takacs, S.; Belton, P. He effects of proteins on the proton N.M.R. transverse relaxation times of water I. Native bovine serum albumin. Mol. Phys. 1989, 67, 903–918. [Google Scholar] [CrossRef]
- Hills, B.; Takacs, S.; Belton, P. The effects of proteins on the proton N.M.R. transverse relaxation time of water—II. Mol. Phys. 1989, 67, 919–937. [Google Scholar] [CrossRef]
- Alinovi, M.; Corredig, M.; Mucchetti, G.; Carini, E. Water status and dynamics of high-moisture Mozzarella cheese as affected by frozen and refrigerated storage. Food Res. Int. 2020, 137, 109415. [Google Scholar] [CrossRef] [PubMed]
- Borgia, G.; Brown, R.; Fantazzini, P. Uniform-Penalty Inversion of Multiexponential Decay Data. J. Magn. Reson. 1998, 132, 65–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McMahon, N.J.; Fife, R.L.; Oberg, C.J. Water Partitioning in Mozzarella Cheese and Its Relationship to Cheese Meltability. J. Dairy Sci. 1999, 82, 1361–1369. [Google Scholar] [CrossRef]
- Gianferri, R.; D’Aiuto, V.; Curini, R.; Delfini, M.; Brosio, E. Proton NMR transverse relaxation measurements to study water dynamic states and age-related changes in Mozzarella di Bufala Campana cheese. Food Chem. 2007, 105, 720–726. [Google Scholar] [CrossRef]
- Vermeir, L.; Declerck, A.; To, C.M.; Kerkaert, B.; Van Der Meeren, P. Water and oil signal assignment in low-moisture mozzarella as determined by time-domain NMR T2 relaxometry. Magn. Reson. Chem. 2019, 57, 674–685. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.J.; Capozzi, F.; Cavani, C.; A Cremonini, M.; Petracci, M.; Placucci, G. Relationships between 1H NMR Relaxation Data and Some Technological Parameters of Meat: A Chemometric Approach. J. Magn. Reson. 2000, 147, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, M.; Capozzi, F.; Cremonini, M.A.; Laghi, L.; Petracci, M.; Placucci, G.; Cavani, C. Influence of the season on the relationships between NMR transverse relaxation data and water-holding capacity of turkey breast meat. J. Sci. Food Agric. 2004, 84, 1535–1540. [Google Scholar] [CrossRef]




| Class | 0% FC | 15% FC | 30% FC | 50% FC | Total |
|---|---|---|---|---|---|
| Number of Observations | |||||
| Training Set | 11 | 13 | 11 | 13 | 48 |
| Test Set | 3 | 3 | 3 | 3 | 12 |
| FC% | I0,1 (%) | T21 (ms) | I0,2 (%) | T22 (ms) | I0,3 (%) | T23 (ms) | I0,4 (%) | T24 (ms) | |
|---|---|---|---|---|---|---|---|---|---|
| Day 2 | 0 | 13.220 ± 2.187 a | 11.944 ± 1.846 a | 43.629 ± 2.740 a | 39.771 ± 3.856 a | 21.906 ± 2.147 a | 135.085 ± 13.323 a | 21.244 ± 2.862 a | 846.984 ± 47.347 a |
| 15 | 16.316 ± 1.667 b | 11.823 ± 1.676 a | 42.418 ± 2.974 a | 38.316 ± 4.485 a | 21.629 ± 0.994 a | 131.194 ± 16.175 a | 19.637 ± 2.180 a | 918.424 ± 168.591 a | |
| 30 | 14.054 ± 2.823 a | 11.277 ± 1.619 a | 43.439 ± 2.294 a | 36.629 ± 4.355 a | 21.541 ± 2.479 a | 127.584 ± 17.635 a | 20.966 ± 2.241 a | 705.069 ± 89.450 b | |
| 50 | 17.924 ± 2.410 b | 11.409 ± 0.725 a | 39.937 ± 1.260 b | 36.804 ± 1.989 a | 20.922 ± 1.767 a | 129.341 ± 9.782 a | 21.216 ± 1.243 a | 807.657 ± 177.846 a | |
| Day 6 | 0 | 12.503 ± 3.623 a | 10.680 ± 1.164 a | 38.806 ± 2.075 a | 34.153 ± 3.021 a | 19.300 ± 1.140 a | 114.991 ± 8.721 a | 29.390 ± 3.776 a | 682.550 ± 88.990 a |
| 15 | 11.556 ± 2.334 a | 11.054 ± 1.601 a | 37.320 ± 3.067 a | 36.040 ± 3.414 a | 21.626 ± 2.100 b | 120.579 ± 10.478 a | 29.498 ± 4.498 a | 661.462 ± 86.248 a | |
| 30 | 9.922 ± 2.180 a,b | 9.797 ± 1.624 a | 39.658 ± 2.067 a | 33.360 ± 1.955 a | 22.172 ± 2.121 b | 114.198 ± 6.283 a | 28.274 ± 3.213 a | 629.142 ± 46.564 a | |
| 50 | 9.091 ± 1.759 b | 9.523 ± 1.116 a | 38.290 ± 1.888 a | 33.366 ± 2.950 a | 22.907 ± 1.690 b | 113.300 ± 9.415 a | 29.711 ± 3.950 a | 542.335 ± 88.236 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mengucci, C.; Rabiti, D.; Urbinati, E.; Picone, G.; Romano, R.; Aiello, A.; Ferranti, P.; Capozzi, F. Spotting Frozen Curd in PDO Buffalo Mozzarella Cheese Through Insights on Its Supramolecular Structure Acquired by 1H TD-NMR Relaxation Experiments. Appl. Sci. 2021, 11, 1466. https://doi.org/10.3390/app11041466
Mengucci C, Rabiti D, Urbinati E, Picone G, Romano R, Aiello A, Ferranti P, Capozzi F. Spotting Frozen Curd in PDO Buffalo Mozzarella Cheese Through Insights on Its Supramolecular Structure Acquired by 1H TD-NMR Relaxation Experiments. Applied Sciences. 2021; 11(4):1466. https://doi.org/10.3390/app11041466
Chicago/Turabian StyleMengucci, Carlo, Davide Rabiti, Eleonora Urbinati, Gianfranco Picone, Raffaele Romano, Alessandra Aiello, Pasquale Ferranti, and Francesco Capozzi. 2021. "Spotting Frozen Curd in PDO Buffalo Mozzarella Cheese Through Insights on Its Supramolecular Structure Acquired by 1H TD-NMR Relaxation Experiments" Applied Sciences 11, no. 4: 1466. https://doi.org/10.3390/app11041466
APA StyleMengucci, C., Rabiti, D., Urbinati, E., Picone, G., Romano, R., Aiello, A., Ferranti, P., & Capozzi, F. (2021). Spotting Frozen Curd in PDO Buffalo Mozzarella Cheese Through Insights on Its Supramolecular Structure Acquired by 1H TD-NMR Relaxation Experiments. Applied Sciences, 11(4), 1466. https://doi.org/10.3390/app11041466