RNA-Seq Analysis of Gene Expression Changes Related to Delay of Flowering Time under Drought Stress in Tropical Maize
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Growth and Drought Conditions
2.2. Total RNA Isolation and RNA Sequence
2.3. Mapping and Differential Gene Expression Analysis
2.4. Gene Ontology Analysis and Functional Annotation
2.5. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Validation
3. Results
3.1. Changes in Anthesis–Silking Interval (ASI) Due to Drought Stress in Ki11
3.2. Analysis of Drought-Responsive Transcriptome
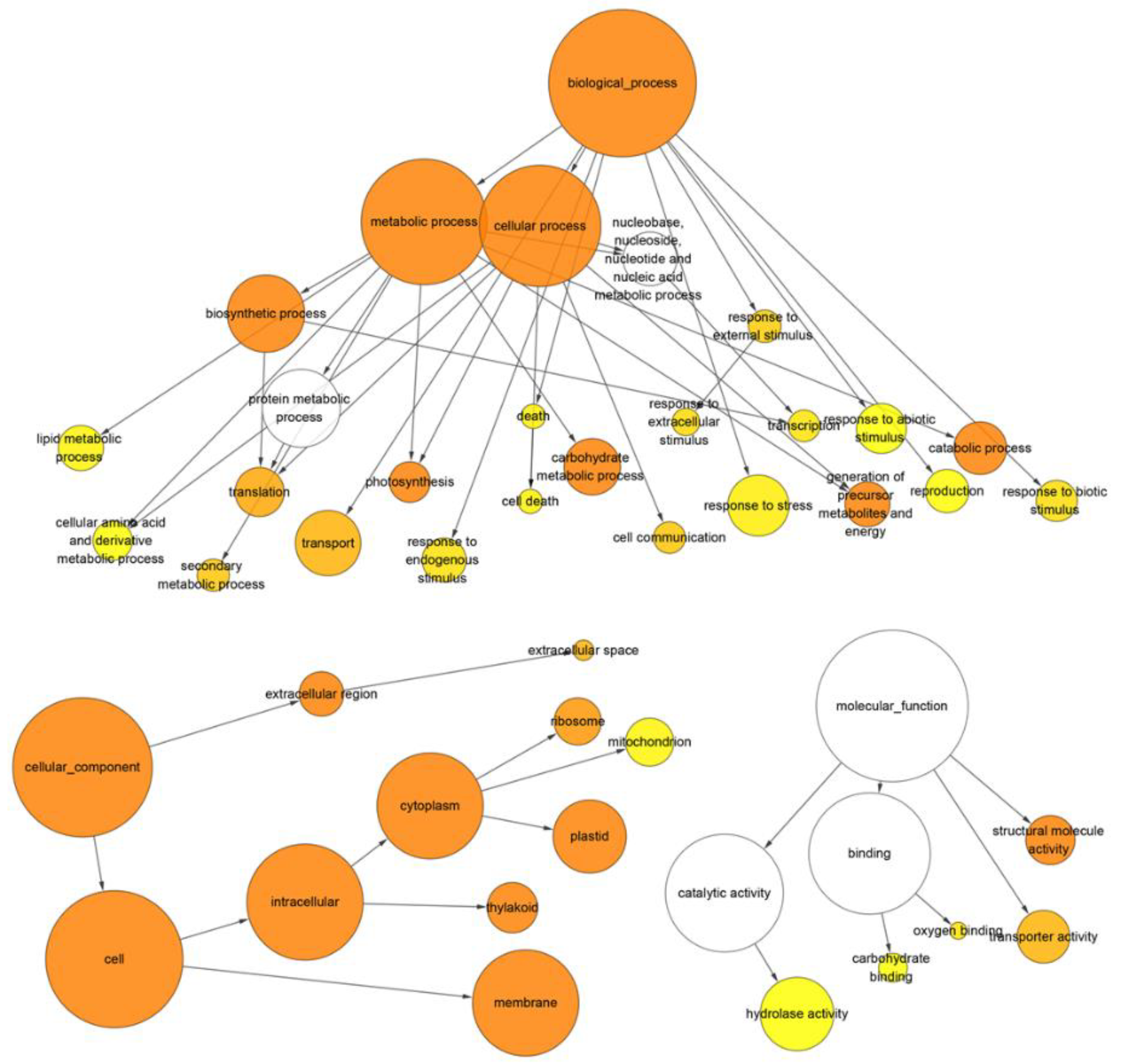
3.3. Gene Ontology Enrichment Analysis of Differentially Expressed Genes and Transcripts
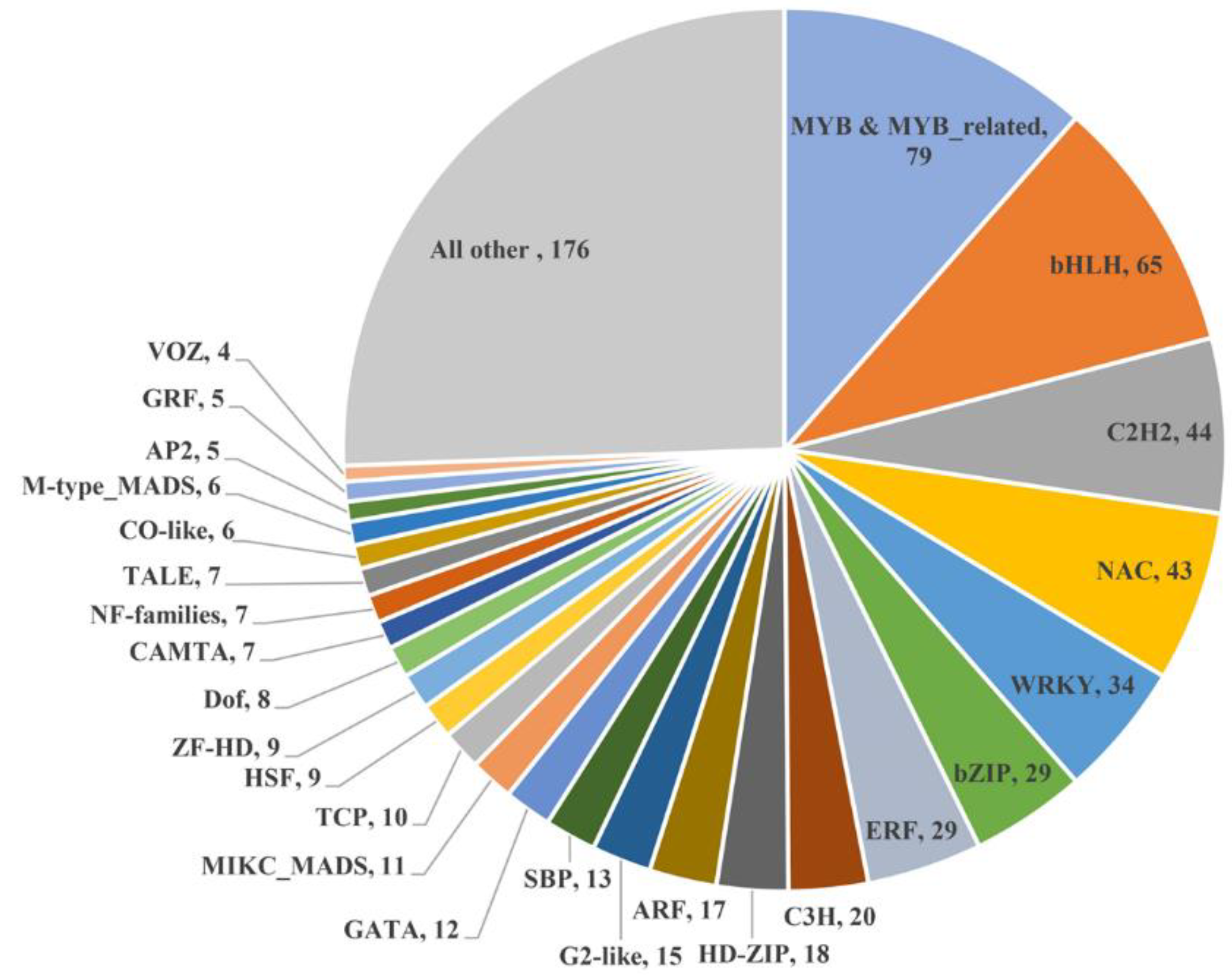
3.4. Transcription Factors That Mediate Drought Tolerance and Flowering Time
3.5. qRT-PCR Validation of Differentially Expressed Transcripts
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Cárcova, J.; Otegui, M.E. Ear temperature and pollination timing effects on maize kernel set. Crop Sci. 2001, 41, 1809–1815. [Google Scholar] [CrossRef]
- Fonseca, A.E.; Westgate, M.E. Relationship between desiccation and viability of maize pollen. Field Crop. Res. 2005, 94, 114–125. [Google Scholar] [CrossRef]
- Byrne, P.F.; Bolanos, J.; Edmeades, G.O.; Eaton, D.L. Gains from selection under drought versus multilocation testing in related tropical maize populations. Crop Sci. 1995, 35, 63–69. [Google Scholar] [CrossRef]
- Bolaños, J.; Edmeades, G.O. The importance of the anthesis-silking interval in breeding for drought tolerance in tropical maize. Field Crop. Res. 1996, 48, 65–80. [Google Scholar] [CrossRef]
- Edmeades, G.O.; Bänziger, M.; Elings, A.; Chapman, S.C.; Ribaut, J.-M. Recent Advances in Breeding for Drought Tolerance in Maize; Springer: Dordrecht, The Netherlands; Berlin, Germany, 1997. [Google Scholar]
- Edmeades, G.O.; Bolaños, J.; Chapman, S.C.; Lafitte, H.R.; Bänziger, M. Selection Improves Drought Tolerance in Tropical Maize Populations. Crop Sci. 1999, 39, 1315–1324. [Google Scholar] [CrossRef]
- Ziyomo, C.; Bernardo, R. Drought tolerance in maize: Indirect selection through secondary traits versus genomewide selection. Crop Sci. 2013, 53, 1269–1275. [Google Scholar] [CrossRef]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef]
- Yu, J.; Holland, J.B.; McMullen, M.D.; Buckler, E.S. Genetic design and statistical power of nested association mapping in maize. Genetics 2008, 178, 539–551. [Google Scholar] [CrossRef]
- Bari, R.; Jones, J.D.G. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. Proc. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef]
- Golldack, D.; Li, C.; Mohan, H.; Probst, N. Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 2014, 5, 151. [Google Scholar] [CrossRef]
- Marino, R.; Ponnaiah, M.; Krajewski, P.; Frova, C.; Gianfranceschi, L.; Pè, M.E.; Sari-Gorla, M. Addressing drought tolerance in maize by transcriptional profiling and mapping. Mol. Genet. Genom. 2009, 281, 163–179. [Google Scholar] [CrossRef]
- Ying, S.; Zhang, D.F.; Fu, J.; Shi, Y.S.; Song, Y.C.; Wang, T.Y.; Li, Y. Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta 2012, 235, 253–266. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, X.; Zhang, D.; Tang, H.; Sun, B.; Li, C.; Hao, L.; Liu, C.; Li, Y.; Shi, Y.; et al. Genome-wide identification of gene expression in contrasting maize inbred lines under field drought conditions reveals the significance of transcription factors in drought tolerance. PLoS ONE 2017, 12, e0179477. [Google Scholar] [CrossRef] [PubMed]
- Miao, Z.; Han, Z.; Zhang, T.; Chen, S.; Ma, C. A systems approach to a spatio-temporal understanding of the drought stress response in maize. Sci. Rep. 2017, 7, 6590. [Google Scholar] [CrossRef] [PubMed]
- Kakumanu, A.; Ambavaram, M.M.R.; Klumas, C.; Krishnan, A.; Batlang, U.; Myers, E.; Grene, R.; Pereira, A. Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol. 2012, 160, 846–867. [Google Scholar] [CrossRef] [PubMed]
- Staiger, D.; Brown, J.W.S. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar] [CrossRef] [PubMed]
- Opitz, N.; Paschold, A.; Marcon, C.; Malik, W.A.; Lanz, C.; Piepho, H.P.; Hochholdinger, F. Transcriptomic complexity in young maize primary roots in response to low water potentials. BMC Genom. 2014, 15, 741. [Google Scholar] [CrossRef]
- Seeve, C.M.; Cho, I.J.; Hearne, L.B.; Srivastava, G.P.; Joshi, T.; Smith, D.O.; Sharp, R.E.; Oliver, M.J. Water-deficit-induced changes in transcription factor expression in maize seedlings. Plant Cell Environ. 2017, 40, 686–701. [Google Scholar] [CrossRef]
- Buckler, E.S.; Holland, J.B.; Bradbury, P.J.; Acharya, C.B.; Brown, P.J.; Browne, C.; Ersoz, E.; Flint-Garcia, S.; Garcia, A.; Glaubitz, J.C.; et al. The genetic architecture of maize flowering time. Science 2009, 325, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.; Müller, A.E. Flowering time control and applications in plant breeding. Trends Plant Sci. 2009, 14, 563–573. [Google Scholar] [CrossRef]
- Thirunavukkarasu, N.; Hossain, F.; Arora, K.; Sharma, R.; Shiriga, K.; Mittal, S.; Mohan, S.; Namratha, P.M.; Dogga, S.; Rani, T.S.; et al. Functional mechanisms of drought tolerance in subtropical maize (Zea mays L.) identified using genome-wide association mapping. BMC Genom. 2014, 15, 1182. [Google Scholar] [CrossRef]
- Li, Y.X.; Li, C.; Bradbury, P.J.; Liu, X.; Lu, F.; Romay, C.M.; Glaubitz, J.C.; Wu, X.; Peng, B.; Shi, Y.; et al. Identification of genetic variants associated with maize flowering time using an extremely large multi-genetic background population. Plant J. 2016, 86, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Setter, T.L.; Yan, J.; Warburton, M.; Ribaut, J.M.; Xu, Y.; Sawkins, M.; Buckler, E.S.; Zhang, Z.; Gore, M.A. Genetic association mapping identifies single nucleotide polymorphisms in genes that affect abscisic acid levels in maize floral tissues during drought. J. Exp. Bot. 2011, 62, 701–716. [Google Scholar] [CrossRef]
- Hung, H.Y.; Shannon, L.M.; Tian, F.; Bradbury, P.J.; Chen, C.; Flint-Garcia, S.A.; McMullen, M.D.; Ware, D.; Buckler, E.S.; Doebley, J.F.; et al. ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc. Natl. Acad. Sci. USA 2012, 109, E1913–E1921. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Kim, H.C.; Shin, S.; Kim, K.H.; Moon, J.C.; Kim, J.Y.; Lee, B.M. Transcriptome analysis of flowering time genes under drought stress in maize leaves. Front. Plant Sci. 2017, 8, 267. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Muszynski, M.G.; Danilevskaya, O.N. The FT-like ZCN8 gene functions as a floral activator and is involved in photoperiod sensitivity in maize. Plant Cell 2011, 23, 942–960. [Google Scholar] [CrossRef]
- Lu, M.; Ying, S.; Zhang, D.F.; Shi, Y.S.; Song, Y.C.; Wang, T.Y.; Li, Y. A maize stress-responsive NAC transcription factor, ZmSNAC1, confers enhanced tolerance to dehydration in transgenic Arabidopsis. Plant Cell Rep. 2012, 31, 1701–1711. [Google Scholar] [CrossRef]
- Zhao, S.; Luo, Y.; Zhang, Z.; Xu, M.; Wang, W.; Zhao, Y.; Zhang, L.; Fan, Y.; Wang, L. ZmSOC1, an MADS-box transcription factor from Zea mays, promotes flowering in Arabidopsis. Int. J. Mol. Sci. 2014, 15, 19987–20003. [Google Scholar] [CrossRef]
- Alter, P.; Bircheneder, S.; Zhou, L.Z.; Schlüter, U.; Gahrtz, M.; Sonnewald, U.; Dresselhaus, T. Flowering time-regulated genes in maize include the transcription factor ZmMADS1. Plant Physiol. 2016, 172, 389–404. [Google Scholar] [CrossRef]
- Komiya, R.; Yokoi, S.; Shimamoto, K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development 2009, 136, 3443–3450. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Chen, H.; Ren, D.; Tang, H.; Qiu, R.; Feng, J.; Long, Y.; Niu, B.; Chen, D.; Zhong, T.; et al. Genetic interactions between diverged alleles of Early heading date 1 (Ehd1) and Heading date 3a (Hd3a)/ RICE FLOWERING LOCUS T1 (RFT1) control differential heading and contribute to regional adaptation in rice (Oryza sativa). New Phytol. 2015, 208, 936–948. [Google Scholar] [CrossRef]
- Hayama, R.; Yokoi, S.; Tamaki, S.; Yano, M.; Shimamoto, K. Adaptation of photoperiodic control pathways produces short-day flowering in rice. Nature 2003, 422, 719–722. [Google Scholar] [CrossRef] [PubMed]
- Itoh, H.; Nonoue, Y.; Yano, M.; Izawa, T. A pair of floral regulators sets critical day length for Hd3a florigen expression in rice. Nat. Genet. 2010, 42, 635–638. [Google Scholar] [CrossRef] [PubMed]
- Goretti, D.; Martignago, D.; Landini, M.; Brambilla, V.; Gómez-Ariza, J.; Gnesutta, N.; Galbiati, F.; Collani, S.; Takagi, H.; Terauchi, R.; et al. Transcriptional and Post-transcriptional Mechanisms Limit Heading Date 1 (Hd1) Function to Adapt Rice to High Latitudes. PLoS Genet. 2017, 13, e1006530. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Jin, M.; Zheng, X.M.; Chen, J.; Yuan, D.; Xin, Y.; Wang, M.; Huang, D.; Zhang, Z.; Zhou, K.; et al. Days to heading 7, a major quantitative locus determining photoperiod sensitivity and regional adaptation in rice. Proc. Natl. Acad. Sci. USA 2014, 111, 16337–16342. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Ariza, J.; Galbiati, F.; Goretti, D.; Brambilla, V.; Shrestha, R.; Pappolla, A.; Courtois, B.; Fornara, F. Loss of floral repressor function adapts rice to higher latitudes in Europe. J. Exp. Bot. 2015, 66, 2027–2039. [Google Scholar] [CrossRef]
- Colasanti, J.; Yuan, Z.; Sundaresan, V. The indeterminate gene encodes a zinc finger protein and regulates a leaf-generated signal required for the transition to flowering in maize. Cell 1998, 93, 593–603. [Google Scholar] [CrossRef]
- Wong, A.Y.M.; Colasanti, J. Maize floral regulator protein INDETERMINATE1 is localized to developing leaves and is not altered by light or the sink/source transition. J. Exp. Bot. 2007, 58, 403–414. [Google Scholar] [CrossRef]
- Danilevskaya, O.N.; Meng, X.; Hou, Z.; Ananiev, E.V.; Simmons, C.R. A genomic and expression compendium of the expanded PEBP gene family from maize. Plant Physiol. 2008, 146, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Muszynski, M.G.; Dam, T.; Li, B.; Shirbroun, D.M.; Hou, Z.; Bruggemann, E.; Archibald, R.; Ananiev, E.V.; Danilevskaya, O.N. delayed flowering1 encodes a basic leucine zipper protein that mediates floral inductive signals at the shoot apex in maize. Plant Physiol. 2006, 142, 1523–1536. [Google Scholar] [CrossRef]
- Miller, T.A.; Muslin, E.H.; Dorweiler, J.E. A maize CONSTANS-like gene, conz1, exhibits distinct diurnal expression patterns in varied photoperiods. Planta 2008, 227, 1377–1388. [Google Scholar] [CrossRef] [PubMed]
- Colasanti, J.; Coneva, V. Mechanisms of floral induction in grasses: Something borrowed, something new. Plant Physiol. 2009, 149, 56–62. [Google Scholar] [CrossRef]
- Bouchet, S.; Servin, B.; Bertin, P.; Madur, D.; Combes, V.; Dumas, F.; Brunel, D.; Laborde, J.; Charcosset, A.; Nicolas, S. Adaptation of Maize to Temperate Climates: Mid-Density Genome-Wide Association Genetics and Diversity Patterns Reveal Key Genomic Regions, with a Major Contribution of the Vgt2 (ZCN8) Locus. PLoS ONE 2013, 8, e71377. [Google Scholar] [CrossRef]
- Jiang, T.; Fountain, J.; Davis, G.; Kemerait, R.; Scully, B.; Lee, R.D.; Guo, B. Root Morphology and Gene Expression Analysis in Response to Drought Stress in Maize (Zea mays). Plant Mol. Biol. Report. 2012, 30, 360–369. [Google Scholar] [CrossRef]
- Hansey, C.N.; Vaillancourt, B.; Sekhon, R.S.; de Leon, N.; Kaeppler, S.M.; Buell, C.R. Maize (Zea mays L.) genome diversity as revealed by rna-sequencing. PLoS ONE 2012, 7, e33071. [Google Scholar] [CrossRef]
- Udomprasert, N.; Kijjanon, J.; Chusri-Iam, K.; Machuay, A. Effects of Water Deficit at Tasseling on Photosynthesis, Development, and Yield of Corn. Kasetsart J. Nat. Sci. 2005, 39, 546–551. [Google Scholar]
- Song, K.; Kim, K.-H.; Kim, H.C.; Moon, J.-C.; Kim, J.Y.; Baek, S.-B.; Kwon, Y.-U.; Lee, B.-M. Evaluation of Drought Tolerance in Maize Seedling using Leaf Rolling. Korean J. Crop Sci. 2015, 60, 8–16. [Google Scholar] [CrossRef]
- Kim, H.C.; Moon, J.-C.; Kim, J.Y.; Song, K.; Kim, K.-H.; Lee, B.-M. Evaluation of Drought Tolerance using Anthesis-silking Interval in Maize. Korean J. Crop Sci. 2017, 62, 24–31. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef]
- Benjamini, Y.; Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001, 29, 1165–1188. [Google Scholar] [CrossRef]
- Anders, S.; McCarthy, D.J.; Chen, Y.; Okoniewski, M.; Smyth, G.K.; Huber, W.; Robinson, M.D. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 2013, 8, 1765–1786. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S. Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int. J. Plant Genom. 2008, 2008, 619832. [Google Scholar] [CrossRef]
- Maere, S.; Heymans, K.; Kuiper, M. BiNGO: A Cytoscape plugin to assess overrepresentation of Gene Ontology categories in Biological Networks. Bioinformatics 2005, 21, 3448–3449. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Edmeades, G.O.; Bolanos, J.; Hernandez, M.; Bello, S. Causes for silk delay in a lowland tropical maize population. Crop Sci. 1993, 33, 1029–1035. [Google Scholar] [CrossRef]
- Thatcher, S.R.; Danilevskaya, O.N.; Meng, X.; Beatty, M.; Zastrow-Hayes, G.; Harris, C.; Van Allen, B.; Habben, J.; Li, B. Genome-wide analysis of alternative splicing during development and drought stress in maize1[OPEN]. Plant Physiol. 2016, 170, 586–599. [Google Scholar] [CrossRef]
- Brambilla, V.; Gomez-Ariza, J.; Cerise, M.; Fornara, F. The importance of being on time: Regulatory networks controlling photoperiodic flowering in cereals. Front. Plant Sci. 2017, 8, 665. [Google Scholar] [CrossRef]
- Tadege, M.; Sheldon, C.C.; Helliwell, C.A.; Upadhyaya, N.M.; Dennis, E.S.; Peacock, W.J. Reciprocal control of flowering time by OsSOC1 in transgenic Arabidopsis and by FLC in transgenic rice. Plant Biotechnol. J. 2003, 1, 361–369. [Google Scholar] [CrossRef]
- Lee, S.; Kim, J.; Han, J.J.; Han, M.J.; An, G. Functional analyses of the flowering time gene OsMADS50, the putative Suppressor of Overexpression of CO 1/Agamous-Like 20 (SOC1/AGL20) ortholog in rice. Plant J. 2004, 38, 754–764. [Google Scholar] [CrossRef]
- Lazakis, C.M.; Coneva, V.; Colasanti, J. ZCN8 encodes a potential orthologue of Arabidopsis FT florigen that integrates both endogenous and photoperiod flowering signals in maize. J. Exp. Bot. 2011, 62, 4833–4842. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yuan, Y.; Xu, Y.; Zhang, G.; Guo, X.; Wu, F.; Wang, Q.; Rong, T.; Pan, G.; Cao, M.; et al. Identification of candidate genes for drought tolerance by whole-genome resequencing in maize. BMC Plant. Biol. 2014, 14, 83. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Rabara, R.C.; Rushton, P.J. A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 2014, 239, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef]
- Han, Y.; Zhang, X.; Wang, Y.; Ming, F. The suppression of WRKY44 by GIGANTEA-miR172 pathway is involved in drought response of Arabidopsis thaliana. PLoS ONE 2013, 8, e73541. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant. Biol. 2006, 57, 781–803. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Seo, P.J.; Kang, S.K.; Park, C.M. miR172 signals are incorporated into the miR156 signaling pathway at the SPL3/4/5 genes in Arabidopsis developmental transitions. Plant. Mol. Biol. 2011, 76, 35–45. [Google Scholar] [CrossRef]
- Wahl, V.; Ponnu, J.; Schlereth, A.; Arrivault, S.; Langenecker, T.; Franke, A.; Feil, R.; Lunn, J.E.; Stitt, M.; Schmid, M. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science 2013, 339, 704–707. [Google Scholar] [CrossRef] [PubMed]
- Hyun, Y.; Richter, R.; Vincent, C.; Martinez-Gallegos, R.; Porri, A.; Coupland, G. Multi-layered Regulation of SPL15 and Cooperation with SOC1 Integrate Endogenous Flowering Pathways at the Arabidopsis Shoot Meristem. Dev. Cell 2016, 37, 254–266. [Google Scholar] [CrossRef] [PubMed]
- Coneva, V.; Zhu, T.; Colasanti, J. Expression differences between normal and indeterminate1 maize suggest downstream targets of ID1, a floral transition regulator in maize. J. Exp. Bot. 2007, 58, 3679–3693. [Google Scholar] [CrossRef]
- Mascheretti, I.; Turner, K.; Brivio, R.S.; Hand, A.; Colasanti, J.; Rossi, V. Florigen-encoding genes of day-neutral and photoperiod-sensitive maize are regulated by different chromatin modifications at the floral transition. Plant. Physiol. 2015, 168, 1351–1363. [Google Scholar] [CrossRef]
- Kojima, S.; Takahashi, Y.; Kobayashi, Y.; Monna, L.; Sasaki, T.; Araki, T.; Yano, M. Hd3a, a rice ortholog of the Arabidopsis FT gene, promotes transition to flowering downstream of Hd1 under short-day conditions. Plant. Cell Physiol. 2002, 43, 1096–1105. [Google Scholar] [CrossRef]
- Ito, S.; Song, Y.H.; Josephson-Day, A.R.; Miller, R.J.; Breton, G.; Olmstead, R.G.; Imaizumi, T. FLOWERING BHLH transcriptional activators control expression of the photoperiodic flowering regulator CONSTANS in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 3582–3587. [Google Scholar] [CrossRef]
- Putterill, J.; Robson, F.; Lee, K.; Simon, R.; Coupland, G. The CONSTANS gene of arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 1995, 80, 847–857. [Google Scholar] [CrossRef]


| Days to Anthesis | SD 1 of DA 2 | Days to Silking | SD of DS 3 | ASI 4 | |
|---|---|---|---|---|---|
| Control | 83.38 | ±2.26 | 85.88 | ±2.29 | 2.50 |
| Drought | 84.80 | ±3.07 | 91.40 | ±4.67 | 6.60 *** |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, K.-H.; Song, K.; Park, J.-M.; Kim, J.-Y.; Lee, B.-M. RNA-Seq Analysis of Gene Expression Changes Related to Delay of Flowering Time under Drought Stress in Tropical Maize. Appl. Sci. 2021, 11, 4273. https://doi.org/10.3390/app11094273
Kim K-H, Song K, Park J-M, Kim J-Y, Lee B-M. RNA-Seq Analysis of Gene Expression Changes Related to Delay of Flowering Time under Drought Stress in Tropical Maize. Applied Sciences. 2021; 11(9):4273. https://doi.org/10.3390/app11094273
Chicago/Turabian StyleKim, Kyung-Hee, Kitae Song, Jeong-Min Park, Jae-Yoon Kim, and Byung-Moo Lee. 2021. "RNA-Seq Analysis of Gene Expression Changes Related to Delay of Flowering Time under Drought Stress in Tropical Maize" Applied Sciences 11, no. 9: 4273. https://doi.org/10.3390/app11094273
APA StyleKim, K.-H., Song, K., Park, J.-M., Kim, J.-Y., & Lee, B.-M. (2021). RNA-Seq Analysis of Gene Expression Changes Related to Delay of Flowering Time under Drought Stress in Tropical Maize. Applied Sciences, 11(9), 4273. https://doi.org/10.3390/app11094273







