Kinetic Analysis Misinterpretations Due to the Occurrence of Enzyme Inhibition by Reaction Product: Comparison between Initial Velocities and Reaction Time Course Methodologies
Abstract
:Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Simulated Data for the Graphical Representation of Product Versus Time
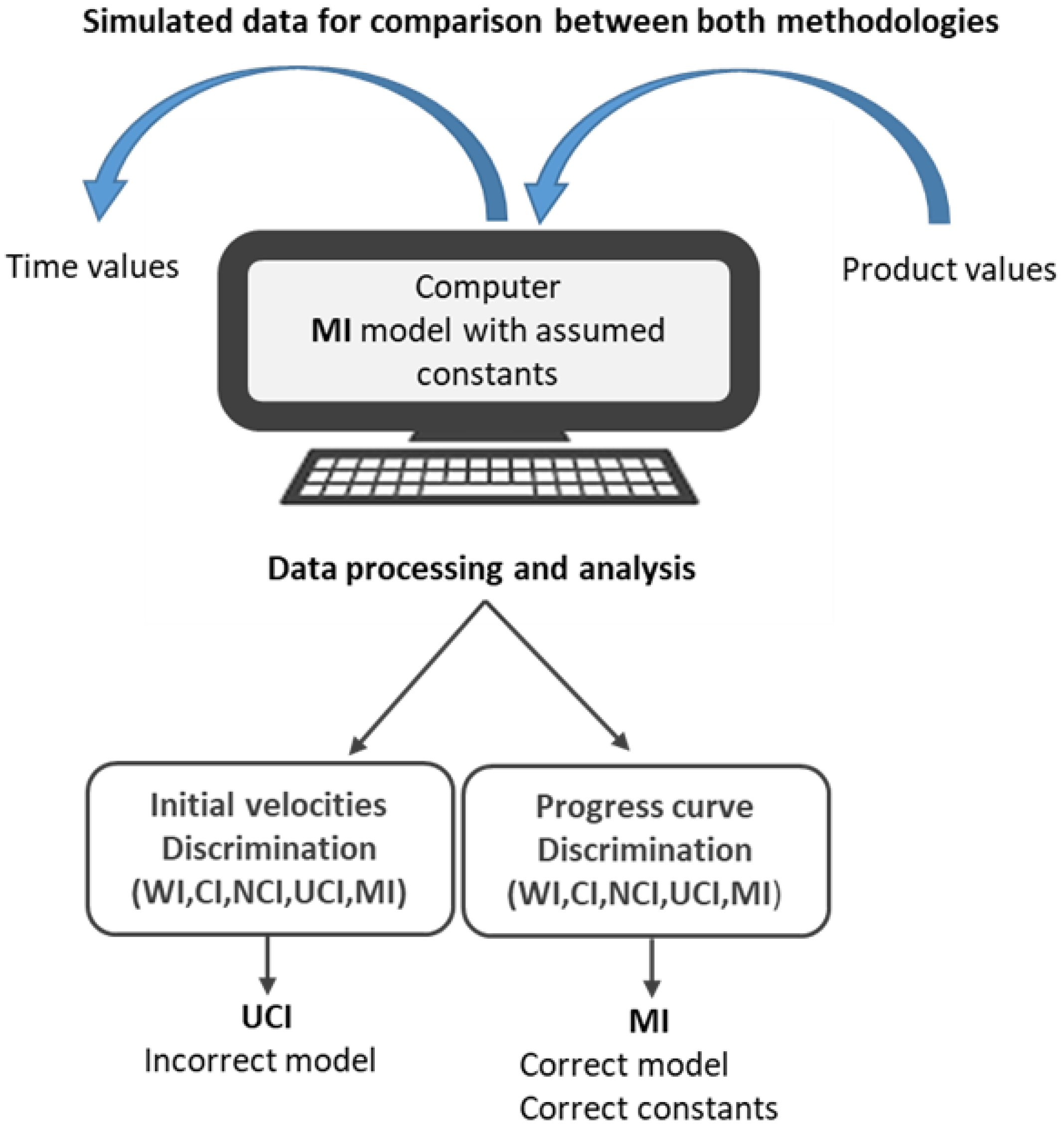
2.2. Simulated Data for Comparison between Both Methodologies
2.3. Data Processing and Analysis
3. Results
3.1. Simulated Data for the Graphical Representation of Product Versus Time
3.2. Kinetic Studies Using MME and IMME Based on Simulated Values
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Adrio, J.L.; Demain, A.L. Microbial enzymes: Tools for biotechnological processes. Biomolecules 2014, 4, 117–139. [Google Scholar] [CrossRef] [Green Version]
- El Harrad, L.; Bourais, I.; Mohammadi, H.; Amine, A. Recent advances in electrochemical biosensors based on enzyme inhibition for clinical and pharmaceutical applications. Sensors 2018, 18, 164. [Google Scholar] [CrossRef] [Green Version]
- Henri, V. Theorie generale de l’action de quelques diástases. Comptes Rendus Biol. 1902, 135, 916–919. [Google Scholar]
- Michaelis, L.; Menten, M. Die kinetik der invertinwirkung. Biochem. Z. 1913, 49, 333–369. [Google Scholar]
- Alam, M.T.; Olin-Sandoval, V.; Stincone, A.; Keller, M.A.; Zelezniak, A.; Luisi, B.F.; Ralser, M. The self-inhibitory nature of metabolic networks and its alleviation through compartmentalization. Nat. Commun. 2017, 8, 16018. [Google Scholar] [CrossRef] [PubMed]
- Andrić, P.; Meyer, A.S.; Jensen, P.A.; Dam-Johansen, K. Reactor design for minimizing product inhibition during enzymatic lignocellulose hydrolysis: II. Quantification of inhibition and suitability of membrane reactors. Biotechnol. Adv. 2010, 28, 407–425. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, N.D.; Peschon, J.J.; Segel, I.H. Kinetics of enzymes subject to very strong product inhibition: Analysis using simplified integrated rate equations and average velocities. J. Theor. Biol. 1983, 100, 597–611. [Google Scholar] [CrossRef]
- Bezerra, R.M.; Pinto, P.A.; Fraga, I.; Dias, A.A. Enzyme inhibition studies by integrated Michaelis–Menten equation considering simultaneous presence of two inhibitors when one of them is a reaction product. Comput. Meth. Prog. Biomed. 2016, 125, 2–7. [Google Scholar] [CrossRef]
- Bezerra, R.M.F.; Pinto, P.A.; Dias, A.A. A kinetic process to determine the interaction type between two compounds, one of which is a reaction product, using alkaline phosphatase inhibition as a case study. Appl. Biochem. Biotechnol. 2020, 191, 657–665. [Google Scholar] [CrossRef]
- Bezerra, R.M.; Fraga, I.; Dias, A.A. Utilization of integrated Michaelis-Menten equations for enzyme inhibition diagnosis and determination of kinetic constants using Solver supplement of Microsoft Office Excel. Comput. Meth. Prog. Biomed. 2013, 109, 26–31. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, R.M.; Dias, A.A.; Fraga, I.; Pereira, A.N. Simultaneous ethanol and cellobiose inhibition of cellulose hydrolysis studied with integrated equations assuming constant or variable substrate concentration. Appl. Biochem. Biotechnol. 2006, 134, 27–38. [Google Scholar] [CrossRef]
- Bezerra, R.M.; Dias, A.A. Utilization of integrated Michaelis-Menten equation to determine kinetic constants. Biochem. Mol. Biol. Educ. 2007, 35, 145–150. [Google Scholar] [CrossRef]
- Dias, A.A.; Pinto, P.A.; Fraga, I.; Bezerra, R.M. Diagnosis of enzyme inhibition using Excel Solver: A combined dry and wet laboratory exercise. J. Chem. Educ. 2014, 91, 1017–1021. [Google Scholar] [CrossRef]
- Pinto, P.A.; Bezerra, R.M.F.; Dias, A.A. Discrimination between rival laccase inhibition models from data sets with one inhibitor concentration using a penalized likelihood analysis and Akaike weights. Biocatal. Biotransform. 2018, 36, 401–407. [Google Scholar] [CrossRef]
- Cao, W.; de La Cruz, E. Quantitative full time course analysis of nonlinear enzyme cycling kinetics. Sci. Rep. 2013, 3, 2658. [Google Scholar] [CrossRef] [Green Version]
- Pinto, M.F.; Ripoll-Rozada, J.; Ramos, H.; Watson, E.E.; Franck, C.; Payne, R.J.; Saraiva, L.; Pereira, P.J.B.; Pastore, A.; Rocha, F.; et al. A simple linearization method unveils hidden enzymatic assay interferences. Biophys. Chem. 2019, 252, 106193. [Google Scholar] [CrossRef]
- Pamidipati, S.; Ahmed, A. A first report on competitive inhibition of laccase enzyme by lignin degradation intermediates. Folia Microbiol. 2020, 65, 431–437. [Google Scholar] [CrossRef]
- Bezerra, R.M.F.; Dias, A.A.; Fraga, I.; Pereira, A.N. Cellulose hydrolysis by cellobiohydrolase Cel7A shows mixedhyperbolic product inhibition. Appl. Biochem. Biotechnol. 2011, 165, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Bezerra, R.M.F.; Dias, A.A. Discrimination among eight modified Michaelis-Menten kinetics models of cellulose hydrolysis with a large range of substrate/enzyme ratios: Inhibition by cellobiose. Appl. Biochem. Biotechnol. 2004, 112, 173–184. [Google Scholar] [CrossRef]
- Bezerra, R.M.F.; Dias, A.A. Enzymatic kinetic of cellulose hydrolysis, Inhibition by ethanol and cellobiose. Appl. Biochem. Biotechnol. 2005, 126, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Prosser, G.A.; de Carvalho, L.P.S. Kinetic mechanism and inhibition of Mycobacterium tuberculosis D-alanine: D-alanine ligase by the antibiotic D-cycloserine. FEBS J. 2013, 280, 1150–1166. [Google Scholar] [CrossRef] [PubMed]
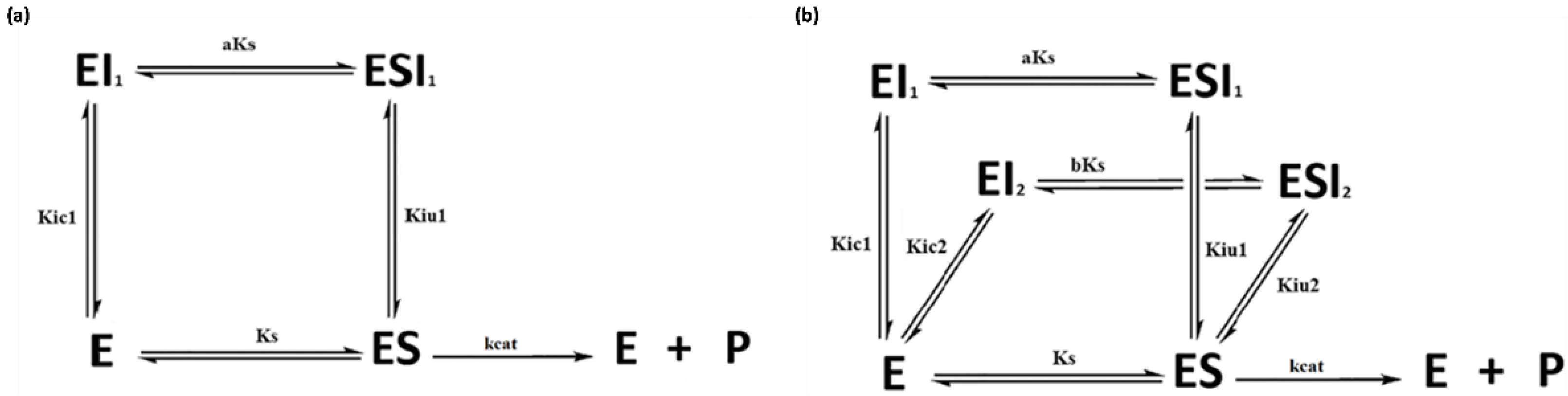
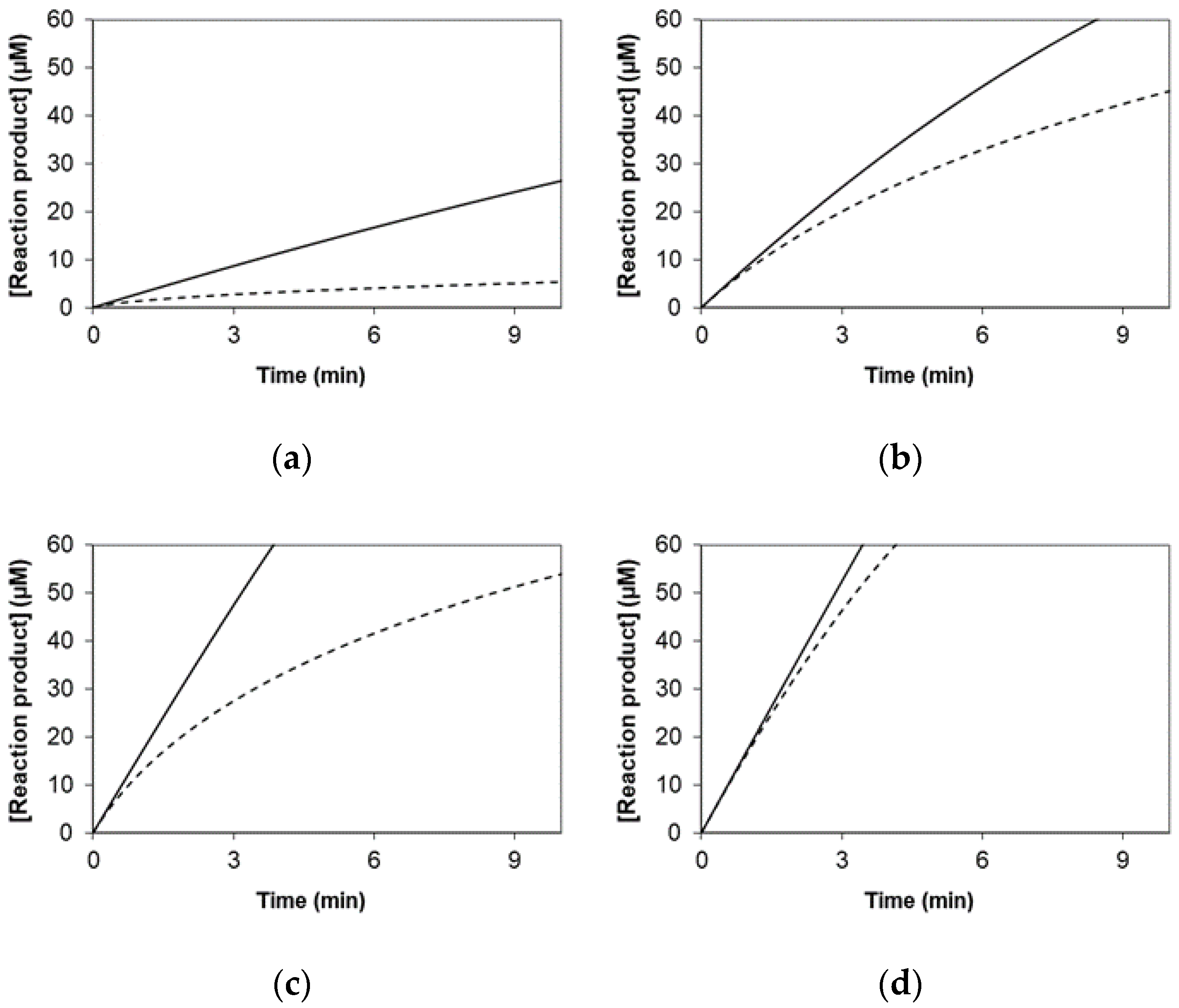
| Kinetic Model * | Equation | |
|---|---|---|
| WI | (1) | |
| CI | (2) | |
| NCI | (3) | |
| UCI | (4) | |
| MI | (5) | |
| Kinetic Model * | Equation | |
|---|---|---|
| CI | (6) | |
| NCI | (7) | |
| UCI | (8) | |
| MI | (9) | |
| WI | CI | NCI | UCI | MI | |
|---|---|---|---|---|---|
| MME | |||||
| SSE | 0.001599 | 0.000340 | 0.000033 | 0.000010 | 0.000007 |
| p | 2 | 3 | 3 | 3 | 4 |
| n | 14 | 14 | 14 | 14 | 14 |
| IMME | |||||
| SSE | 946.511 | 339.962 | 19.814 | 3.064 | 0.0000006 |
| p | 2 | 4 | 4 | 4 | 6 |
| n | 432 | 432 | 432 | 432 | 432 |
| Models A/B | SSEA | SSEB | n | PA + 1 | PB + 1 | AICA | AICB | AICcA | AICcB | Δ | Probability B Correct |
|---|---|---|---|---|---|---|---|---|---|---|---|
| WI/UCI | 0.001599 | 0.000010 | 14 | 3 | 4 | −121.1 | −190.2 | −118.7 | −185.7 | −67.0 | 1.00 |
| UCI/MI | 0.000010 | 0.000007 | 14 | 4 | 5 | −190.2 | −192.4 | −185.7 | −184.9 | 0.8 | 0.397 |
| Method (Model) | Km (μM) | Kic1 (μM) | Kiu1 (μM) | Kic2 (μM) | Kiu2 (μM) | Vmax (μmol min−1 mg−1) |
|---|---|---|---|---|---|---|
| MME (UCI) | 344.4 (±17.3) | - | 7.3 (±0.3) | 0.077 (±0.001) | ||
| IMME (MI) | 499.99 (±0.001) | 99.98 (±0.001) | 50.0 (±0.000) | 50.0 (±0.000) | 5.0 (±0.000) | 0.121 (±0.000) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes, J.M.C.; Dias, A.A.; Bezerra, R.M.F. Kinetic Analysis Misinterpretations Due to the Occurrence of Enzyme Inhibition by Reaction Product: Comparison between Initial Velocities and Reaction Time Course Methodologies. Appl. Sci. 2022, 12, 102. https://doi.org/10.3390/app12010102
Fernandes JMC, Dias AA, Bezerra RMF. Kinetic Analysis Misinterpretations Due to the Occurrence of Enzyme Inhibition by Reaction Product: Comparison between Initial Velocities and Reaction Time Course Methodologies. Applied Sciences. 2022; 12(1):102. https://doi.org/10.3390/app12010102
Chicago/Turabian StyleFernandes, Joana M. C., Albino A. Dias, and Rui M. F. Bezerra. 2022. "Kinetic Analysis Misinterpretations Due to the Occurrence of Enzyme Inhibition by Reaction Product: Comparison between Initial Velocities and Reaction Time Course Methodologies" Applied Sciences 12, no. 1: 102. https://doi.org/10.3390/app12010102
APA StyleFernandes, J. M. C., Dias, A. A., & Bezerra, R. M. F. (2022). Kinetic Analysis Misinterpretations Due to the Occurrence of Enzyme Inhibition by Reaction Product: Comparison between Initial Velocities and Reaction Time Course Methodologies. Applied Sciences, 12(1), 102. https://doi.org/10.3390/app12010102








