Figure 1.
Differences of 1D-Convolution and 1D-Atrous Convolution. (a) 1D-Convolution. (b) 1D-Atrous Convolution. The gray regions are not included in the calculation when calculating one kernel in the convolution. In general, the 1D-Convolution calculates only limited neighbors, but the 1D-Atrous Convolution calculates far distances as well.
Figure 1.
Differences of 1D-Convolution and 1D-Atrous Convolution. (a) 1D-Convolution. (b) 1D-Atrous Convolution. The gray regions are not included in the calculation when calculating one kernel in the convolution. In general, the 1D-Convolution calculates only limited neighbors, but the 1D-Atrous Convolution calculates far distances as well.
Figure 2.
Examples of paint spectral of YU dataset acquired in an outdoor environment. This shows how difficult it is to classify spectral information.
Figure 2.
Examples of paint spectral of YU dataset acquired in an outdoor environment. This shows how difficult it is to classify spectral information.
Figure 3.
Basic concept of Atrous convolution in 2D image processing.
Figure 3.
Basic concept of Atrous convolution in 2D image processing.
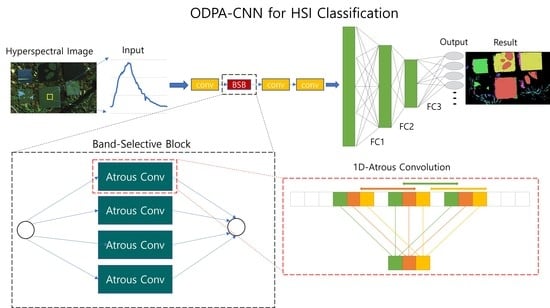
Figure 4.
Proposed CNN Model, ODPA-CNN: AtConv is 1D-Atrous Convolution. Conv is normal 1D-Convolution.
Table 1 has detailed parameters of each convolution. In this model, AtConv is processed in parallel to extract features for bands separated by various distances.
Figure 4.
Proposed CNN Model, ODPA-CNN: AtConv is 1D-Atrous Convolution. Conv is normal 1D-Convolution.
Table 1 has detailed parameters of each convolution. In this model, AtConv is processed in parallel to extract features for bands separated by various distances.
Figure 5.
Concept of spectral feature extraction using 1D-Atrous Convolution: Example has 3 kernel size and 3 rate size.
Figure 5.
Concept of spectral feature extraction using 1D-Atrous Convolution: Example has 3 kernel size and 3 rate size.
Figure 6.
Graph of ReLU6 and Hard-swish: (a) ReLU6 (b) Hard-swish.
Figure 6.
Graph of ReLU6 and Hard-swish: (a) ReLU6 (b) Hard-swish.
Figure 7.
Indian Pines dataset: Left is the names of classes, Center is ground truth, Right is the RGB image.
Figure 7.
Indian Pines dataset: Left is the names of classes, Center is ground truth, Right is the RGB image.
Figure 8.
Salinas dataset: Left is the names of classes, Center is ground truth, Right is the RGB image.
Figure 8.
Salinas dataset: Left is the names of classes, Center is ground truth, Right is the RGB image.
Figure 9.
Pavia University dataset: Left is the names of classes, Center is ground truth, Right is the RGB image.
Figure 9.
Pavia University dataset: Left is the names of classes, Center is ground truth, Right is the RGB image.
Figure 10.
Acquisition of the YU paint data.
Figure 10.
Acquisition of the YU paint data.
Figure 11.
YU Paint dataset: Left is the names of classes, Top is ground truth, Bottom is the RGB image.
Figure 11.
YU Paint dataset: Left is the names of classes, Top is ground truth, Bottom is the RGB image.
Figure 12.
HSI datasets applied random spatial information: (a) Indian Pines, (b) Pavia University, (c) Salinas.
Figure 12.
HSI datasets applied random spatial information: (a) Indian Pines, (b) Pavia University, (c) Salinas.
Figure 13.
Classification results of Indian Pines: (a) ground truth, (b) Luo model, (c) Li model, (d) Hamida model, (e) Chen model, (f) Hu model, (g) proposed ODPA-CNN.
Figure 13.
Classification results of Indian Pines: (a) ground truth, (b) Luo model, (c) Li model, (d) Hamida model, (e) Chen model, (f) Hu model, (g) proposed ODPA-CNN.
Figure 14.
Classification results of Salinas: (a) ground truth, (b) Luo model, (c) Li model, (d) Hamida model, (e) Chen model, (f) Hu model, (g) proposed ODPA-CNN.
Figure 14.
Classification results of Salinas: (a) ground truth, (b) Luo model, (c) Li model, (d) Hamida model, (e) Chen model, (f) Hu model, (g) proposed ODPA-CNN.
Figure 15.
Classification results of Pavia University: (a) ground truth, (b) Luo model, (c) Li model, (d) Hamida model, (e) Chen model, (f) Hu model, (g) proposed ODPA-CNN.
Figure 15.
Classification results of Pavia University: (a) ground truth, (b) Luo model, (c) Li model, (d) Hamida model, (e) Chen model, (f) Hu model, (g) proposed ODPA-CNN.
Figure 16.
Classification results of YU Paint and Comparison of 3D-CNNs and the proposed ODPA-CNN: (a) ground truth, (b) Luo model, (c) Li model, (d) Hamida model, (e) Chen model, (f) Hu model, (g) proposed ODPA-CNN.
Figure 16.
Classification results of YU Paint and Comparison of 3D-CNNs and the proposed ODPA-CNN: (a) ground truth, (b) Luo model, (c) Li model, (d) Hamida model, (e) Chen model, (f) Hu model, (g) proposed ODPA-CNN.
Table 1.
The parameters of ODPA-CNN.
Table 1.
The parameters of ODPA-CNN.
| Layers | Kernel Size | Feature Maps | Dilated Rate | Stride | Padding |
|---|
| Conv1 | 1 | 1 | 1 | 1 | 1 |
| Conv2_1 | 1 | 32 | 1 | 1 | 0 |
| Conv2_2 | 3 | 32 | 6 | 1 | 6 |
| Conv2_3 | 3 | 32 | 12 | 1 | 12 |
| Conv2_4 | 3 | 32 | 18 | 1 | 18 |
| Conv3 | 1 | 64 | 1 | 1 | same |
| Conv4 | 3 | 32 | 1 | 1 | same |
| Conv5 | 3 | 32 | 1 | 1 | same |
| FC1 | Input: feature size, Output: feature size/2 |
| FC2 | Output: 128 |
| FC3 | Output: number of classes |
Table 2.
Number of train and test sample in the Indian Pines dataset.
Table 2.
Number of train and test sample in the Indian Pines dataset.
| # | Class Name | Total | Train | Test |
|---|
| 1 | Alfalfa | 46 | 4 | 42 |
| 2 | Corn-notill | 1428 | 142 | 1286 |
| 3 | Corn-mintill | 830 | 83 | 747 |
| 4 | Corn | 237 | 23 | 214 |
| 5 | Grass-pasture | 483 | 48 | 435 |
| 6 | Grass-trees | 730 | 73 | 657 |
| 7 | Grass-pasture-mowed | 28 | 2 | 26 |
| 8 | Hay-windrowed | 478 | 47 | 431 |
| 9 | Oats | 20 | 2 | 18 |
| 10 | Soybean-notill | 972 | 97 | 875 |
| 11 | Soybean-mintill | 2455 | 245 | 2210 |
| 12 | Soybean-clean | 593 | 59 | 534 |
| 13 | Wheat | 205 | 20 | 185 |
| 14 | Woods | 1265 | 126 | 1139 |
| 15 | Buildings-Grass-Trees-Drives | 386 | 38 | 348 |
| 16 | Stone-Steel-Towers | 93 | 9 | 84 |
Table 3.
Number of train and test sample in the Salinas dataset.
Table 3.
Number of train and test sample in the Salinas dataset.
| # | Class Name | Total | Train | Test |
|---|
| 1 | Brocoli_green_weeds_1 | 2009 | 200 | 1809 |
| 2 | Brocoli_green_weeds_2 | 3726 | 372 | 3354 |
| 3 | Fallow | 1976 | 197 | 1779 |
| 4 | Fallow_rough_plow | 1394 | 139 | 1255 |
| 5 | Fallow_smooth | 2678 | 267 | 2411 |
| 6 | Stubble | 3959 | 395 | 3564 |
| 7 | Celery | 3579 | 357 | 3222 |
| 8 | Grapes_untrained | 11,271 | 1127 | 10,144 |
| 9 | Soil_vinyard_develop | 6203 | 620 | 5583 |
| 10 | Corn_senesced_green_weeds | 3278 | 327 | 2951 |
| 11 | Lettuce_romaine_4wk | 1068 | 106 | 962 |
| 12 | Lettuce_romaine_5wk | 1927 | 192 | 1735 |
| 13 | Lettuce_romaine_6wk | 916 | 91 | 825 |
| 14 | Lettuce_romaine_7wk | 1070 | 107 | 963 |
| 15 | Vinyard_untrained | 7268 | 726 | 6542 |
| 16 | Vinyard_vertical_trellis | 1807 | 180 | 1627 |
Table 4.
Number of train and test sample in the Pavia University dataset.
Table 4.
Number of train and test sample in the Pavia University dataset.
| # | Class Name | Total | Train | Test |
|---|
| 1 | Asphalt | 6631 | 663 | 5968 |
| 2 | Meadows | 18,649 | 1864 | 16,785 |
| 3 | Gravel | 2099 | 209 | 1890 |
| 4 | Trees | 3064 | 306 | 2758 |
| 5 | Painted metal sheets | 1345 | 134 | 1211 |
| 6 | Bare Soil | 5029 | 502 | 4527 |
| 7 | Bitumen | 1330 | 133 | 1197 |
| 8 | Self-Blocking Bricks | 3682 | 368 | 3314 |
| 9 | Shadows | 947 | 94 | 853 |
Table 5.
Number of train and test sample in the YU Paint dataset.
Table 5.
Number of train and test sample in the YU Paint dataset.
| # | Class Name | Total | Train | Test |
|---|
| 1 | bright paint | 26,734 | 50 | 26,684 |
| 2 | dark paint | 28,737 | 50 | 28,687 |
| 3 | aluminum | 10,977 | 50 | 10,927 |
| 4 | grass | 3516 | 50 | 3466 |
| 5 | fallen leaves | 1326 | 50 | 1276 |
| 6 | shadow | 2616 | 50 | 2566 |
Table 6.
Classification results of the Indian Pines dataset to which random spatial information is not applied.
Table 6.
Classification results of the Indian Pines dataset to which random spatial information is not applied.
| | Luo Model | Li Model | Hamida Model |
|---|
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy |
| 1 | 0.000 | 0.00% | 0.189 | 12.20% | 0.735 | 60.98% |
| 2 | 0.002 | 0.08% | 0.623 | 61.79% | 0.744 | 76.34% |
| 3 | 0.000 | 0.00% | 0.539 | 50.47% | 0.759 | 71.89% |
| 4 | 0.000 | 0.00% | 0.448 | 41.78% | 0.803 | 83.10% |
| 5 | 0.000 | 0.00% | 0.878 | 81.84% | 0.918 | 89.89% |
| 6 | 0.516 | 79.76% | 0.882 | 88.58% | 0.972 | 97.11% |
| 7 | 0.000 | 0.00% | 0.783 | 72.00% | 0.851 | 80.00% |
| 8 | 0.364 | 32.56% | 0.939 | 98.60% | 0.978 | 99.07% |
| 9 | 0.000 | 0.00% | 0.389 | 38.89% | 0.882 | 83.33% |
| 10 | 0.000 | 0.00% | 0.610 | 62.86% | 0.766 | 75.77% |
| 11 | 0.536 | 96.97% | 0.725 | 73.21% | 0.819 | 82.17% |
| 12 | 0.000 | 0.00% | 0.563 | 57.30% | 0.683 | 64.04% |
| 13 | 0.000 | 0.00% | 0.945 | 96.76% | 0.992 | 98.38% |
| 14 | 0.816 | 99.21% | 0.951 | 94.64% | 0.970 | 97.37% |
| 15 | 0.000 | 0.00% | 0.551 | 50.43% | 0.765 | 70.03% |
| 16 | 0.000 | 0.00% | 0.851 | 75.00% | 0.957 | 92.86% |
| OA(%) | 42.688% | 61.34% | 82.851% |
| AA(%) | 19.29% | 66.02% | 82.64% |
| Kappa | 0.297 | 0.7010 | 0.804 |
| | Chen Model | Hu Model | Proposed Model |
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy |
| 1 | 0.963 | 95.12% | 0.000 | 0.00% | 0.714 | 59.52% |
| 2 | 0.911 | 89.73% | 0.482 | 36.50% | 0.783 | 79.32% |
| 3 | 0.515 | 35.48% | 0.506 | 45.78% | 0.731 | 71.08% |
| 4 | 0.427 | 28.64% | 0.417 | 40.38% | 0.639 | 68.22% |
| 5 | 0.699 | 56.09% | 0.140 | 8.28% | 0.912 | 89.89% |
| 6 | 0.959 | 96.04% | 0.819 | 97.11% | 0.936 | 93.15% |
| 7 | 0.902 | 92.00% | 0.312 | 20.00% | 0.833 | 76.92% |
| 8 | 0.739 | 59.07% | 0.933 | 98.37% | 0.964 | 99.54% |
| 9 | 0.636 | 77.78% | 0.000 | 0.00% | 0.703 | 72.22% |
| 10 | 0.856 | 80.46% | 0.523 | 46.06% | 0.761 | 76.46% |
| 11 | 0.908 | 86.15% | 0.685 | 86.97% | 0.823 | 81.76% |
| 12 | 0.748 | 63.48% | 0.450 | 35.96% | 0.760 | 74.91% |
| 13 | 0.966 | 99.46% | 0.862 | 92.97% | 0.923 | 94.05% |
| 14 | 0.842 | 74.28% | 0.840 | 96.49% | 0.931 | 92.27% |
| 15 | 0.332 | 19.88% | 0.332 | 22.77% | 0.641 | 66.09% |
| 16 | 0.642 | 51.19% | 0.976 | 95.24% | 0.897 | 98.81% |
| OA(%) | 73.420% | 64.455% | 82.342% |
| AA(%) | 69.05% | 51.43% | 80.89% |
| Kappa | 0.704 | 0.585 | 0.799 |
Table 7.
Classification results of the Salinas dataset to which random spatial information is not applied.
Table 7.
Classification results of the Salinas dataset to which random spatial information is not applied.
| | Luo Model | Li Model | Hamida Model |
|---|
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy |
| 1 | 0.000 | 11.06% | 0.904 | 89.11% | 0.983 | 96.74% |
| 2 | 0.002 | 0.18% | 0.947 | 94.28% | 1 | 100.00% |
| 3 | 0.000 | 39.24% | 0.842 | 82.41% | 0.993 | 99.72% |
| 4 | 0.000 | 99.84% | 0.958 | 94.34% | 0.993 | 99.92% |
| 5 | 0.000 | 94.73% | 0.893 | 90.17% | 0.995 | 99.50% |
| 6 | 0.516 | 98.15% | 0.990 | 98.74% | 0.996 | 99.27% |
| 7 | 0.000 | 99.29% | 0.984 | 97.30% | 0.995 | 99.22% |
| 8 | 0.364 | 98.58% | 0.818 | 80.58% | 0.92 | 93.66% |
| 9 | 0.000 | 97.21% | 0.953 | 95.24% | 0.998 | 99.77% |
| 10 | 0.000 | 55.66% | 0.871 | 84.85% | 0.978 | 96.07% |
| 11 | 0.536 | 0.00% | 0.762 | 71.00% | 0.974 | 96.36% |
| 12 | 0.000 | 32.76% | 0.876 | 86.57% | 0.989 | 98.04% |
| 13 | 0.000 | 0.00% | 0.867 | 84.48% | 0.99 | 98.42% |
| 14 | 0.816 | 7.17% | 0.864 | 83.28% | 0.987 | 97.92% |
| 15 | 0.000 | 0.00% | 0.740 | 72.67% | 0.867 | 86.37% |
| 16 | 0.000 | 12.18% | 0.940 | 90.29% | 0.947 | 90.77% |
| OA(%) | 59.605% | 86.512% | 95.269% |
| AA(%) | 46.63% | 87.21% | 96.99% |
| Kappa | 0.539 | 0.850 | 0.947 |
| | Chen Model | Hu Model | Proposed Model |
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy |
| 1 | 0.963 | 80.20% | 0.985 | 100% | 0.967 | 93.68% |
| 2 | 0.911 | 98.87% | 0.989 | 97.95% | 0.980 | 99.69% |
| 3 | 0.515 | 94.83% | 0.930 | 95.88% | 0.989 | 98.99% |
| 4 | 0.427 | 92.83% | 0.988 | 98.73% | 0.988 | 97.66% |
| 5 | 0.699 | 93.40% | 0.959 | 93.56% | 0.983 | 98.66% |
| 6 | 0.959 | 91.05% | 0.999 | 99.94% | 0.998 | 99.94% |
| 7 | 0.902 | 91.24% | 0.993 | 99.13% | 0.999 | 100% |
| 8 | 0.739 | 84.39% | 0.761 | 78.94% | 0.862 | 80.6% |
| 9 | 0.636 | 96.53% | 0.992 | 99.09% | 0.995 | 99.04% |
| 10 | 0.856 | 88.85% | 0.890 | 85.64% | 0.967 | 97.52% |
| 11 | 0.908 | 87.72% | 0.908 | 92.54% | 0.966 | 98.38% |
| 12 | 0.748 | 86.97% | 0.983 | 97.34% | 0.986 | 97.31% |
| 13 | 0.966 | 89.21% | 0.964 | 96.25% | 0.973 | 95.34% |
| 14 | 0.842 | 86.81% | 0.954 | 95.81% | 0.964 | 97.15% |
| 15 | 0.332 | 99.29% | 0.667 | 64.73% | 0.744 | 85.93% |
| 16 | 0.642 | 39.54% | 0.979 | 97.68% | 0.994 | 99.81% |
| OA(%) | 87.005% | 88.66% | 92.94% |
| AA(%) | 87.61% | 93.32% | 96.23% |
| Kappa | 0.857 | 0.874 | 0.9212 |
Table 8.
Classification results of the Pavia University dataset to which random spatial information is not applied.
Table 8.
Classification results of the Pavia University dataset to which random spatial information is not applied.
| | Luo Model | Li Model | Hamida Model |
|---|
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy |
| 1 | 0.837 | 95.16% | 0.965 | 97.69% | 0.982 | 97.45% |
| 2 | 0.846 | 95.04% | 0.973 | 94.77% | 0.998 | 95.84% |
| 3 | 0.000 | 0.00% | 0.926 | 91.05% | 0.979 | 93.17% |
| 4 | 0.660 | 52.14% | 0.979 | 98.15% | 0.989 | 96.63% |
| 5 | 0.995 | 99.26% | 0.99 | 99.83% | 0.988 | 99.83% |
| 6 | 0.213 | 13.19% | 0.979 | 99.87% | 0.995 | 98.12% |
| 7 | 0.000 | 0.00% | 0.938 | 89.06% | 0.996 | 97.83% |
| 8 | 0.726 | 89.38% | 0.962 | 94.33% | 0.889 | 96.29% |
| 9 | 0.985 | 97.07% | 0.997 | 99.53% | 0.996 | 99.18% |
| OA(%) | 74.433% | 95.932% | 96.584% |
| AA(%) | 60.14% | 96.03% | 97.15% |
| Kappa | 0.642 | 0.947 | 0.955 |
| | Chen Model | Hu Model | Proposed Model |
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy |
| 1 | 0.963 | 78.74% | 0.804 | 76.35% | 0.942 | 94.44% |
| 2 | 0.911 | 76.38% | 0.867 | 79.48% | 0.958 | 94.52% |
| 3 | 0.515 | 74.01% | 0.058 | 25.41% | 0.805 | 75.89% |
| 4 | 0.427 | 88.32% | 0.787 | 78.32% | 0.930 | 97.34% |
| 5 | 0.699 | 100.00% | 0.976 | 96.15% | 0.996 | 99.59% |
| 6 | 0.959 | 92.97% | 0.355 | 81.65% | 0.876 | 89.77% |
| 7 | 0.902 | 99.58% | 0.055 | 11.78% | 0.864 | 92.64% |
| 8 | 0.739 | 98.61% | 0.747 | 62.28% | 0.854 | 86.62% |
| 9 | 0.636 | 95.31% | 0.996 | 99.3% | 0.998 | 99.77% |
| OA(%) | 83.233% | 76.71% | 92.69% |
| AA(%) | 89.32% | 67.86% | 92.29 |
| Kappa | 0.791 | 0.6678 | 0.9026 |
Table 9.
Classification results of the YU Paint dataset to which random spatial information is not applied.
Table 9.
Classification results of the YU Paint dataset to which random spatial information is not applied.
| | Luo Model | Li Model | Hamida Model | | | |
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy | | | |
| 1 | 0.000 | 0% | 0.942 | 99.33% | 0.997 | 99.89% | | | |
| 2 | 0.000 | 0% | 0.697 | 83.69% | 0.985 | 99.61% | | | |
| 3 | 0.000 | 0% | 0.936 | 99.20% | 0.997 | 99.91% | | | |
| 4 | 0.090 | 4.71% | 0.858 | 83.27% | 0.955 | 97.71% | | | |
| 5 | 0.000 | 0% | 0.887 | 97.35% | 0.983 | 98.73% | | | |
| 6 | 0.000 | 0% | 0.293 | 17.23% | 0.888 | 81.79% | | | |
| OA(%) | 4.694% | 68.58% | 98.25% | | | |
| AA(%) | 0.79% | 80.01% | 96.27% | | | |
| Kappa | 0.000 | 0.7010 | 0.9748 | | | |
| | Chen Model | Hu Model | Proposed Model | | |
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy | | | |
| 1 | 0.514 | 37.36% | 0.052 | 95.34% | 0.996 | 99.44% | | | |
| 2 | 0.000 | 0% | 0.799 | 99.29% | 0.998 | 99.90% | | | |
| 3 | 0.000 | 0% | 0.451 | 29.23% | 0.990 | 99.63% | | | |
| 4 | 0.090 | 0% | 0.815 | 99.87% | 0.989 | 97.46% | | | |
| 5 | 0.000 | 0% | 0.806 | 67.53% | 0.990 | 98.08% | | | |
| 6 | 0.000 | 0% | 0.343 | 20.69% | 0.980 | 81.49% | | | |
| OA(%) | 35.54% | 50.136% | 99.47% | | | |
| AA(%) | 6.23% | 68.66% | 98.67% | | | |
| Kappa | 0.016 | 0.386 | 0.992 | | | |
Table 10.
Classification results of Indian Pines dataset to which random spatial information is applied.
Table 10.
Classification results of Indian Pines dataset to which random spatial information is applied.
| | Luo Model | Li Model | Hamida Model |
|---|
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy |
| 1 | 0.000 | 0.00% | 0.319 | 26.83% | 0.527 | 58.54% |
| 2 | 0.000 | 0.00% | 0.477 | 48.79% | 0.575 | 55.49% |
| 3 | 0.000 | 0.00% | 0.325 | 28.11% | 0.401 | 38.02% |
| 4 | 0.000 | 0.00% | 0.330 | 32.86% | 0.241 | 22.54% |
| 5 | 0.000 | 0.00% | 0.448 | 39.31% | 0.616 | 52.64% |
| 6 | 0.000 | 0.00% | 0.746 | 74.28% | 0.856 | 86.15% |
| 7 | 0.000 | 0.00% | 0.105 | 8.00% | 0.474 | 36.00% |
| 8 | 0.000 | 0.00% | 0.808 | 75.35% | 0.829 | 77.21% |
| 9 | 0.000 | 0.00% | 0.000 | 0.00% | 0.435 | 27.78% |
| 10 | 0.000 | 0.00% | 0.403 | 39.20% | 0.505 | 50.17% |
| 11 | 0.000 | 100.00% | 0.600 | 61.90% | 0.678 | 71.45% |
| 12 | 0.397 | 0.00% | 0.300 | 26.40% | 0.438 | 37.08% |
| 13 | 0.000 | 0.00% | 0.706 | 64.32% | 0.866 | 89.19% |
| 14 | 0.000 | 0.00% | 0.785 | 80.68% | 0.834 | 83.76% |
| 15 | 0.000 | 0.00% | 0.546 | 51.30% | 0.554 | 53.31% |
| 16 | 0.000 | 0.00% | 0.809 | 67.86% | 0.863 | 78.57% |
| OA(%) | 0.000 | 54.504% | 62.829% |
| AA(%) | 6.25% | 45.33% | 57.37% |
| Kappa | 0.010 | 0.481 | 0.575 |
| | Chen Model | Hu Model | Proposed Model |
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy |
| 1 | 0.000 | 0.00% | 0 | 0.00% | 0.791 | 82.93% |
| 2 | 0.424 | 0.00% | 0.424 | 34.63% | 0.810 | 82.57% |
| 3 | 0.126 | 0.00% | 0.126 | 11.51% | 0.788 | 76.97% |
| 4 | 0.000 | 0.00% | 0 | 0.00% | 0.673 | 68.08% |
| 5 | 0.000 | 0.00% | 0 | 0.00% | 0.886 | 87.59% |
| 6 | 0.663 | 0.00% | 0.663 | 97.87% | 0.935 | 96.65% |
| 7 | 0.000 | 0.00% | 0 | 0.00% | 0.683 | 56.00% |
| 8 | 0.873 | 0.00% | 0.873 | 98.60% | 0.965 | 98.37% |
| 9 | 0.000 | 0.00% | 0 | 0.00% | 0.606 | 55.56% |
| 10 | 0.071 | 0.00% | 0.071 | 4.80% | 0.805 | 76.69% |
| 11 | 0.583 | 64.89% | 0.583 | 82.94% | 0.854 | 85.20% |
| 12 | 0.000 | 0.00% | 0 | 0.00% | 0.803 | 85.77% |
| 13 | 0.000 | 0.00% | 0 | 0.00% | 0.933 | 94.05% |
| 14 | 0.836 | 0.00% | 0.836 | 98.60% | 0.938 | 95.96% |
| 15 | 0.000 | 0.00% | 0 | 0.00% | 0.602 | 51.59% |
| 16 | 0.000 | 0.00% | 0 | 0.00% | 0.958 | 94.05% |
| OA(%) | 49.821% | 49.821% | 84.715% |
| AA(%) | 26.81% | 26.81% | 80.50% |
| Kappa | 0.009 | 0.402 | 0.826 |
Table 11.
Classification results of the Salinas dataset to which random spatial information is applied.
Table 11.
Classification results of the Salinas dataset to which random spatial information is applied.
| | Luo Model | Li Model | Hamida Model |
|---|
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy |
| 1 | 0.000 | 0.00% | 0.935 | 91.54% | 0.948 | 93.69% |
| 2 | 0.737 | 94.04% | 0.956 | 95.35% | 0.930 | 90.10% |
| 3 | 0.000 | 0.00% | 0.868 | 86.40% | 0.825 | 79.65% |
| 4 | 0.961 | 96.02% | 0.938 | 90.92% | 0.955 | 92.99% |
| 5 | 0.689 | 96.14% | 0.892 | 87.51% | 0.903 | 89.42% |
| 6 | 0.978 | 95.87% | 0.979 | 95.93% | 0.974 | 95.17% |
| 7 | 0.911 | 96.49% | 0.984 | 97.33% | 0.961 | 93.11% |
| 8 | 0.677 | 93.36% | 0.809 | 79.11% | 0.813 | 80.80% |
| 9 | 0.723 | 98.46% | 0.949 | 94.68% | 0.965 | 97.49% |
| 10 | 0.085 | 6.20% | 0.893 | 88.03% | 0.897 | 90.81% |
| 11 | 0.000 | 0.00% | 0.754 | 68.68% | 0.827 | 77.00% |
| 12 | 0.000 | 0.00% | 0.912 | 92.10% | 0.900 | 90.02% |
| 13 | 0.000 | 0.00% | 0.904 | 88.61% | 0.910 | 90.79% |
| 14 | 0.581 | 41.12% | 0.893 | 87.23% | 0.905 | 90.65% |
| 15 | 0.068 | 3.93% | 0.731 | 71.84% | 0.740 | 73.05% |
| 16 | 0.000 | 0.00% | 0.956 | 93.23% | 0.977 | 97.23% |
| OA(%) | 59.534% | 86.508% | 87.126% |
| AA(%) | 45.10% | 88.03% | 88.87% |
| Kappa | 0.537 | 0.850 | 0.857 |
| | Chen Model | Hu Model | Proposed Model |
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy |
| 1 | 0.632 | 64.16% | 0.976 | 95.24% | 0.997 | 99.45% |
| 2 | 0.673 | 62.34% | 0.985 | 99.88% | 0.998 | 99.82% |
| 3 | 0.375 | 29.45% | 0.897 | 91.74% | 0.992 | 99.61% |
| 4 | 0.149 | 12.03% | 0.985 | 99.60% | 0.993 | 99.52% |
| 5 | 0.258 | 20.54% | 0.941 | 92.49% | 0.985 | 98.46% |
| 6 | 0.582 | 52.43% | 0.998 | 99.55% | 0.999 | 99.97% |
| 7 | 0.513 | 45.89% | 0.981 | 99.22% | 0.998 | 99.81% |
| 8 | 0.655 | 76.47% | 0.791 | 91.01% | 0.868 | 88.64% |
| 9 | 0.736 | 69.59% | 0.97 | 96.76% | 0.997 | 99.98% |
| 10 | 0.479 | 48.34% | 0.865 | 87.59% | 0.971 | 96.71% |
| 11 | 0.271 | 23.52% | 0.808 | 85.85% | 0.985 | 99.69% |
| 12 | 0.289 | 23.36% | 0.967 | 99.65% | 0.998 | 100.00% |
| 13 | 0.141 | 14.18% | 0.947 | 97.58% | 0.998 | 99.64% |
| 14 | 0.199 | 17.34% | 0.931 | 92.32% | 0.987 | 99.17% |
| 15 | 0.602 | 64.44% | 0.535 | 39.93% | 0.788 | 76.00% |
| 16 | 0.569 | 43.36% | 0.955 | 92.13% | 0.983 | 97.60% |
| OA(%) | 51.438% | 87.241% | 93.953% |
| AA(%) | 41.71% | 91.28% | 97.13% |
| Kappa | 0.465 | 0.857 | 0.933 |
Table 12.
Classification results of the Pavia University dataset to which random spatial information is applied.
Table 12.
Classification results of the Pavia University dataset to which random spatial information is applied.
| | Luo Model | Li Model | Hamida Model |
|---|
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy |
| 1 | 0.647 | 97.17% | 0.796 | 79.74% | 0.872 | 87.08% |
| 2 | 0.834 | 95.17% | 0.881 | 89.54% | 0.912 | 90.97% |
| 3 | 0.000 | 0.00% | 0.477 | 45.61% | 0.698 | 68.71% |
| 4 | 0.487 | 38.76% | 0.83 | 79.94% | 0.877 | 84.34% |
| 5 | 0.839 | 72.50% | 0.968 | 95.46% | 0.982 | 97.03% |
| 6 | 0.026 | 1.33% | 0.631 | 58.45% | 0.780 | 75.32% |
| 7 | 0.000 | 0.00% | 0.478 | 43.36% | 0.671 | 62.99% |
| 8 | 0.198 | 13.88% | 0.661 | 68.23% | 0.773 | 76.10% |
| 9 | 0.889 | 80.05% | 0.953 | 92.97% | 0.986 | 97.42% |
| OA(%) | 64.734% | 78.511% | 85.142% |
| AA(%) | 44.32% | 72.59% | 82.22% |
| Kappa | 0.497 | 0.714 | 0.804 |
| | Chen Model | Hu Model | Proposed Model |
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy |
| 1 | 0.241 | 25.49% | 0.871 | 88.79% | 0.959 | 95.53% |
| 2 | 0.659 | 64.23% | 0.913 | 98.47% | 0.978 | 97.96% |
| 3 | 0.143 | 11.22% | 0.607 | 58.71% | 0.843 | 82.42% |
| 4 | 0.143 | 15.08% | 0.875 | 80.78% | 0.958 | 96.01% |
| 5 | 0.378 | 28.32% | 0.993 | 98.76% | 0.995 | 99.75% |
| 6 | 0.311 | 26.14% | 0.629 | 48.06% | 0.938 | 93.28% |
| 7 | 0.116 | 9.44% | 0.693 | 61.65% | 0.914 | 91.40% |
| 8 | 0.161 | 15.12% | 0.827 | 83.61% | 0.886 | 90.37% |
| 9 | 0.073 | 6.46% | 0.999 | 99.88% | 0.998 | 99.65% |
| OA(%) | 39.284% | 85.444% | 95.369% |
| AA(%) | 22.39% | 79.86% | 94.04% |
| Kappa | 0.210 | 0.801 | 0.939 |
Table 13.
Classification results of the Indian Pines dataset to which random spatial information is not applied for training and random spatial information to test.
Table 13.
Classification results of the Indian Pines dataset to which random spatial information is not applied for training and random spatial information to test.
| | Luo Model | Li Model | Hamida Model |
|---|
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy |
| 1 | 0.000 | 0.00% | 0.000 | 0.00% | 0.000 | 0.00% |
| 2 | 0.070 | 4.41% | 0.071 | 4.20% | 0.079 | 4.90% |
| 3 | 0.000 | 0.00% | 0.141 | 33.49% | 0.088 | 6.63% |
| 4 | 0.000 | 0.00% | 0.046 | 16.03% | 0.070 | 64.14% |
| 5 | 0.000 | 0.00% | 0.094 | 8.49% | 0.182 | 21.12% |
| 6 | 0.158 | 56.03% | 0.079 | 6.44% | 0.123 | 7.67% |
| 7 | 0.000 | 0.00% | 0.018 | 7.14% | 0.036 | 10.71% |
| 8 | 0.084 | 25.10% | 0.120 | 10.88% | 0.118 | 6.69% |
| 9 | 0.000 | 0.00% | 0.000 | 0.00% | 0.000 | 0.00% |
| 10 | 0.000 | 0.00% | 0.049 | 3.09% | 0.029 | 1.54% |
| 11 | 0.361 | 36.25% | 0.126 | 8.07% | 0.229 | 15.32% |
| 12 | 0.000 | 0.00% | 0.079 | 6.41% | 0.062 | 4.55% |
| 13 | 0.000 | 0.00% | 0.043 | 2.93% | 0.270 | 25.85% |
| 14 | 0.279 | 17.55% | 0.192 | 11.94% | 0.558 | 60.63% |
| 15 | 0.000 | 0.00% | 0.123 | 31.09% | 0.063 | 10.88% |
| 16 | 0.000 | 0.00% | 0.000 | 0.00% | 0.000 | 0.00% |
| OA(%) | 16.626% | 10.352% | 17.074% |
| AA(%) | 8.71% | 9.39% | 15.04% |
| Kappa | 0.06478 | 0.0376 | 0.113 |
| | Chen Model | Hu Model | Proposed Model |
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy |
| 1 | 0.000 | 0.00% | 0.000 | 0.00% | 0.744 | 63.04% |
| 2 | 0.000 | 0.00% | 0.483 | 36.41% | 0.802 | 81.23% |
| 3 | 0.006 | 0.30% | 0.512 | 46.27% | 0.757 | 73.86% |
| 4 | 0.001 | 0.07% | 0.432 | 41.77% | 0.671 | 71.31% |
| 5 | 0.000 | 0.00% | 0.150 | 8.90% | 0.921 | 90.89% |
| 6 | 0.000 | 0.00% | 0.821 | 97.40% | 0.942 | 93.84% |
| 7 | 0.000 | 0.00% | 0.286 | 17.86% | 0.846 | 78.57% |
| 8 | 0.242 | 31.17% | 0.933 | 98.33% | 0.967 | 99.58% |
| 9 | 0.000 | 0.00% | 0.000 | 0.00% | 0.714 | 75.00% |
| 10 | 0.101 | 22.88% | 0.521 | 45.68% | 0.783 | 78.60% |
| 11 | 0.000 | 0.00% | 0.686 | 87.54% | 0.840 | 83.42% |
| 12 | 0.030 | 1.92% | 0.467 | 37.10% | 0.782 | 77.23% |
| 13 | 0.029 | 25.22% | 0.862 | 93.17% | 0.930 | 94.63% |
| 14 | 0.011 | 0.84% | 0.841 | 96.60% | 0.937 | 92.96% |
| 15 | 0.001 | 0.03% | 0.331 | 22.54% | 0.671 | 68.91% |
| 16 | 0.000 | 0.00% | 0.972 | 94.62% | 0.906 | 98.92% |
| OA(%) | 8.403% | 64.719% | 83.969% |
| AA(%) | 5.15% | 51.51% | 82.63% |
| Kappa | −0.0028 | 0.5878 | 0.817 |
Table 14.
Classification results of the Salinas dataset to which random spatial information is not applied for training and random spatial information to test.
Table 14.
Classification results of the Salinas dataset to which random spatial information is not applied for training and random spatial information to test.
| | Luo Model | Li Model | Hamida Model |
|---|
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy |
| 1 | 0.000 | 0.00% | 0.000 | 0.00% | 0.000 | 0.00% |
| 2 | 0.029 | 1.48% | 0.000 | 0.00% | 0.089 | 5.07% |
| 3 | 0.067 | 8.76% | 0.015 | 0.76% | 0.018 | 0.96% |
| 4 | 0.048 | 2.94% | 0.008 | 0.43% | 0.003 | 0.14% |
| 5 | 0.082 | 5.97% | 0.053 | 3.25% | 0.039 | 2.65% |
| 6 | 0.012 | 0.63% | 0.052 | 2.80% | 0.219 | 14.85% |
| 7 | 0.063 | 4.36% | 0.000 | 0.00% | 0.072 | 4.47% |
| 8 | 0.370 | 73.33% | 0.336 | 54.64% | 0.368 | 54.68% |
| 9 | 0.102 | 5.59% | 0.060 | 3.77% | 0.192 | 14.19% |
| 10 | 0.133 | 22.94% | 0.173 | 24.86% | 0.142 | 10.37% |
| 11 | 0.000 | 0.00% | 0.014 | 0.84% | 0.030 | 2.25% |
| 12 | 0.040 | 4.88% | 0.075 | 7.58% | 0.083 | 10.59% |
| 13 | 0.000 | 0.00% | 0.049 | 20.63% | 0.045 | 27.07% |
| 14 | 0.021 | 3.46% | 0.024 | 6.92% | 0.033 | 9.35% |
| 15 | 0.000 | 0.00% | 0.163 | 13.11% | 0.132 | 9.42% |
| 16 | 0.000 | 0.00% | 0.022 | 1.16% | 0.061 | 3.76% |
| OA(%) | 18.668% | 16.292% | 17.996% |
| AA(%) | 8.40% | 8.80% | 10.61% |
| Kappa | 0.0469 | 0.0432 | 0.0753 |
| | Chen Model | Hu Model | Proposed Model |
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy |
| 1 | 0.000 | 0.00% | 0.971 | 97.76% | 0.9988 | 99.75% |
| 2 | 0.051 | 3.08% | 0.980 | 97.45% | 0.9973 | 99.95% |
| 3 | 0.028 | 2.05% | 0.871 | 80.97% | 0.9832 | 97.82% |
| 4 | 0.000 | 0.00% | 0.979 | 98.21% | 0.9932 | 99.00% |
| 5 | 0.056 | 5.59% | 0.923 | 96.86% | 0.9860 | 99.70% |
| 6 | 0.110 | 26.58% | 0.997 | 99.67% | 0.9990 | 99.80% |
| 7 | 0.000 | 0.00% | 0.975 | 98.71% | 0.9976 | 99.58% |
| 8 | 0.090 | 22.38% | 0.774 | 94.28% | 0.8598 | 85.36% |
| 9 | 0.000 | 0.00% | 0.963 | 97.37% | 0.9944 | 99.53% |
| 10 | 0.002 | 0.10% | 0.854 | 81.73% | 0.9555 | 94.69% |
| 11 | 0.094 | 5.42% | 0.751 | 72.94% | 0.9653 | 98.97% |
| 12 | 0.000 | 0.00% | 0.946 | 99.90% | 0.9979 | 99.95% |
| 13 | 0.038 | 2.93% | 0.938 | 98.91% | 0.9851 | 97.60% |
| 14 | 0.035 | 1.82% | 0.927 | 90.37% | 0.9718 | 98.13% |
| 15 | 0.000 | 0.00% | 0.401 | 26.50% | 0.7933 | 80.09% |
| 16 | 0.000 | 0.00% | 0.950 | 92.81% | 0.9922 | 98.84% |
| OA(%) | 5.385% | 85.281% | 93.589% |
| AA(%) | 4.37% | 89.03% | 96.80% |
| Kappa | 0.0067 | 0.8348 | 0.9286 |
Table 15.
Classification results of the Paviua University dataset to which random spatial information is not applied for training and random spatial information to test.
Table 15.
Classification results of the Paviua University dataset to which random spatial information is not applied for training and random spatial information to test.
| | Luo Model | Li Model | Hamida Model |
|---|
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy |
| 1 | 0.212 | 17.78% | 0.202 | 17.24% | 0.056 | 3.17% |
| 2 | 0.640 | 86.61% | 0.069 | 3.64% | 0.471 | 41.79% |
| 3 | 0.000 | 0.00% | 0.003 | 0.14% | 0.006 | 0.29% |
| 4 | 0.001 | 0.07% | 0.172 | 21.54% | 0.073 | 4.47% |
| 5 | 0.004 | 0.22% | 0.019 | 0.97% | 0.000 | 0.00% |
| 6 | 0.151 | 13.78% | 0.244 | 73.35% | 0.262 | 63.65% |
| 7 | 0.000 | 0.00% | 0.003 | 0.15% | 0.000 | 0.00% |
| 8 | 0.081 | 6.06% | 0.221 | 30.53% | 0.190 | 26.67% |
| 9 | 0.002 | 0.11% | 0.002 | 0.11% | 0.000 | 0.00% |
| OA(%) | 42.67% | 17.098% | 28.824% |
| AA(%) | 13.85% | 16.41% | 15.56% |
| Kappa | 0.1100 | 0.061 | 0.0897 |
| | Chen Model | Hu Model | Proposed Model |
| # | F1-Score | Accuracy | F1-Score | Accuracy | F1-Score | Accuracy |
| 1 | 0.157 | 12.74% | 0.867 | 87.92% | 0.950 | 94.60% |
| 2 | 0.110 | 6.11% | 0.912 | 97.30% | 0.979 | 98.48% |
| 3 | 0.000 | 0.00% | 0.341 | 27.11% | 0.833 | 83.18% |
| 4 | 0.097 | 8.78% | 0.881 | 85.15% | 0.965 | 95.56% |
| 5 | 0.113 | 11.23% | 0.991 | 98.88% | 0.994 | 99.85% |
| 6 | 0.207 | 55.44% | 0.639 | 50.07% | 0.937 | 92.54% |
| 7 | 0.000 | 0.00% | 0.661 | 57.22% | 0.904 | 91.58% |
| 8 | 0.145 | 18.31% | 0.781 | 87.86% | 0.881 | 87.81% |
| 9 | 0.002 | 0.11% | 0.997 | 99.68% | 0.996 | 99.26% |
| OA(%) | 13.72% | 84.02% | 95.149% |
| AA(%) | 12.52% | 76.80% | 93.65% |
| Kappa | 0.0274 | 0.7824 | 0.9355 |