Prebiotic Isomaltooligosaccharide Provides an Advantageous Fitness to the Probiotic Bacillus subtilis CU1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Spectrometric Analyses of IMOS Components
2.2. Strains and Culture Conditions
2.3. Growth Experiments
2.4. Determination of Substrate Utilization in the Culture Medium
2.5. Determination of Glucosidase Activity
2.6. Determination of Lactate and Short-Chain Fatty Acid (SCFA) Production
2.7. Proteomic Analysis
2.7.1. Preparation of Protein Samples
2.7.2. Isoelectrofocalization (IEF)
2.7.3. SDS-PAGE
2.7.4. Protein Digestion
2.7.5. MALDI-TOF/TOF-MS Analysis for Protein Identification
2.7.6. Peptide Identification
2.8. Tolerance to Bile
2.9. Statistical Analysis
3. Results
3.1. Growth Ability of B. subtilis Probiotic Strains on Prebiotics
3.2. Chemical Analysis of Commercial IMOS Structure
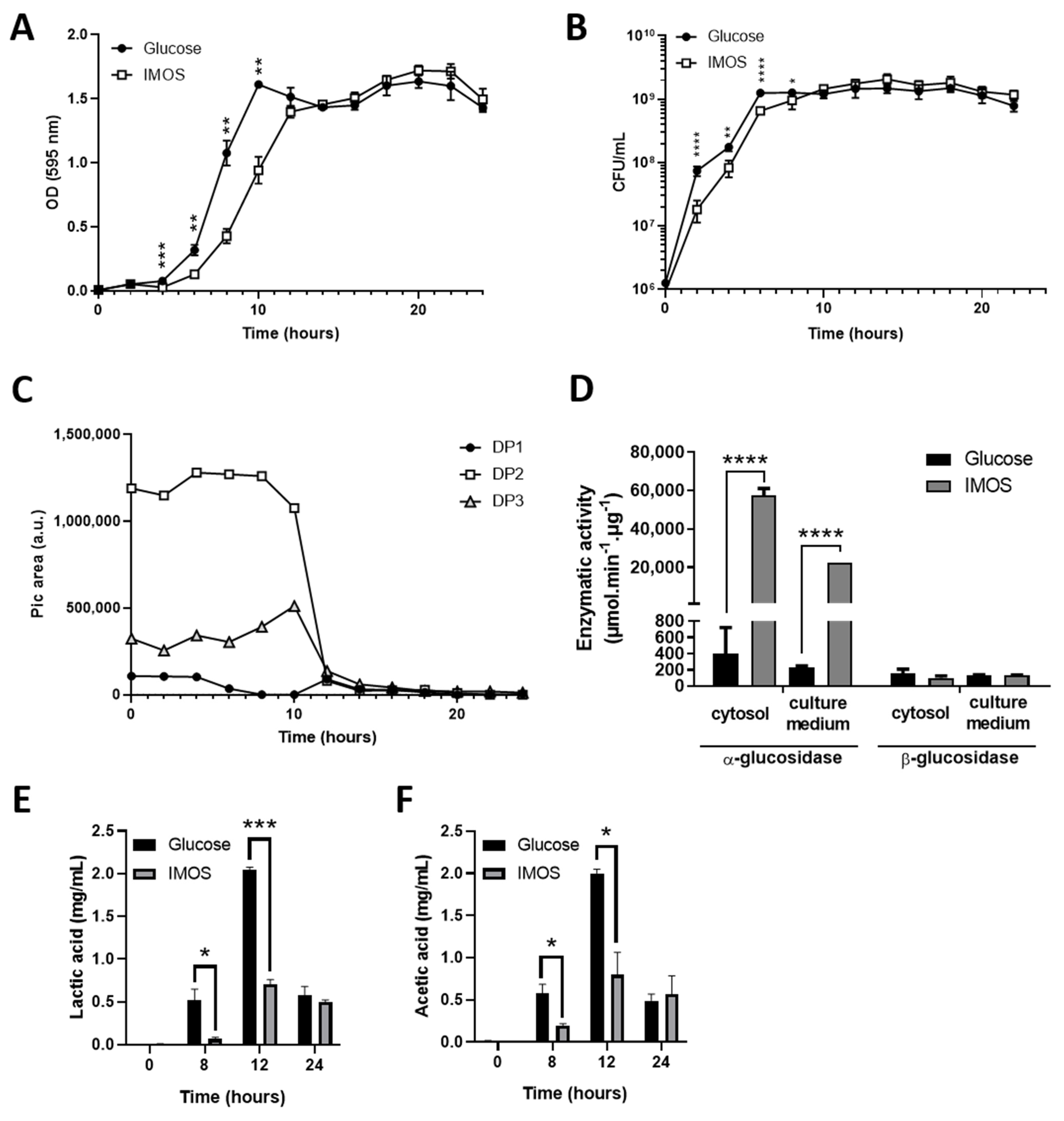
3.3. B. subtilis CU1 Is Able to Grow on IMOSs as a Unique Carbon Source
3.4. IMOSs Are Hydrolyzed in the Extracellular Medium
3.5. IMOS Induces α-Glucosidase Activity in B. subtilis CU1 and Reduces Lactic and Acetic Acid Production
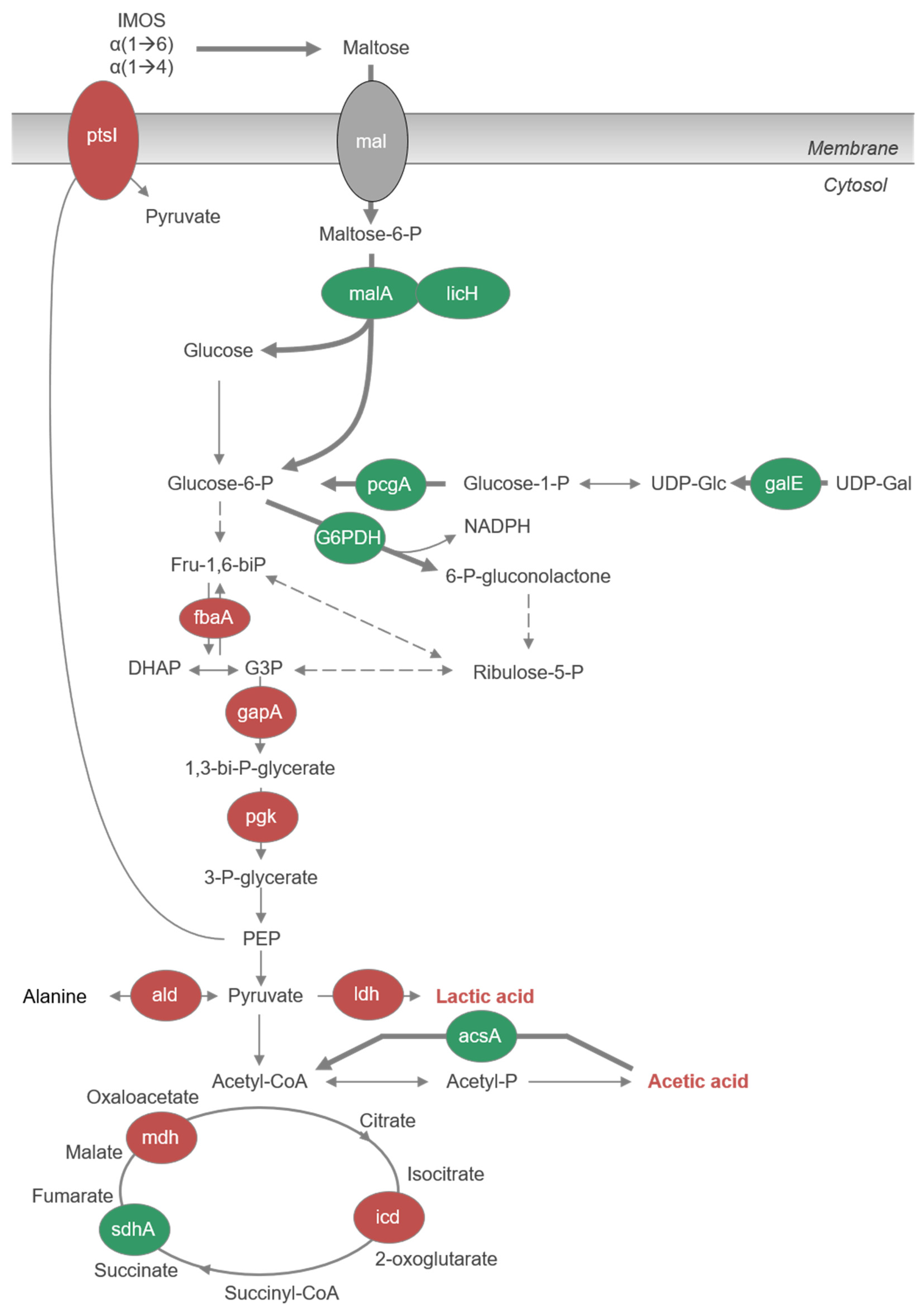
3.6. Proteome Analysis after Growth on IMOS
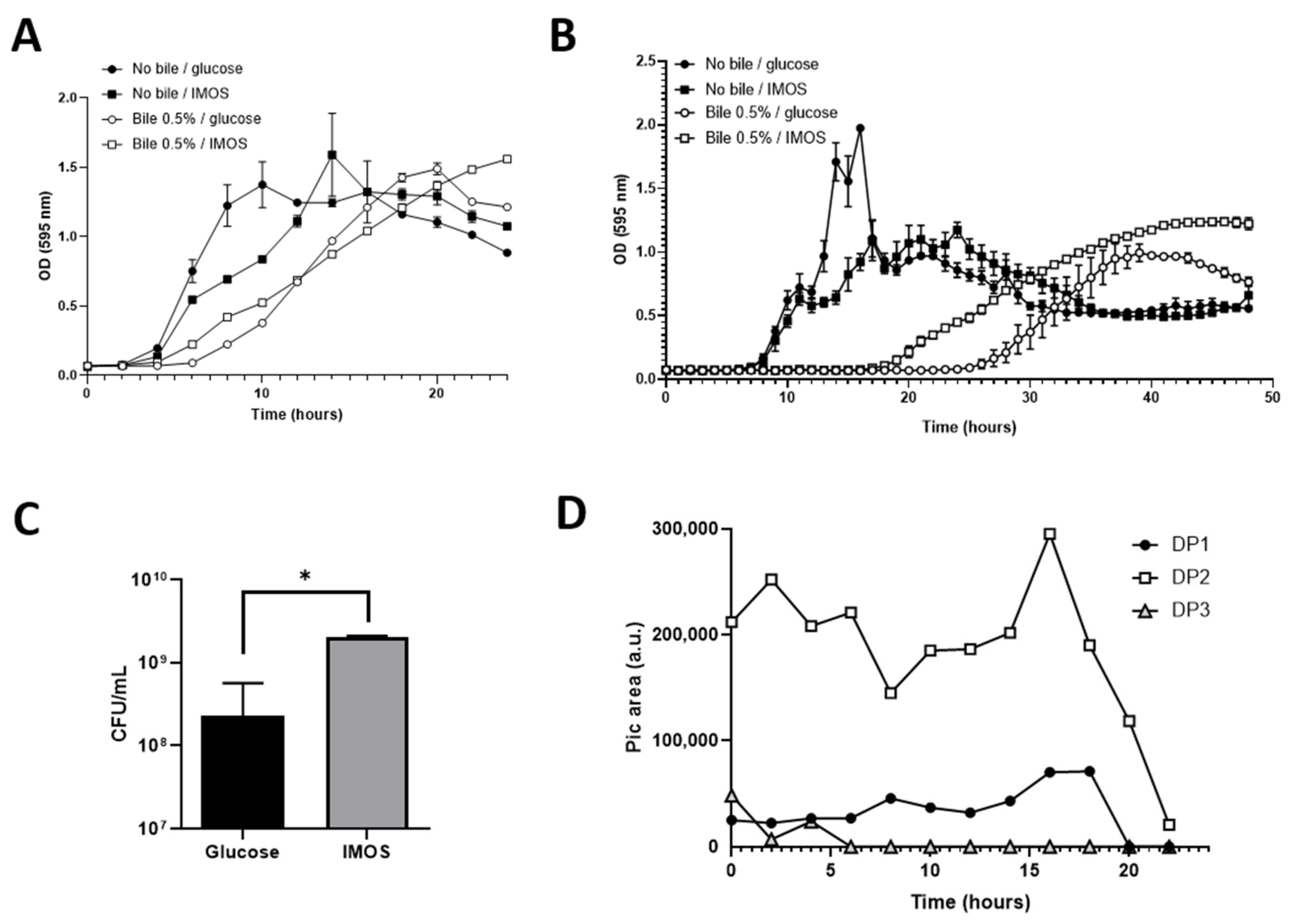
3.7. IMOSs Provide a Fitness Advantage to B. subtilis CU1 for Bile Tolerance
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert Consensus Document. The International Scientific Association for Probiotics and Prebiotics Consensus Statement on the Scope and Appropriate Use of the Term Probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suez, J.; Zmora, N.; Segal, E.; Elinav, E. The Pros, Cons, and Many Unknowns of Probiotics. Nat. Med. 2019, 25, 716–729. [Google Scholar] [CrossRef] [PubMed]
- Wilkins, T.; Sequoia, J. Probiotics for Gastrointestinal Conditions: A Summary of the Evidence. Am. Fam. Physician 2017, 96, 170–178. [Google Scholar] [PubMed]
- Lee, N.-K.; Kim, W.-S.; Paik, H.-D. Bacillus Strains as Human Probiotics: Characterization, Safety, Microbiome, and Probiotic Carrier. Food Sci. Biotechnol. 2019, 28, 1297–1305. [Google Scholar] [CrossRef]
- Spinosa, M.R.; Braccini, T.; Ricca, E.; De Felice, M.; Morelli, L.; Pozzi, G.; Oggioni, M.R. On the Fate of Ingested Bacillus Spores. Res. Microbiol. 2000, 151, 361–368. [Google Scholar] [CrossRef]
- Barbosa, T.M.; Serra, C.R.; La Ragione, R.M.; Woodward, M.J.; Henriques, A.O. Screening for Bacillus Isolates in the Broiler Gastrointestinal Tract. Appl. Environ. Microbiol. 2005, 71, 968–978. [Google Scholar] [CrossRef] [Green Version]
- Casula, G.; Cutting, S.M. Bacillus Probiotics: Spore Germination in the Gastrointestinal Tract. Appl. Environ. Microbiol. 2002, 68, 2344–2352. [Google Scholar] [CrossRef] [Green Version]
- Duc, L.H.; Hong, H.A.; Cutting, S.M. Germination of the Spore in the Gastrointestinal Tract Provides a Novel Route for Heterologous Antigen Delivery. Vaccine 2003, 21, 4215–4224. [Google Scholar] [CrossRef]
- Ramirez-Peralta, A.; Zhang, P.; Li, Y.; Setlow, P. Effects of Sporulation Conditions on the Germination and Germination Protein Levels of Bacillus subtilis Spores. Appl. Environ. Microbiol. 2012, 78, 2689–2697. [Google Scholar] [CrossRef] [Green Version]
- Piewngam, P.; Zheng, Y.; Nguyen, T.H.; Dickey, S.W.; Joo, H.-S.; Villaruz, A.E.; Glose, K.A.; Fisher, E.L.; Hunt, R.L.; Li, B.; et al. Pathogen Elimination by Probiotic Bacillus via Signalling Interference. Nature 2018, 562, 532–537. [Google Scholar] [CrossRef]
- Urdaci, M.C.; Bressollier, P.; Pinchuk, I. Bacillus clausii Probiotic Strains: Antimicrobial and Immunomodulatory Activities. J. Clin. Gastroenterol. 2004, 38, S86–S90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villéger, R.; Saad, N.; Grenier, K.; Falourd, X.; Foucat, L.; Urdaci, M.C.; Bressollier, P.; Ouk, T.-S. Characterization of Lipoteichoic Acid Structures from Three Probiotic Bacillus Strains: Involvement of d-Alanine in Their Biological Activity. Antonie Van Leeuwenhoek 2014, 106, 693–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lefevre, M.; Racedo, S.M.; Ripert, G.; Housez, B.; Cazaubiel, M.; Maudet, C.; Jüsten, P.; Marteau, P.; Urdaci, M.C. Probiotic Strain Bacillus subtilis CU1 Stimulates Immune System of Elderly during Common Infectious Disease Period: A Randomized, Double-Blind Placebo-Controlled Study. Immun. Ageing A 2015, 12, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urdaci, M.C.; Lefevre, M.; Lafforgue, G.; Cartier, C.; Rodriguez, B.; Fioramonti, J. Antidiarrheal Action of Bacillus subtilis CU1 CNCM I-2745 and Lactobacillus plantarum CNCM I-4547 in Mice. Front. Microbiol. 2018, 9, 1537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibson, G.R.; Roberfroid, M.B. Dietary Modulation of the Human Colonic Microbiota: Introducing the Concept of Prebiotics. J. Nutr. 1995, 125, 1401–1412. [Google Scholar] [CrossRef] [PubMed]
- Laparra, J.M.; Sanz, Y. Interactions of Gut Microbiota with Functional Food Components and Nutraceuticals. Pharmacol. Res. 2010, 61, 219–225. [Google Scholar] [CrossRef] [Green Version]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert Consensus Document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) Consensus Statement on the Definition and Scope of Prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [Green Version]
- Cencic, A.; Chingwaru, W. The Role of Functional Foods, Nutraceuticals, and Food Supplements in Intestinal Health. Nutrients 2010, 2, 611–625. [Google Scholar] [CrossRef]
- Kaneko, T.; Yokoyama, A.; Suzuki, M. Digestibility Characteristics of Isomaltooligosaccharides in Comparison with Several Saccharides Using the Rat Jejunum Loop Method. Biosci. Biotechnol. Biochem. 1995, 59, 1190–1194. [Google Scholar] [CrossRef] [Green Version]
- Yen, C.-H.; Tseng, Y.-H.; Kuo, Y.-W.; Lee, M.-C.; Chen, H.-L. Long-Term Supplementation of Isomalto-Oligosaccharides Improved Colonic Microflora Profile, Bowel Function, and Blood Cholesterol Levels in Constipated Elderly People—A Placebo-Controlled, Diet-Controlled Trial. Nutrition 2011, 27, 445–450. [Google Scholar] [CrossRef]
- Ketabi, A.; Dieleman, L.A.; Gänzle, M.G. Influence of Isomalto-Oligosaccharides on Intestinal Microbiota in Rats. J. Appl. Microbiol. 2011, 110, 1297–1306. [Google Scholar] [CrossRef] [PubMed]
- Rycroft, C.E.; Jones, M.R.; Gibson, G.R.; Rastall, R.A. A Comparative in Vitro Evaluation of the Fermentation Properties of Prebiotic Oligosaccharides. J. Appl. Microbiol. 2001, 91, 878–887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Q.; Tan, B.; Mai, K.; Zhang, W.; Ma, H.; Ai, Q.; Wang, X.; Liufu, Z. Dietary Administration of Bacillus (B. licheniformis and B. subtilis) and Isomaltooligosaccharide Influences the Intestinal Microflora, Immunological Parameters and Resistance against Vibrio Alginolyticus in Shrimp, Penaeus japonicus (Decapoda: Penaeidae). Aquac. Res. 2011, 42, 943–952. [Google Scholar] [CrossRef]
- Ropartz, D.; Bodet, P.-E.; Przybylski, C.; Gonnet, F.; Daniel, R.; Fer, M.; Helbert, W.; Bertrand, D.; Rogniaux, H. Performance Evaluation on a Wide Set of Matrix-Assisted Laser Desorption Ionization Matrices for the Detection of Oligosaccharides in a High-Throughput Mass Spectrometric Screening of Carbohydrate Depolymerizing Enzymes. Rapid Commun. Mass Spectrom. RCM 2011, 25, 2059–2070. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ding, X.; Xia, L.; Sun, Y.; Yuan, C.; Yin, J. Proteomic Analysis of Bacillus thuringiensis Strain 4.0718 at Different Growth Phases. Sci. World J. 2012, 2012, e798739. [Google Scholar] [CrossRef] [Green Version]
- Doan, T.; Aymerich, S. Regulation of the Central Glycolytic Genes in Bacillus subtilis: Binding of the Repressor CggR to Its Single DNA Target Sequence Is Modulated by Fructose-1,6-Bisphosphate. Mol. Microbiol. 2003, 47, 1709–1721. [Google Scholar] [CrossRef]
- Chai, Y.; Beauregard, P.B.; Vlamakis, H.; Losick, R.; Kolter, R. Galactose Metabolism Plays a Crucial Role in Biofilm Formation by Bacillus subtilis. mBio 2012, 3, e00184-12. [Google Scholar] [CrossRef] [Green Version]
- Soldo, B.; Scotti, C.; Karamata, D.; Lazarevic, V. The Bacillus subtilis Gne (GneA, GalE) Protein Can Catalyse UDP-Glucose as Well as UDP-N-Acetylglucosamine 4-Epimerisation. Gene 2003, 319, 65–69. [Google Scholar] [CrossRef]
- Stöver, A.G.; Driks, A. Secretion, Localization, and Antibacterial Activity of TasA, a Bacillus subtilis Spore-Associated Protein. J. Bacteriol. 1999, 181, 1664–1672. [Google Scholar] [CrossRef] [Green Version]
- Cenci, G.; Trotta, F.; Caldini, G. Tolerance to Challenges Miming Gastrointestinal Transit by Spores and Vegetative Cells of Bacillus clausii. J. Appl. Microbiol. 2006, 101, 1208–1215. [Google Scholar] [CrossRef]
- Cutting, S.M. Bacillus Probiotics. Food Microbiol. 2011, 28, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Saad, N.; Villéger, R.; Ouk, T.-S.; Delattre, C.; Urdaci, M.; Bressollier, P. Probiotics, Prebiotics, and Synbiotics for Gut Health Benefices. In Beneficial Microbes in Fermented and Functional Foods; CRC Press: Boca Raton, FL, USA, 2014; ISBN 978-0-429-18974-6. [Google Scholar]
- Lefevre, M.; Racedo, S.M.; Denayrolles, M.; Ripert, G.; Desfougères, T.; Lobach, A.R.; Simon, R.; Pélerin, F.; Jüsten, P.; Urdaci, M.C. Safety Assessment of Bacillus subtilis CU1 for Use as a Probiotic in Humans. Regul. Toxicol. Pharmacol. RTP 2017, 83, 54–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohmoto, T.; Tsuji, K.; Kaneko, T.; Shiota, M.; Fukui, F.; Takaku, H.; Nakagawa, Y.; Ichikawa, T.; Kobayash, S. Metabolism of (13)C-Isomaltooligosaccharides in Healthy Men. Biosci. Biotechnol. Biochem. 1992, 56, 937–940. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.; Goyal, A. Functional Oligosaccharides: Production, Properties and Applications. World J. Microbiol. Biotechnol. 2011, 27, 1119–1128. [Google Scholar] [CrossRef]
- Lan, J.; Wang, K.; Chen, G.; Cao, G.; Yang, C. Effects of Inulin and Isomalto-Oligosaccharide on Diphenoxylate-Induced Constipation, Gastrointestinal Motility-Related Hormones, Short-Chain Fatty Acids, and the Intestinal Flora in Rats. Food Funct. 2020, 11, 9216–9225. [Google Scholar] [CrossRef]
- Logtenberg, M.J.; Akkerman, R.; Hobé, R.G.; Donners, K.M.H.; Van Leeuwen, S.S.; Hermes, G.D.A.; de Haan, B.J.; Faas, M.M.; Buwalda, P.L.; Zoetendal, E.G.; et al. Structure-Specific Fermentation of Galacto-Oligosaccharides, Isomalto-Oligosaccharides and Isomalto/Malto-Polysaccharides by Infant Fecal Microbiota and Impact on Dendritic Cell Cytokine Responses. Mol. Nutr. Food Res. 2021, 65, e2001077. [Google Scholar] [CrossRef]
- Hong, H.A.; Duc, L.H.; Cutting, S.M. The Use of Bacterial Spore Formers as Probiotics. FEMS Microbiol. Rev. 2005, 29, 813–835. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.-L.; Li, W.-S.; Xu, D.-N.; Zheng, W.-W.; Liu, Y.; Chen, J.; Qiu, Z.-B.; Dorfman, R.G.; Zhang, J.; Liu, J. Mucosa-Reparing and Microbiota-Balancing Therapeutic Effect of Bacillus subtilis Alleviates Dextrate Sulfate Sodium-Induced Ulcerative Colitis in Mice. Exp. Ther. Med. 2016, 12, 2554–2562. [Google Scholar] [CrossRef] [Green Version]
- Tsukahara, T.; Kimura, Y.; Inoue, R.; Iwata, T. Preliminary Investigation of the Use of Dietary Supplementation with Probiotic Bacillus subtilis Strain QST713 Shows That It Attenuates Antimicrobial-Induced Dysbiosis in Weaned Piglets. Anim. Sci. J. 2020, 91, e13475. [Google Scholar] [CrossRef]
- Marzorati, M.; Van den Abbeele, P.; Bubeck, S.S.; Bayne, T.; Krishnan, K.; Young, A.; Mehta, D.; DeSouza, A. Bacillus subtilis HU58 and Bacillus coagulans SC208 Probiotics Reduced the Effects of Antibiotic-Induced Gut Microbiome Dysbiosis in an M-SHIME® Model. Microorganisms 2020, 8, 1028. [Google Scholar] [CrossRef]
- Li, J.; Tan, B.; Mai, K. Dietary Probiotic Bacillus OJ and Isomaltooligosaccharides Influence the Intestine Microbial Populations, Immune Responses and Resistance to White Spot Syndrome Virus in Shrimp (Litopenaeus vannamei). Aquaculture 2009, 291, 35–40. [Google Scholar] [CrossRef]
- Gu, X.L.; Li, H.; Song, Z.H.; Ding, Y.N.; He, X.; Fan, Z.Y. Effects of Isomaltooligosaccharide and Bacillus Supplementation on Sow Performance, Serum Metabolites, and Serum and Placental Oxidative Status. Anim. Reprod. Sci. 2019, 207, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Chen, J.; Li, H.; Song, Z.; Chang, L.; He, X.; Fan, Z. Isomaltooligosaccharide and Bacillus Regulate the Duration of Farrowing and Weaning-Estrous Interval in Sows during the Perinatal Period by Changing the Gut Microbiota of Sows. Anim. Nutr. 2021, 7, 72–83. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Ketabi, A.; Buchko, A.; Gänzle, M.G. Metabolism of Isomalto-Oligosaccharides by Lactobacillus reuteri and Bifidobacteria. Lett. Appl. Microbiol. 2013, 57, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Böger, M.; Hekelaar, J.; van Leeuwen, S.S.; Dijkhuizen, L.; Lammerts van Bueren, A. Structural and Functional Characterization of a Family GH53 β-1,4-Galactanase from Bacteroides Thetaiotaomicron That Facilitates Degradation of Prebiotic Galactooligosaccharides. J. Struct. Biol. 2019, 205, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Endo, A.; Tanno, H.; Kadowaki, R.; Fujii, T.; Tochio, T. Extracellular Fructooligosaccharide Degradation in Anaerostipes Hadrus for Co-Metabolism with Non-Fructooligosaccharide Utilizers. Biochem. Biophys. Res. Commun. 2022, 613, 81–86. [Google Scholar] [CrossRef]
- Garcia, C.A.; Gardner, J.G. Bacterial α-Diglucoside Metabolism: Perspectives and Potential for Biotechnology and Biomedicine. Appl. Microbiol. Biotechnol. 2021, 105, 4033–4052. [Google Scholar] [CrossRef]
- Yamamoto, H.; Serizawa, M.; Thompson, J.; Sekiguchi, J. Regulation of the Glv Operon in Bacillus subtilis: YfiA (GlvR) Is a Positive Regulator of the Operon That Is Repressed through CcpA and Cre. J. Bacteriol. 2001, 183, 5110–5121. [Google Scholar] [CrossRef] [Green Version]
- Zamboni, N.; Fischer, E.; Laudert, D.; Aymerich, S.; Hohmann, H.-P.; Sauer, U. The Bacillus subtilis YqjI Gene Encodes the NADP+-Dependent 6-P-Gluconate Dehydrogenase in the Pentose Phosphate Pathway. J. Bacteriol. 2004, 186, 4528–4534. [Google Scholar] [CrossRef] [Green Version]
- Leser, T.D.; Knarreborg, A.; Worm, J. Germination and Outgrowth of Bacillus subtilis and Bacillus licheniformis Spores in the Gastrointestinal Tract of Pigs. J. Appl. Microbiol. 2008, 104, 1025–1033. [Google Scholar] [CrossRef]
- Setlow, P. Germination of Spores of Bacillus Species: What We Know and Do Not Know. J. Bacteriol. 2014, 196, 1297–1305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hornstra, L.M.; de Vries, Y.P.; de Vos, W.M.; Abee, T. Influence of Sporulation Medium Composition on Transcription of Ger Operons and the Germination Response of Spores of Bacillus cereus ATCC 14579. Appl. Environ. Microbiol. 2006, 72, 3746–3749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez, B.; Champomier-Vergès, M.-C.; Anglade, P.; Baraige, F.; de Los Reyes-Gavilán, C.G.; Margolles, A.; Zagorec, M. Proteomic Analysis of Global Changes in Protein Expression during Bile Salt Exposure of Bifidobacterium longum NCIMB 8809. J. Bacteriol. 2005, 187, 5799–5808. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, B.; de los Reyes-Gavilán, C.G.; Margolles, A. The F1F0-ATPase of Bifidobacterium animalis Is Involved in Bile Tolerance. Environ. Microbiol. 2006, 8, 1825–1833. [Google Scholar] [CrossRef] [Green Version]
- Sánchez, B.; Champomier-Vergès, M.-C.; Stuer-Lauridsen, B.; Ruas-Madiedo, P.; Anglade, P.; Baraige, F.; de los Reyes-Gavilán, C.G.; Johansen, E.; Zagorec, M.; Margolles, A. Adaptation and Response of Bifidobacterium animalis Subsp. Lactis to Bile: A Proteomic and Physiological Approach. Appl. Environ. Microbiol. 2007, 73, 6757–6767. [Google Scholar] [CrossRef] [Green Version]
- Sanchez, B.; Ruiz, L.; de los Reyes-Gavilan, C.G.; Margolles, A. Proteomics of Stress Response in Bifidobacterium. Front. Biosci. J. Virtual Libr. 2008, 13, 6905–6919. [Google Scholar] [CrossRef] [Green Version]
- Perrin, S.; Grill, J.P.; Schneider, F. Effects of Fructooligosaccharides and Their Monomeric Components on Bile Salt Resistance in Three Species of Bifidobacteria. J. Appl. Microbiol. 2000, 88, 968–974. [Google Scholar] [CrossRef] [Green Version]
- Ruas-Madiedo, P.; Hernández-Barranco, A.; Margolles, A.; de los Reyes-Gavilán, C.G. A Bile Salt-Resistant Derivative of Bifidobacterium animalis Has an Altered Fermentation Pattern When Grown on Glucose and Maltose. Appl. Environ. Microbiol. 2005, 71, 6564–6570. [Google Scholar] [CrossRef] [Green Version]
- Hamon, E.; Horvatovich, P.; Izquierdo, E.; Bringel, F.; Marchioni, E.; Aoudé-Werner, D.; Ennahar, S. Comparative Proteomic Analysis of Lactobacillus plantarum for the Identification of Key Proteins in Bile Tolerance. BMC Microbiol. 2011, 11, 63. [Google Scholar] [CrossRef] [Green Version]

| Minimal Medium | Glucose | Inuline | FOS | GOS | XOS | AXOS | Lactulose | SOS | IMOS | |
|---|---|---|---|---|---|---|---|---|---|---|
| B. subtilis CU1 | - | ++ | + | ++ | ++ | ++ | + | - | ++ | ++ |
| B. subtilis 2-4/2 | - | ++ | + | ++ | ++ | + | + | - | ++ | ++ |
| B. licheniformis BL31 | - | + | + | + | ++ | + | - | - | ++ | + |
| B. amyloliquefaciens 26D | - | ++ | ++ | ++ | ++ | ++ | ++ | - | ++ | ++ |
| B. cereus CH | - | - | - | - | - | - | - | - | + | - |
| m/z | Intensity | Structure |
|---|---|---|
| 527.17 | 6157.68 | Hexose DP3 [M + Na]+ |
| 689.214 | 2223.32 | Hexose DP4 [M + Na]+ |
| 851.26 | 1450.49 | Hexose DP5 [M + Na]+ |
| 1013.31 | 2968.06 | Hexose DP6 [M + Na]+ |
| 1175.391 | 2780 | Hexose DP7 [M + Na]+ |
| 1337.498 | 990.45 | Hexose DP8 [M + Na]+ |
| 1499.634 | 306.84 | Hexose DP9 [M + Na]+ |
| 1661.8 | 123.6 | Hexose DP10 [M + Na]+ |
| 1823.997 | 60.99 | Hexose DP11 [M + Na]+ |
| 1986.217 | 32.69 | Hexose DP12 [M + Na]+ |
| 2148.556 | 19.39 | Hexose DP13 [M + Na]+ |
| m/z | Intensity | Structure | m/z | Intensity | Structure |
|---|---|---|---|---|---|
| 2304.9 | 191.8 | Hexose DP14 [M + Na]+ | 5716.9 | 357.89 | Hexose DP35 [M + Na]+ |
| 2467.5 | 2417.54 | Hexose DP15 [M + Na]+ | 5879.2 | 289.48 | Hexose DP36 [M + Na]+ |
| 2630.1 | 5320.01 | Hexose DP16 [M + Na]+ | 6041.2 | 241.18 | Hexose DP37 [M + Na]+ |
| 2792.6 | 6904.94 | Hexose DP17 [M + Na]+ | 6203.5 | 171.66 | Hexose DP38 [M + Na]+ |
| 2955.2 | 7446.49 | Hexose DP18 [M + Na]+ | 6366 | 149.94 | Hexose DP39 [M + Na]+ |
| 3117.8 | 6954.73 | Hexose DP19 [M + Na]+ | 6528 | 95.96 | Hexose DP40 [M + Na]+ |
| 3280.3 | 6237.11 | Hexose DP20 [M + Na]+ | 6690.7 | 77.76 | Hexose DP41 [M + Na]+ |
| 3442.9 | 5472.17 | Hexose DP21 [M + Na]+ | 6852.7 | 60.27 | Hexose DP42 [M + Na]+ |
| 3605.4 | 4478.89 | Hexose DP22 [M + Na]+ | 7015.1 | 42.01 | Hexose DP43 [M + Na]+ |
| 3768 | 3701.86 | Hexose DP23 [M + Na]+ | 7177.7 | 48.18 | Hexose DP44 [M + Na]+ |
| 3930.3 | 3221.14 | Hexose DP24 [M + Na]+ | 7340 | 28.32 | Hexose DP45 [M + Na]+ |
| 4092.8 | 2474.79 | Hexose DP25 [M + Na]+ | 7501.3 | 23.11 | Hexose DP46 [M + Na]+ |
| 4255.5 | 2011.96 | Hexose DP26 [M + Na]+ | 7663.1 | 20.08 | Hexose DP47 [M + Na]+ |
| 4417.9 | 1667.19 | Hexose DP27 [M + Na]+ | 7826.8 | 16.6 | Hexose DP48 [M + Na]+ |
| 4580.1 | 1462.97 | Hexose DP28 [M + Na]+ | 7988.4 | 16.03 | Hexose DP49 [M + Na]+ |
| 4742.8 | 1206.39 | Hexose DP29 [M + Na]+ | 8150.6 | 7.11 | Hexose DP50 [M + Na]+ |
| 4905.1 | 973.39 | Hexose DP30 [M + Na]+ | 8312 | 14.43 | Hexose DP51 [M + Na]+ |
| 5067.5 | 748.39 | Hexose DP31 [M + Na]+ | 8474.8 | 9.34 | Hexose DP52 [M + Na]+ |
| 5229.9 | 573.19 | Hexose DP32 [M + Na]+ | 8799.6 | 4.14 | Hexose DP54 [M + Na]+ |
| 5392.1 | 487.02 | Hexose DP33 [M + Na]+ | 8961.1 | 6.26 | Hexose DP55 [M + Na]+ |
| 5554.6 | 388.19 | Hexose DP34 [M + Na]+ | 9121 | 5.09 | Hexose DP56 [M + Na]+ |
| Identified Proteins | Access Number | Name | Spots | Fold Change | p-Value |
|---|---|---|---|---|---|
| Amino acid & protein synthesis | |||||
| Serine-glycine hydroxymethyltransferase | P39148 | glyA | 949 | −4.0 | <0.001 |
| Elongation factor G | P80868 | fusA | 412 | −3.3 | <0.001 |
| 60 kDa chaperonin | P28598 | groEL | 692 | −2.1 | 0.016 |
| Elongation factor Tu | P33166 | tufA | 1022 | −3.3 | <0.001 |
| Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B | O30509 | gatB | 600 | 1.7 | 0.001 |
| Chaperone protein DnaK | P17820 | dnaK | 483 | −1.7 | 0.005 |
| Methionine--tRNA ligase | P37465 | metS | 1076 | 2.1 | 0.023 |
| Phenylalanine--tRNA ligase beta subunit | P17922 | pheT | 326 | 1.9 | 0.035 |
| Energy metabolism | |||||
| Cytidylate kinase | P38493 | cmk | 867 | 2.2 | 0.002 |
| Acetyl-coenzyme A synthetase | P39062 | acsA | 1020 | 2.2 | 0.002 |
| ATP synthase subunit beta | P37809 | atpD | 878 | −2.1 | 0.003 |
| Carbohydrate metabolism & transport | |||||
| Methylmalonate semialdehyde dehydrogenase | P42412 | iolA | 586 | 2.1 | <0.001 |
| Alanine dehydrogenase | Q08352 | ald | 845 | −1.5 | 0.004 |
| Isocitrate dehydrogenase | P39126 | icd | 894 | −2.9 | <0.001 |
| Maltose-6-phosphate glucosidase | P54716 | malA | 609 | 2.2 | <0.001 |
| Probable 6-phospho-beta-glucosidase | P46320 | licH | 1047 | 2 | 0.002 |
| UDP-glucose 4-epimerase | P55180 | galE | 741 | 1.5 | 0.002 |
| Succinate dehydrogenase flavoprotein subunit | P08065 | sdhA | 518 | 1.5 | 0.002 |
| Glyceraldehyde-3-phosphate dehydrogenase | P09124 | gapA | 1042 | −2 | 0.003 |
| Phosphoglucomutase | P18159 | pgcA | 458 | 1.4 | 0.02 |
| Probable fructose-bisphosphate aldolase | P13243 | fbaA | 834 | −2.4 | <0.001 |
| Phosphoglycerate kinase | P40924 | pgk | 706 | −1.6 | 0.006 |
| Malate dehydrogenase | P49814 | mdh | 760 | −1.8 | 0.005 |
| L-lactate dehydrogenase | P13714 | ldh | 729 | −1.5 | 0.019 |
| Phosphoenolpyruvate-protein phosphotransferase | P08838 | ptsI | 1073 | −1.6 | 0.022 |
| Cell-wall synthesis | |||||
| UDP-N-acetylmuramoyl-L-alanyl-D-glutamate--2,6-diaminopimelate ligase | Q03523 | murE | 1012 | 2.1 | 0.003 |
| Spore coat-associated protein N | P54507 | tasA | 1062 | −1.4 | 0.015 |
| Antibiotic synthesis | |||||
| Polyketide biosynthesis 3-hydroxy-3-methylglutaryl-ACP synthase | P40830 | pksG | 601 | 1.6 | 0.007 |
| Other functions | |||||
| Pyridoxal biosynthesis lyase | P37527 | pdxS | 923 | −1.9 | 0.002 |
| DNA-directed RNA polymerase subunit alpha | P20429 | rpoA | 893 | −1.4 | 0.012 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villéger, R.; Pinault, E.; Vuillier-Devillers, K.; Grenier, K.; Landolt, C.; Ropartz, D.; Sol, V.; Urdaci, M.C.; Bressollier, P.; Ouk, T.-S. Prebiotic Isomaltooligosaccharide Provides an Advantageous Fitness to the Probiotic Bacillus subtilis CU1. Appl. Sci. 2022, 12, 6404. https://doi.org/10.3390/app12136404
Villéger R, Pinault E, Vuillier-Devillers K, Grenier K, Landolt C, Ropartz D, Sol V, Urdaci MC, Bressollier P, Ouk T-S. Prebiotic Isomaltooligosaccharide Provides an Advantageous Fitness to the Probiotic Bacillus subtilis CU1. Applied Sciences. 2022; 12(13):6404. https://doi.org/10.3390/app12136404
Chicago/Turabian StyleVilléger, Romain, Emilie Pinault, Karine Vuillier-Devillers, Karine Grenier, Cornelia Landolt, David Ropartz, Vincent Sol, Maria C. Urdaci, Philippe Bressollier, and Tan-Sothéa Ouk. 2022. "Prebiotic Isomaltooligosaccharide Provides an Advantageous Fitness to the Probiotic Bacillus subtilis CU1" Applied Sciences 12, no. 13: 6404. https://doi.org/10.3390/app12136404






