The Role of Iron in DNA and Genomic Instability in Cancer, a Target for Iron Chelators That Can Induce ROS
Abstract
1. Introduction
2. Iron Chelators Design and Redox Activity of Iron
2.1. Redox Activity
2.2. Thermodynamics, Stability, Sensitivity and Lipophilicity
3. Brief Overview of Iron Metabolism, the Labile Iron Pool and How It Is Altered in Cancer
3.1. Cellular Iron Homeostasis
Mitochondrial Iron Metabolism and Fe-S Biosynthesis
3.2. Altered Iron Metabolism in Cancer
4. Genomic Instability, Fe-S Clusters and Iron Chelators
4.1. Iron Is Vital for DNA Replication and Repair
DNA Helicases and Polymerases
4.2. Ribonucleotide Reductases
Iron Chelators and Their Activity against RNR
4.3. Iron Chelators and The Cell Cycle
4.4. Could Iron Chelators Passively or Actively Target Fe-S Clusters
5. Iron, ROS and Iron Chelators That Can Cause DNA Damage in Cancer
5.1. Iron Chelators and ROS
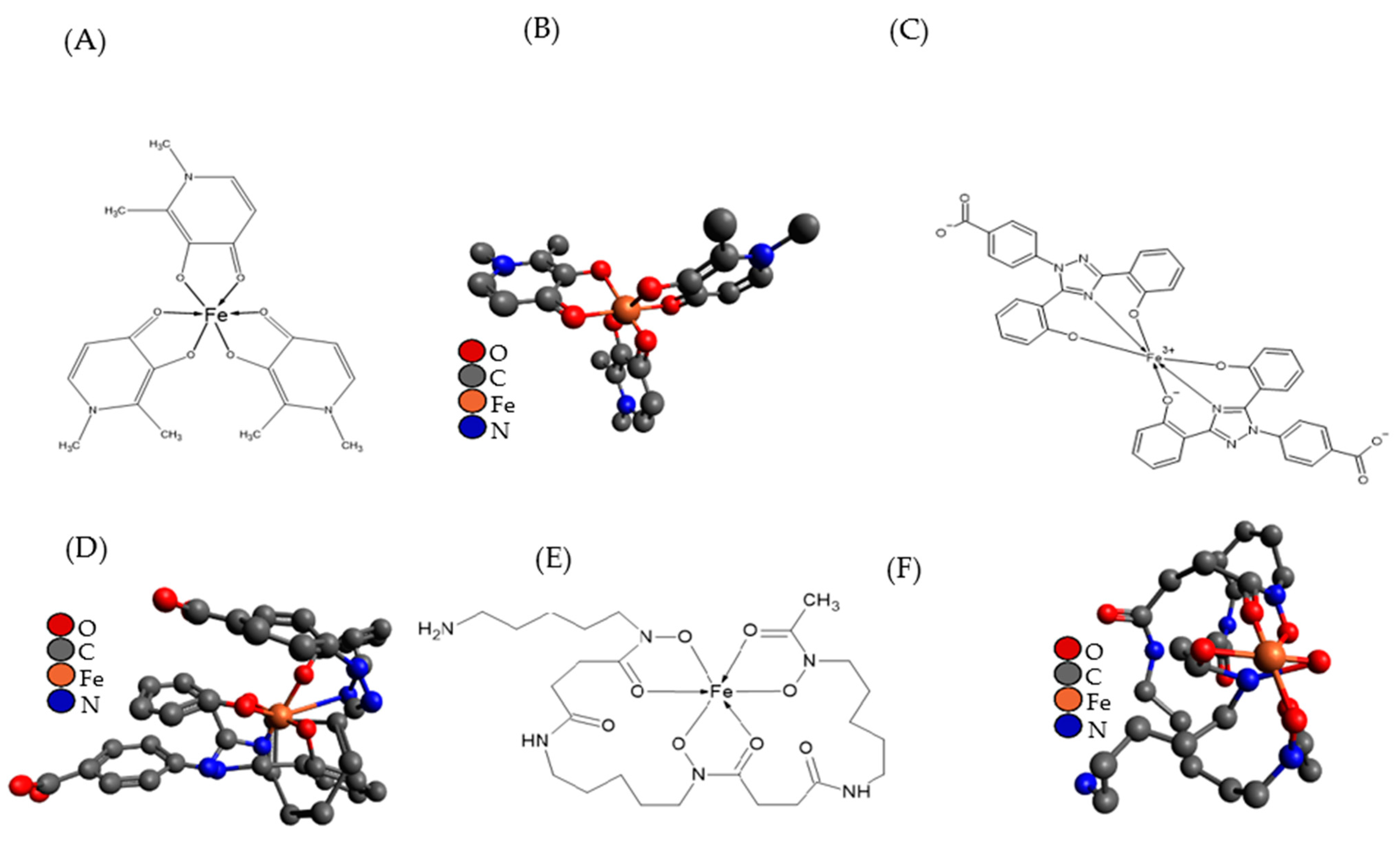
5.1.1. Mimosine and Analogues
5.1.2. Di-2-Pyridylketone Isonicotinoyl Hydrazone (PKIH)
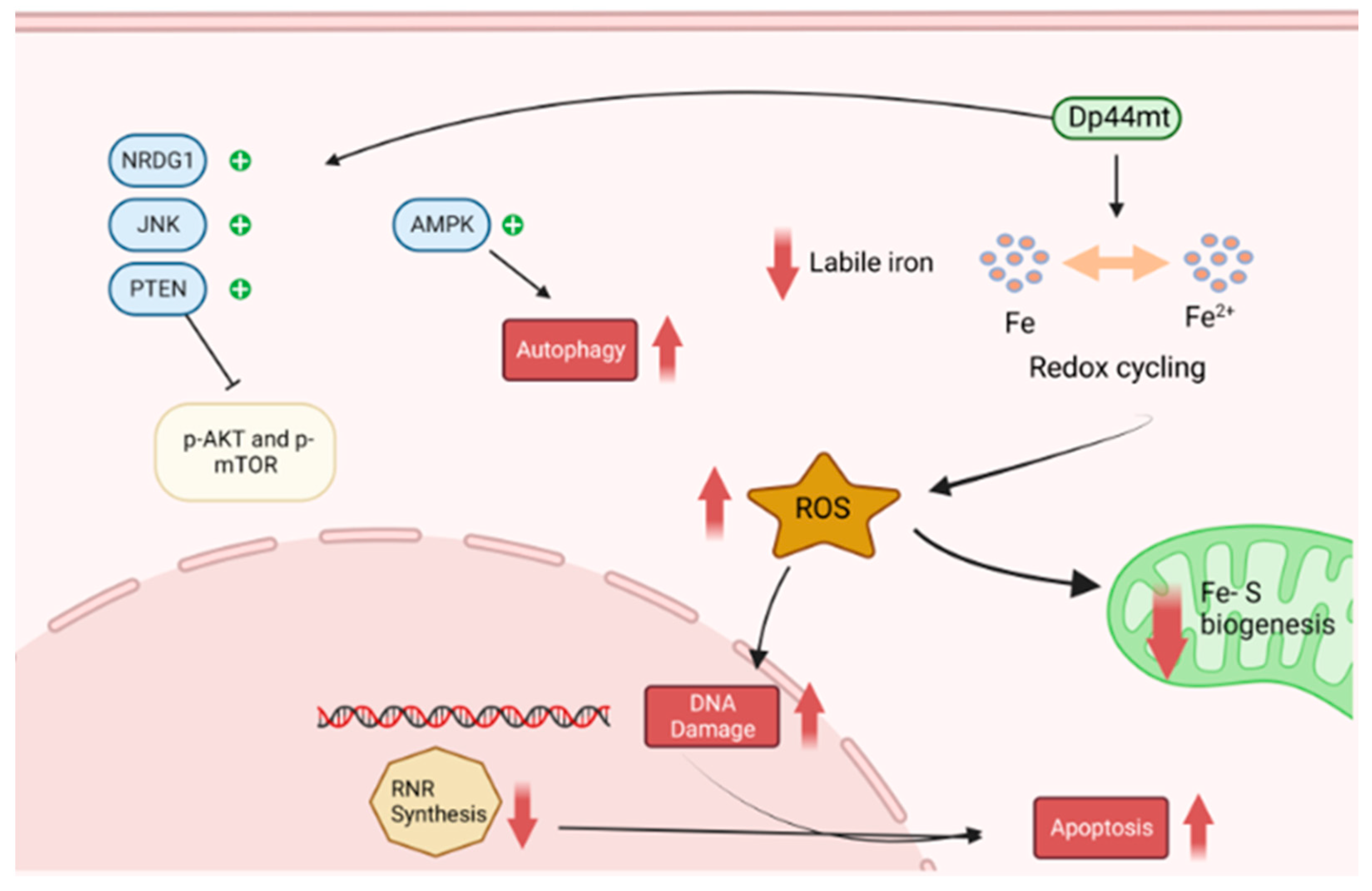
5.1.3. DP44mT and Analogues
5.1.4. Di-2-Pyridineketone Hydrazone Dithiocarbamate (DpdtC)
5.1.5. KS10076
5.1.6. Triapine and Deferiprone
6. Perspectives and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| reactive oxygen species | ROS |
| ribonucleotide reductase | RnR |
| transferrin receptor 1 | TfR1 |
| divalent metal transporter1 | DMT1 |
| ferritin | Ft |
| ferroportin | FPN1 |
| iron regulating protein one and iron regulating protein two | IRP1 and IRP2 |
| iron-responsive element | IRE |
| outer mitochondrial membrane | OMM |
| inner mitochondrial membrane | IMM |
| Glutaredoxin-related protein 5 | GLRX5 |
| Iron sulphur cluster | ISC |
| DNA damage response | DDR |
| Nutrient-deprivation autophagy factor-1 | NAF-1 |
| transcription factor II H | TFIIH |
| Fanconi Anemia | FA |
| base excision repair | BER |
| uridine diphosphate | dUDP |
| pyridoxal isonicotinoyl hydrazone | PIH |
| triapine | 3-AP |
| cyclin dependant kinases | CDKs |
| hypoxia-inducible factor-1a | HIF-1a |
| N-myc downstream regulatory gene-1 | Ndrg-1 |
| 2-hydroxy-1-naphthylaldehydeisonicotinoyl hydrazone | 311 |
| 4-[3,5-bis-(hydroxyphenyl)-1,2,4-triazol-1-yl]-benzoic acid | ICL670 |
| deferoxamine | DFO |
| hydrogen atom transfer | HAT |
| β-N-3-hydroxy-4-pyridine-a-amino-propionic acid | Mimosine |
| N-acetyl cysteine | NAC |
| Jun-N-terminal protein kinase | JNK |
| Di-2-Pyridylketone Isonicotinoyl Hydrazone | PKIH |
| di-2-pyridyl ketone-4,4,-dimethyl-3-thiosemicarbazone | Dp44mT |
| di-2-pyridylketone 4-cyclohexyl-4-methyl-3-thiosemicarbazone | DpC |
| Di-2-pyridineketone hydrazone dithiocarbamate | DpdtC |
| aldehyde dehydrogenase isoform 1-positive | ALDH1+ |
References
- Theil, E.C.; Goss, D.J. Living with Iron (and Oxygen): Questions and Answers about Iron Homeostasis. Chem. Rev. 2009, 109, 4568–4579. [Google Scholar] [CrossRef] [PubMed]
- Porter, C.J.H.; Trevaskis, N.L.; Charman, W.N. Lipids and lipid-based formulations: Optimizing the oral delivery of lipophilic drugs. Nat. Rev. Drug Discov. 2007, 6, 231–248. [Google Scholar] [CrossRef] [PubMed]
- Pelicano, H.; Carney, D.; Huang, P. ROS stress in cancer cells and therapeutic implications. Drug Resist. Update 2004, 7, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Beinert, H.; Holm, R.H.; Munck, E. Iron-sulfur clusters: Nature’s modular, multipurpose structures. Science 1997, 277, 653–659. [Google Scholar] [CrossRef]
- Richardson, D.R.; Baker, E. The uptake of iron and transferrin by the human-malignant melanoma cell. Biochim. Biophys. Acta 1990, 1053, 1–12. [Google Scholar] [CrossRef]
- Chekhun, S.V.; Lukyanova, N.Y.; Shvets, Y.V.; Burlaka, A.P.; Buchynska, L.G. Significance of ferritin expression in formation of malignant phenotype of human breast cancer cells. Exp. Oncol. 2014, 36, 179–183. [Google Scholar]
- Simoes, R.V.; Veeraperumal, S.; Serganova, I.S.; Kruchevsky, N.; Varshavsky, J.; Blasberg, R.G.; Ackerstaff, E.; Koutcher, J.A. Inhibition of prostate cancer proliferation by Deferiprone. Nmr. Biomed. 2017, 30, 11. [Google Scholar] [CrossRef]
- Perillo, B.; Di Donato, M.; Pezone, A.; Di Zazzo, E.; Giovannelli, P.; Galasso, G.; Castoria, G.; Migliaccio, A. ROS in cancer therapy: The bright side of the moon. Exp. Mol. Med. 2020, 52, 192–203. [Google Scholar] [CrossRef]
- Seebacher, N.A.; Richardson, D.R.; Jansson, P.J. A mechanism for overcoming P-glycoprotein-mediated drug resistance: Novel combination therapy that releases stored doxorubicin from lysosomes via lysosomal permeabilization using Dp44mT or DpC. Cell Death Dis. 2016, 7, e2510. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Zhang, L.P.; Chen, H.; Song, H.Q.; Chen, S.G.; Xiao, H.H. Enhancing the chemotherapeutic efficacy of platinum prodrug nanoparticles and inhibiting cancer metastasis by targeting iron homeostasis. Nanoscale Horiz. 2020, 5, 999–1015. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Ohara, T.; Xing, B.Y.; Qi, J.P.; Noma, K.; Matsukawa, A. A Promising New Anti-Cancer Strategy: Iron Chelators Targeting CSCs. Acta Med. Okayama 2020, 74, 1–6. [Google Scholar] [PubMed]
- Recalcati, S.; Gammella, E.; Cairo, G. Dysregulation of iron metabolism in cancer stem cells. Free Radic. Biol. Med. 2019, 133, 216–220. [Google Scholar] [CrossRef]
- Finkel, T. Reactive oxygen species and signal transduction. IUBMB Life 2001, 52, 3–6. [Google Scholar] [CrossRef]
- Trachootham, D.; Lu, W.Q.; Ogasawara, M.A.; Valle, N.R.D.; Huang, P. Redox regulation of cell survival. Antioxid. Redox Signal. 2008, 10, 1343–1374. [Google Scholar] [CrossRef]
- Andrews, N.C. Iron homeostasis: Insights from genetics and animal models. Nat. Rev. Genet. 2000, 1, 208–217. [Google Scholar] [CrossRef]
- Halliwell, B.; Gutteridge, J.M.C. Oxygen-toxicity, oxygen radicals, transition-metals and disease. Biochem. J. 1984, 219, 1–14. [Google Scholar] [CrossRef]
- Stohner, J.; Quack, M. Education about the Use of Quantities, Units and Symbols in Chemistry: The Earlier the Better. In Chemistry Education in the Ict Age; Springer: Dordrecht, The Netherlands, 2009; pp. 339–344. [Google Scholar] [CrossRef]
- Koppenol, W.H. The Haber-Weiss cycle—70 years later. Redox Rep. 2001, 6, 229–234. [Google Scholar] [CrossRef]
- Koppenol, W.H. Thermodynamics of reactions involving oxyradicals and hydrogen-peroxide. Bioelectrochem. Bioenerg. 1987, 18, 3–11. [Google Scholar] [CrossRef]
- Crichton, R.R.; Ward, R.J. An overview of iron metabolism: Molecular and cellular criteria for the selection of iron chelators. Curr. Med. Chem. 2003, 10, 997–1004. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.F.; Pignatello, J.J. Activation of hydrogen-peroxide by iron(III) chelates for abiotic degradation of herbicides and insecticides in water. J. Agric. Food Chem. 1993, 41, 308–312. [Google Scholar] [CrossRef]
- Salgado, P.; Melin, V.; Contreras, D.; Moreno, Y.; Mansilla, H.D. Fenton reaction driven by iron ligands. J. Chil. Chem. Soc. 2013, 58, 2096–2101. [Google Scholar] [CrossRef]
- Shao, J.M.; Zhou, B.S.; Di Bilio, A.J.; Zhu, L.J.; Wang, T.L.; Qi, C.; Shih, J.; Yen, Y. A ferrous-Triapine complex mediates formation of reactive oxygen species chat inactivate human ribonucleotide reductase. Mol. Cancer Ther. 2006, 5, 586–592. [Google Scholar] [CrossRef]
- Lehmann, C.; Islam, S.; Jarosch, S.; Zhou, J.; Hoskin, D.; Greenshields, A.; Al-Banna, N.; Sharawy, N.; Sczcesniak, A.; Kelly, M.; et al. The Utility of Iron Chelators in the Management of Inflammatory Disorders. Mediat. Inflamm. 2015, 2015, 12. [Google Scholar] [CrossRef]
- Hancock, R.D.; Martell, A.E. Ligand design for selective complexation of metal-ions in aqueous-solution. Chem. Rev. 1989, 89, 1875–1914. [Google Scholar] [CrossRef]
- Tilbrook, G.S.; Hider, R.C. Iron chelators for clinical use. In Metal Ions in Biological Systems; Iron Transport and Storage in Microorganisms, Plants, and Animals; Sigel, A., Sigel, H., Eds.; Metal Ions in Biological Systems; Marcel Dekker, Inc.: New York, NY, USA; Basel, Switzerland, 1998; Volume 35, pp. 691–730. [Google Scholar]
- Hider, R.C.; Zhou, T. The design of orally active iron chelators. In Proceedings of the Cooley’s Anemia: Eighth Symposium, Lake Buena Vista, FL, USA, 17–19 March 2005; Volume 1054, pp. 141–154. [Google Scholar] [CrossRef]
- Liu, Z.D.; Hider, R.C. Design of clinically useful iron(III)-selective chelators. Med. Res. Rev. 2002, 22, 26–64. [Google Scholar] [CrossRef]
- Zhou, T.; Winkelmann, G.; Dai, Z.Y.; Hider, R.C. Design of clinically useful macromolecular iron chelators. J. Pharm. Pharmacol. 2011, 63, 893–903. [Google Scholar] [CrossRef]
- Pearson, R.G. Hard and soft acids and bases. J. Am. Chem. Soc. 1963, 85, 3533–3539. [Google Scholar] [CrossRef]
- Kalinowski, D.S.; Richardson, D.R. Future of toxicology-iron chelators and differing modes of action and toxicity: The changing face of iron chelation therapy. Chem. Res. Toxicol. 2007, 20, 715–720. [Google Scholar] [CrossRef]
- Helsel, M.E.; Franz, K.J. Pharmacological activity of metal binding agents that alter copper bioavailability. Dalton Trans. 2015, 44, 8760–8770. [Google Scholar] [CrossRef] [PubMed]
- Crisponi, G.; Remelli, M. Iron chelating agents for the treatment of iron overload. Coord. Chem. Rev. 2008, 252, 1225–1240. [Google Scholar] [CrossRef]
- Lobo, S. Is there enough focus on lipophilicity in drug discovery? Expert. Opin. Drug Discov. 2020, 15, 261–263. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, M.; Gomez-Orellana, I. Challenges for the oral delivery of macromolecules. Nat. Rev. Drug Discov. 2003, 2, 289–295. [Google Scholar] [CrossRef]
- Charman, W.N.; Stella, V.J. Transport of lipophilic molecules by the intestinal lymphatic-system. Adv. Drug Deliv. Rev. 1991, 7, 1–14. [Google Scholar] [CrossRef]
- Ahn, H.; Park, J.-H. Liposomal delivery systems for intestinal lymphatic drug transport. Biomater. Res. 2016, 20, 36. [Google Scholar] [CrossRef]
- Kararli, T.T. GASTROINTESTINAL ABSORPTION OF DRUGS. Crit. Rev. Ther. Drug Carr. Syst. 1989, 6, 39–86. [Google Scholar]
- Hodges, Y.K.; Antholine, W.E.; Horwitz, L.D. Effect on ribonucleotide reductase of novel lipophilic iron chelators: The desferri-exochelins. Biochem. Biophys. Res. Commun. 2004, 315, 595–598. [Google Scholar] [CrossRef]
- Lipinski, C.A.; Lombardo, F.; Dominy, B.W.; Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2012, 64, 4–17. [Google Scholar] [CrossRef]
- Davis, B.A.; Porter, J.B. Long-term outcome of continuous 24-hour deferoxamine infusion via indwelling intravenous catheters in high-risk beta-thalassemia. Blood 2000, 95, 1229–1236. [Google Scholar] [CrossRef]
- Richardson, D.R.; Lane, D.J.R.; Becker, E.M.; Huang, M.L.H.; Whitnall, M.; Rahmanto, Y.S.; Sheftel, A.D.; Ponka, P. Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc. Natl. Acad. Sci. USA 2010, 107, 10775–10782. [Google Scholar] [CrossRef] [PubMed]
- Fleming, M.D.; Romano, M.A.; Su, M.A.; Garrick, L.M.; Garrick, M.D.; Andrews, N.C. Nramp2 is mutated in the anemic Belgrade (b) rat: Evidence of a role for Nramp2 in endosomal iron transport. Proc. Natl. Acad. Sci. USA 1998, 95, 1148–1153. [Google Scholar] [CrossRef] [PubMed]
- Petronek, M.S.; Spitz, D.R.; Buettner, G.R.; Allen, B.G. Linking Cancer Metabolic Dysfunction and Genetic Instability through the Lens of Iron Metabolism. Cancers 2019, 11, 1077. [Google Scholar] [CrossRef] [PubMed]
- Hider, R.C.; Kong, X.L. Iron speciation in the cytosol: An overview. Dalton Trans. 2013, 42, 3220–3229. [Google Scholar] [CrossRef] [PubMed]
- Lv, H.H.; Shang, P. The significance, trafficking and determination of labile iron in cytosol, mitochondria and lysosomes. Metallomics 2018, 10, 899–916. [Google Scholar] [CrossRef]
- Hilton, R.J.; Andros, N.D.; Watt, R.K. The ferroxidase center is essential for ferritin iron loading in the presence of phosphate and minimizes side reactions that form Fe(III)-phosphate colloids. Biometals 2012, 25, 259–273. [Google Scholar] [CrossRef]
- Cairo, G.; Pietrangelo, A. Iron regulatory proteins in pathobiology. Biochem. J. 2000, 352, 241–250. [Google Scholar] [CrossRef]
- Brown, R.A.M.; Richardson, K.L.; Kabir, T.D.; Trinder, D.; Ganss, R.; Leedman, P.J. Altered Iron Metabolism and Impact in Cancer Biology, Metastasis, and Immunology. Front. Oncol. 2020, 10, 25. [Google Scholar] [CrossRef]
- Wilkinson, N.; Pantopoulos, K. The IRP/IRE system in vivo: Insights from mouse models. Front. Pharmacol. 2014, 5, 15. [Google Scholar] [CrossRef]
- Ganz, T.; Nemeth, E. Hepcidin and iron homeostasis. Biochim. Biophys. Acta-Mol. Cell Res. 2012, 1823, 1434–1443. [Google Scholar] [CrossRef]
- Thompson, J.W.; Bruick, R.K. Protein degradation and iron homeostasis. Biochim. Biophys. Acta-Mol. Cell Res. 2012, 1823, 1484–1490. [Google Scholar] [CrossRef] [PubMed]
- Lambe, T.; Simpson, R.J.; Dawson, S.; Bouriez-Jones, T.; Crockford, T.L.; Lepherd, M.; Latunde-Dada, G.O.; Robinson, H.; Raja, K.B.; Campagna, D.R.; et al. Identification of a Steap3 endosomal targeting motif essential for normal iron metabolism. Blood 2009, 113, 1805–1808. [Google Scholar] [CrossRef] [PubMed]
- Hamdi, A.; Roshan, T.M.; Kahawita, T.M.; Mason, A.B.; Sheftel, A.D.; Ponka, P. Erythroid cell mitochondria receive endosomal iron by a “kiss-and-run” mechanism. Biochim. Biophys. Acta-Mol. Cell Res. 2016, 1863, 2859–2867. [Google Scholar] [CrossRef]
- Christenson, E.T.; Gallegos, A.S.; Banerjee, A. In vitro reconstitution, functional dissection, and mutational analysis of metal ion transport by mitoferrin-1. J. Biol. Chem. 2018, 293, 3819–3828. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Arimura, H.; Fukushige, T.; Minami, K.; Nishizawa, Y.; Tanimoto, A.; Kanekura, T.; Nakagawa, M.; Akiyama, S.; Furukawa, T. Abcb10 Role in Heme Biosynthesis In Vivo: Abcb10 Knockout in Mice Causes Anemia with Protoporphyrin IX and Iron Accumulation. Mol. Cell. Biol. 2014, 34, 1077–1084. [Google Scholar] [CrossRef]
- Cronin, S.J.F.; Woolf, C.J.; Weiss, G.; Penninger, J.M. The Role of Iron Regulation in Immunometabolism and Immune-Related Disease. Front. Mol. Biosci. 2019, 6, 19. [Google Scholar] [CrossRef]
- Wachnowsky, C.; Fidai, I.; Cowan, J.A. Iron-sulfur cluster biosynthesis and trafficking—Impact on human disease conditions. Metallomics 2018, 10, 9–29. [Google Scholar] [CrossRef]
- Braymer, J.J.; Lill, R. Iron-sulfur cluster biogenesis and trafficking in mitochondria. J. Biol. Chem. 2017, 292, 12754–12763. [Google Scholar] [CrossRef]
- Lill, R.; Freibert, S.A. Mechanisms of Mitochondrial Iron-Sulfur Protein Biogenesis. In Annual Review of Biochemistry; Kornberg, R.D., Ed.; Annual Reviews: San Mateo, CA, USA, 2020; Volume 89, pp. 471–499. [Google Scholar]
- Paul, V.D.; Lill, R. Biogenesis of cytosolic and nuclear iron-sulfur proteins and their role in genome stability. Biochim. Biophys. Acta-Mol. Cell Res. 2015, 1853, 1528–1539. [Google Scholar] [CrossRef]
- Shen, Y.; Li, X.; Dong, D.D.; Zhang, B.; Xue, Y.R.; Shang, P. Transferrin receptor 1 in cancer: A new sight for cancer therapy. Am. J. Cancer Res. 2018, 8, 916–931. [Google Scholar]
- Daniels, T.R.; Bernabeu, E.; Rodriguez, J.A.; Patel, S.; Kozman, M.; Chiappetta, D.A.; Holler, E.; Ljubimova, J.Y.; Helguera, G.; Penichet, M.L. The transferrin receptor and the targeted delivery of therapeutic agents against cancer. Biochim. Biophys. Acta-Gen. Subj. 2012, 1820, 291–317. [Google Scholar] [CrossRef] [PubMed]
- Torti, S.V.; Torti, F.M. Iron: The cancer connection. Mol. Asp. Med. 2020, 75, 100860. [Google Scholar] [CrossRef] [PubMed]
- Shah, Y.M.; Xie, L.W. Hypoxia-Inducible Factors Link Iron Homeostasis and Erythropoiesis. Gastroenterology 2014, 146, 630–642. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.Y.; Manz, D.H.; Torti, S.V.; Torti, F.M. Iron-responsive element-binding protein 2 plays an essential role in regulating prostate cancer cell growth. Oncotarget 2017, 8, 82231–82243. [Google Scholar] [CrossRef] [PubMed]
- Boult, J.; Roberts, K.; Brookes, M.J.; Hughes, S.; Bury, J.P.; Cross, S.S.; Anderson, G.J.; Spychal, R.; Lqbal, T.; Tselepis, C. Overexpression of cellular iron import proteins is associated with malignant progression of Esophageal adenocarcinoma. Clin. Cancer Res. 2008, 14, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Gomes, I.M.; Maia, C.J.; Santos, C.R. STEAP Proteins: From Structure to Applications in Cancer Therapy. Mol. Cancer Res. 2012, 10, 573–587. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.Y.; Jiang, J.N.; Fang, X.D.; Ji, F.J. STEAP1 Regulates Tumorigenesis and Chemoresistance During Peritoneal Metastasis of Gastric Cancer. Front. Physiol. 2018, 9, 7. [Google Scholar] [CrossRef]
- Whiteland, H.; Spencer-Harty, S.; Morgan, C.; Kynaston, H.; Thomas, D.H.; Bose, P.; Fenn, N.; Lewis, P.; Jenkins, S.; Doak, S.H. A role for STEAP2 in prostate cancer progression. Clin. Exp. Metastasis 2014, 31, 909–920. [Google Scholar] [CrossRef]
- Babu, K.R.; Muckenthaler, M.U. miR-20a regulates expression of the iron exporter ferroportin in lung cancer. J. Mol. Med. 2016, 94, 347–359. [Google Scholar] [CrossRef]
- Basuli, D.; Tesfay, L.; Deng, Z.; Paul, B.; Yamamoto, Y.; Ning, G.; Xian, W.; McKeon, F.; Lynch, M.; Crum, C.P.; et al. Iron addiction: A novel therapeutic target in ovarian cancer. Oncogene 2017, 36, 4089–4099. [Google Scholar] [CrossRef]
- Gu, Z.M.; Wang, H.; Xia, J.L.; Yang, Y.; Jin, Z.D.; Xu, H.W.; Shi, J.M.; De Domenico, I.; Tricot, G.; Zhan, F.H. Decreased Ferroportin Promotes Myeloma Cell Growth and Osteoclast Differentiation. Cancer Res. 2015, 75, 2211–2221. [Google Scholar] [CrossRef] [PubMed]
- Xue, D.; Zhou, C.X.; Shi, Y.B.; Lu, H.; He, X.Z. Decreased expression of ferroportin in prostate cancer. Oncol. Lett. 2015, 10, 913–916. [Google Scholar] [CrossRef] [PubMed]
- Genereux, J.C.; Boal, A.K.; Barton, J.K. DNA-Mediated Charge Transport in Redox Sensing and Signaling. J. Am. Chem. Soc. 2010, 132, 891–905. [Google Scholar] [CrossRef] [PubMed]
- Gee, H.E.; Ivan, C.; Calin, G.A.; Ivan, M. HypoxamiRs and Cancer: From Biology to Targeted Therapy. Antioxid. Redox Signal. 2014, 21, 1220–1238. [Google Scholar] [CrossRef] [PubMed]
- Darash-Yahana, M.; Pozniak, Y.; Lu, M.Y.; Sohn, Y.S.; Karmi, O.; Tamir, S.; Bai, F.; Song, L.H.; Jennings, P.A.; Pikarsky, E.; et al. Breast cancer tumorigenicity is dependent on high expression levels of NAF-1 and the lability of its Fe-S clusters. Proc. Natl. Acad. Sci. USA 2016, 113, 10890–10895. [Google Scholar] [CrossRef]
- Chen, F.Y.; Tang, Q.; Ma, H.; Bian, K.; Seeram, N.P.; Li, D.Y. Hydrolyzable Tannins Are Iron Chelators That Inhibit DNA Repair Enzyme ALKBH2. Chem. Res. Toxicol. 2019, 32, 1082–1086. [Google Scholar] [CrossRef]
- Matson, S.W.; Bean, D.W.; George, J.W. DNA helicases—Enzymes with essential roles in all aspects of dna metabolism. Bioessays 1994, 16, 13–22. [Google Scholar] [CrossRef]
- Datta, A.; Brosh, R.M. New Insights Into DNA Helicases as Druggable Targets for Cancer Therapy. Front. Mol. Biosci. 2018, 5, 21. [Google Scholar] [CrossRef]
- Fan, L.; Fuss, J.O.; Cheng, Q.J.; Arvai, A.S.; Hammel, M.; Roberts, V.A.; Cooper, P.K.; Tainer, J.A. XPD helicase structures and activities: Insights into the cancer and aging phenotypes from XPD mutations. Cell 2008, 133, 789–800. [Google Scholar] [CrossRef]
- Liu, H.; Rudolf, J.; Johnson, K.A.; McMahon, S.A.; Oke, M.; Carter, L.; McRobbie, A.M.; Brown, S.E.; Naismith, J.H.; White, M.F. Structure of the DNA repair helicase XPD. Cell 2008, 133, 801–812. [Google Scholar] [CrossRef]
- Wu, Y.; Suhasini, A.N.; Brosh, R.M. Welcome the Family of FANCJ-like Helicases to the Block of Genome Stability Maintenance Proteins. Cell. Mol. Life Sci. 2009, 66, 1209–1222. [Google Scholar] [CrossRef] [PubMed]
- Nath, S.; Somyajit, K.; Mishra, A.; Scully, R.; Nagaraju, G. FANCJ helicase controls the balance between short- and long-tract gene conversions between sister chromatids. Nucleic Acids Res. 2017, 45, 8886–8900. [Google Scholar] [CrossRef] [PubMed]
- Brosh, R.M.; Cantor, S.B. Molecular and cellular functions of the FANCJ DNA helicase defective in cancer and in Fanconi anemia. Front. Genet. 2014, 5, 14. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.L.; Shin-ya, K.; Brosh, R.M. FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 2008, 28, 4116–4128. [Google Scholar] [CrossRef]
- Simon, A.K.; Kummer, S.; Wild, S.; Lezaja, A.; Teloni, F.; Jozwiakowski, S.K.; Altmeyer, M.; Gari, K. The iron-sulfur helicase DDX11 promotes the generation of single-stranded DNA for CHK1 activation. Life Sci. Alliance 2020, 3, 15. [Google Scholar] [CrossRef]
- Farina, A.; Shin, J.H.; Kim, D.H.; Bermudez, V.P.; Kelman, Z.; Seo, Y.S.; Hurwitz, J. Studies with the human cohesin establishment factor, ChlR1—Association of ChlR1 with Ctf18-RFC and Fen1. J. Biol. Chem. 2008, 283, 20925–20936. [Google Scholar] [CrossRef]
- Shah, N.; Inoue, A.; Lee, S.W.; Beishline, K.; Lahti, J.M.; Noguchi, E. Roles of ChlR1 DNA helicase in replication recovery from DNA damage. Exp. Cell Res. 2013, 319, 2244–2253. [Google Scholar] [CrossRef]
- Miyazawa, M.; Bogdan, A.R.; Tsuji, Y. Perturbation of Iron Metabolism by Cisplatin through Inhibition of Iron Regulatory Protein 2. Cell Chem. Biol. 2019, 26, 85–97.e4. [Google Scholar] [CrossRef]
- Pisani, F.M.; Napolitano, E.; Napolitano, L.M.R.; Onesti, S. Molecular and Cellular Functions of the Warsaw Breakage Syndrome DNA Helicase DDX11. Genes 2018, 9, 564. [Google Scholar] [CrossRef]
- Wu, Y.L.; Brosh, R.M. DNA helicase and helicase-nuclease enzymes with a conserved iron-sulfur cluster. Nucleic Acids Res. 2012, 40, 4247–4260. [Google Scholar] [CrossRef]
- Pokharel, S.; Campbell, J.L. Cross talk between the nuclease and helicase activities of DNA2: Role of an essential iron-sulfur cluster domain. Nucleic Acids Res. 2012, 40, 7821–7830. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Zhou, M.A.; Guo, Z.G.; Lu, H.M.; Qian, L.M.; Dai, H.F.; Qiu, J.Z.; Yakubovskaya, E.; Bogenhagen, D.F.; Demple, B.; et al. Human DNA2 Is a Mitochondrial Nuclease/Helicase for Efficient Processing of DNA Replication and Repair Intermediates. Mol. Cell 2008, 32, 325–336. [Google Scholar] [CrossRef] [PubMed]
- Mariotti, L.; Wild, S.; Brunoldi, G.; Piceni, A.; Ceppi, I.; Kummer, S.; Lutz, R.E.; Cejka, P.; Gari, K. The iron-sulphur cluster in human DNA2 is required for all biochemical activities of DNA2. Commun. Biol. 2020, 3, 322. [Google Scholar] [CrossRef] [PubMed]
- Thierbach, R.; Drewes, G.; Fusser, M.; Voigt, A.; Kuhlow, D.; Blume, U.; Schulz, T.J.; Reiche, C.; Glatt, H.; Epe, B.; et al. The Friedreich’s ataxia protein frataxin modulates DNA base excision repair in prokaryotes and mammals. Biochem. J. 2010, 432, 165–172. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Baranovskiy, A.G.; Siebler, H.M.; Pavlov, Y.I.; Tahirov, T.H. Iron-Sulfur Clusters in DNA Polymerases and Primases of Eukaryotes. In Fe-S Cluster Enzymes, Pt B; David, S.S., Ed.; Methods in Enzymology; Elsevier Academic Press Inc.: San Diego, CA, USA, 2018; Volume 599, pp. 1–20. [Google Scholar]
- Puig, S.; Ramos-Alonso, L.; Romero, A.M.; Martinez-Pastor, M. The elemental role of iron in DNA synthesis and repair. Metallomics 2017, 9, 1483–1500. [Google Scholar] [CrossRef]
- Wang, X.; Ira, G.; Tercero, J.A.; Holmes, A.M.; Diffley, J.F.X.; Haber, J.E. Role of DNA replication proteins in double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 2004, 24, 6891–6899. [Google Scholar] [CrossRef]
- ter Beek, J.; Parkash, V.; Bylund, G.O.; Osterman, P.; Sauer-Eriksson, A.E.; Johansson, E. Structural evidence for an essential Fe-S cluster in the catalytic core domain of DNA polymerase epsilon. Nucleic Acids Res. 2019, 47, 5712–5722. [Google Scholar] [CrossRef]
- Jozwiakowski, S.K.; Kummer, S.; Gari, K. Human DNA polymerase delta requires an iron-sulfur cluster for high-fidelity DNA synthesis. Life Sci. Alliance 2019, 2, 321. [Google Scholar] [CrossRef]
- Chanet, R.; Baille, D.; Golinelli-Cohen, M.-P.; Riquier, S.; Guittet, O.; Lepoivre, M.; Huang, M.-E.; Vernis, L. Fe-S coordination defects in the replicative DNA polymerase delta cause deleterious DNA replication in vivo and subsequent DNA damage in the yeast Saccharomyces cerevisiae. G3 Genes Genomes Genet. 2021, 11, jkab124. [Google Scholar] [CrossRef]
- Holt, M.E.; Salay, L.E.; Chazin, W.J. A Polymerase with Potential: The Fe-S Cluster in Human DNA Primase. In Fe-S Cluster Enzymes, Pt A; David, S.S., Ed.; Methods in Enzymology; Elsevier Academic Press Inc.: San Diego, CA, USA, 2017; Volume 595, pp. 361–390. [Google Scholar]
- Stubble, J. Di-iron-tyrosyl radical ribonucleotide reductases. Curr. Opin. Chem. Biol. 2003, 7, 183–188. [Google Scholar] [CrossRef]
- Elford, H.L.; Freese, M.; Passamani, E.; Morris, H.P. Ribonucleotide reductase and cell proliferation. I. Variations of ribonucleotide reductase activity with tumor growth rate in a series of rat hepatomas. J. Biol. Chem. 1970, 245, 5228–5233. [Google Scholar] [CrossRef]
- Cotruvo, J.A.; Stubbe, J. Class I Ribonucleotide Reductases: Metallocofactor Assembly and Repair In Vitro and In Vivo. In Annual Review of Biochemistry; Kornberg, R.D., Raetz, C.R.H., Rothman, J.E., Thorner, J.W., Eds.; Annual Reviews: San Mateo, CA, USA, 2011; Volume 80, pp. 733–767. [Google Scholar]
- Kolberg, M.; Strand, K.R.; Graff, P.; Andersson, K.K. Structure, function, and mechanism of ribonucleotide reductases. Biochim. Biophys. Acta-Proteins Proteom. 2004, 1699, 1–34. [Google Scholar] [CrossRef]
- Nyholm, S.; Thelander, L.; Graslund, A. Reduction and loss of the iron center in the reaction of the small-subunit of mouse ribonucleotide reductase with hydroxyurea. Biochemistry 1993, 32, 11569–11574. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.M.; Yeh, M.T.; Tsao, N.; Chen, C.W.; Gao, Q.Z.; Chang, C.Y.; Lee, M.H.; Fang, J.M.; Sheu, S.Y.; Lin, C.J.; et al. Tumor Cells Require Thymidylate Kinase to Prevent dUTP Incorporation during DNA Repair. Cancer Cell 2012, 22, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Viberg, J.; Nilsson, A.K.; Chabes, A. Highly mutagenic and severely imbalanced dNTP pools can escape detection by the S-phase checkpoint. Nucleic Acids Res. 2010, 38, 3975–3983. [Google Scholar] [CrossRef]
- Mathews, C.K. DNA precursor metabolism and genomic stability. Faseb J. 2006, 20, 1300–1314. [Google Scholar] [CrossRef]
- Cerqueira, N.; Fernandes, P.A.; Ramos, M.J. Ribonucleotide reductase: A critical enzyme for cancer chemotherapy and antiviral agents. Recent Patents Anti-Canc. Drug Discov. 2007, 2, 11–29. [Google Scholar] [CrossRef]
- Miyajima, H.; Takahashi, Y.; Kamata, T.; Shimizu, H.; Sakai, N.; Gitlin, J.D. Use of desferrioxamine in the treatment of aceruloplasminemia. Ann. Neurol. 1997, 41, 404–407. [Google Scholar] [CrossRef]
- Lassmann, G.; Thelander, L.; Graslund, A. Epr stopped-flow studies of the reaction of the tyrosyl radical of protein-r2 from ribonucleotide reductase with hydroxyurea. Biochem. Biophys. Res. Commun. 1992, 188, 879–887. [Google Scholar] [CrossRef]
- Zeidner, J.F.; Karp, J.E.; Blackford, A.L.; Smith, B.D.; Gojo, I.; Gore, S.D.; Levis, M.J.; Carraway, H.E.; Greer, J.M.; Ivy, S.P.; et al. A phase II trial of sequential ribonucleotide reductase inhibition in aggressive myeloproliferative neoplasms. Haematologica 2014, 99, 672–678. [Google Scholar] [CrossRef]
- Ponka, P.; Richardson, D.R.; Edward, J.T.; Chubb, F.L. Iron chelators of the pyridoxal isonicotinoyl hydrazone class—Relationship of the lipophilicity of the apochelator to its ability to mobilize iron from reticulocytes in-vitro. Can. J. Physiol. Pharmacol. 1994, 72, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Offenbacher, A.R.; Vassiliev, I.R.; Seyedsayamdost, M.R.; Stubbe, J.; Barry, B.A. Redox-Linked Structural Changes in Ribonucleotide Reductase. J. Am. Chem. Soc. 2009, 131, 7496–7497. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Anand, S.; Verma, H.; Kumar, L.; Singh, N. Induction of apoptosis in chronic myelogenous leukemia lymphocytes by hydroxyurea and adriamycin. Cancer Lett. 1995, 88, 101–105. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, G.; Huang, M. Ribonucleotide reductase metallocofactor: Assembly, maintenance and inhibition. Front. Biol. 2014, 9, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Glickstein, H.; Ben El, R.; Shvartsman, M.; Cabantchik, Z.I. Intracellular labile iron pools as direct targets of iron chelators: A fluorescence study of chelator action in living cells. Blood 2005, 106, 3242–3250. [Google Scholar] [CrossRef] [PubMed]
- Bajbouj, K.; Shafarin, J.; Hamad, M. High-Dose Deferoxamine Treatment Disrupts Intracellular Iron Homeostasis, Reduces Growth, and Induces Apoptosis in Metastatic and Nonmetastatic Breast Cancer Cell Lines. Technol. Cancer Res. Treat. 2018, 17, 11. [Google Scholar] [CrossRef] [PubMed]
- Fryknas, M.; Zhang, X.N.; Bremberg, U.; Senkowski, W.; Olofsson, M.H.; Brandt, P.; Persson, I.; D’Arcy, P.; Gullbo, J.; Nygren, P.; et al. Iron chelators target both proliferating and quiescent cancer cells. Sci. Rep. 2016, 6, 11. [Google Scholar] [CrossRef]
- Dayani, P.N.; Bishop, M.C.; Black, K.; Zeltzer, P.M. Desferoxamine (DFO)—mediated iron chelation: Rationale for a novel approach to therapy for brain cancer. J. Neuro-Oncol. 2004, 67, 367–377. [Google Scholar] [CrossRef]
- Keberle, H. Biochemistry of Desferrioxamine + its Relation to Iron Metabolism. Annal. N. Y. Acad. Sci. 1964, 119, 758–768. [Google Scholar] [CrossRef]
- Lovejoy, D.B.; Richardson, D.R. Iron chelators as anti-neoplastic agents: Current developments and promise of the PIH class of chelators. Curr. Med. Chem. 2003, 10, 1035–1049. [Google Scholar] [CrossRef][Green Version]
- Hatcher, H.C.; Singh, R.N.; Torti, F.M.; Torti, S.V. Synthetic and natural iron chelators: Therapeutic potential and clinical use. Future Med. Chem. 2009, 1, 1643–1670. [Google Scholar] [CrossRef] [PubMed]
- Richardson, D.R.; Ponka, P. The Iron-Metabolism Of The Human Neuroblastoma Cell—Lack Of Relationship Between The Efficacy Of Iron Chelation And The Inhibition Of Dna-Synthesis. J. Lab. Clin. Med. 1994, 124, 660–671. [Google Scholar] [PubMed]
- Buss, J.L.; Hermes-Lima, M.; Ponka, P. Pyridoxal isonicotinoyl hydrazone and its analogues. Adv.Exp.Med.Biol. 2002, 509, 205–229. [Google Scholar] [PubMed]
- Buss, J.L.; Arduini, E.; Ponka, P. Mobilization of intracellular iron by analogs of pyridoxal. isonicotinoyl hydrazone (PIH) is determined by the membrane permeability of the iron-chelator complexes. Biochem. Pharmacol. 2002, 64, 1689–1701. [Google Scholar] [CrossRef]
- Richardson, D.R.; Tran, E.H.; Bradshaw, D.; Ponka, P. Potential of iron chelators of the pyridoxal isonicotinoyl hydrazone class as effective antiproliferative agents. Blood 1994, 84, A613. [Google Scholar] [CrossRef]
- Green, D.A.; Antholine, W.E.; Wong, S.J.; Richardson, D.R.; Chitambar, C.R. Inhibition of malignant cell growth by 311, a novel iron chelator of the pyridoxal isonicotinoyl hydrazone class: Effect on the R2 subunit of ribonucleotide reductase. Clin. Cancer Res. 2001, 7, 3574–3579. [Google Scholar]
- Richardson, D.R.; Bernhardt, P.V. Crystal and molecular structure of 2-hydroxy-1-naphthaldehyde isonicotinoyl hydrazone (NIH) and its iron(III) complex: An iron chelator with anti-tumour activity. J. Biol. Inorg. Chem. 1999, 4, 266–273. [Google Scholar] [CrossRef]
- Kunos, C.A.; Ivy, S.P. Triapine Radiochemotherapy in Advanced Stage Cervical Cancer. Front. Oncol. 2018, 8, 7. [Google Scholar] [CrossRef]
- Mortazavi, A.; Ling, Y.H.; Martin, L.K.; Wei, L.; Phelps, M.A.; Liu, Z.F.; Harper, E.J.; Ivy, S.P.; Wu, X.; Zhou, B.S.; et al. A phase I study of prolonged infusion of triapine in combination with fixed dose rate gemcitabine in patients with advanced solid tumors. Investig. New Drugs 2013, 31, 685–695. [Google Scholar] [CrossRef]
- Finch, R.A.; Liu, M.C.; Cory, A.H.; Cory, J.G.; Sartorelli, A.C. Triapine (3-aminopyridine-2-carboxaldehyde thiosemicarbazone; 3-AP), an inhibitor of ribonucleotide reductase with antineoplastic activity. Adv. Enzym. Regul. 1999, 39, 3–12. [Google Scholar] [CrossRef]
- Popovic-Bijelic, A.; Kowol, C.R.; Lind, M.E.S.; Luo, J.H.; Himo, F.; Enyedy, E.A.; Arion, V.B.; Graslund, A. Ribonucleotide reductase inhibition by metal complexes of Triapine (3-aminopyridine-2-carboxaldehyde thiosemicarbazone): A combined experimental and theoretical study. J. Inorg. Biochem. 2011, 105, 1422–1431. [Google Scholar] [CrossRef] [PubMed]
- Nyholm, S.; Mann, G.J.; Johansson, A.G.; Bergeron, R.J.; Graslund, A.; Thelander, L. Role of ribonucleotide reductase in inhibition of mammalian-cell growth by potent iron chelators. J. Biol. Chem. 1993, 268, 26200–26205. [Google Scholar] [CrossRef]
- Shakya, B.; Yadav, P.N. Thiosemicarbazones as Potent Anticancer Agents and their Modes of Action. Mini-Rev. Med. Chem. 2020, 20, 638–661. [Google Scholar] [CrossRef] [PubMed]
- Ertas, M.; Sahin, Z.; Bulbul, E.F.; Bender, C.; Biltekin, S.N.; Berk, B.; Yurttas, L.; Nalbur, A.M.; Celik, H.; Demirayak, S. Potent ribonucleotide reductase inhibitors: Thiazole-containing thiosemicarbazone derivatives. Archiv. Der. Pharm. 2019, 352, 1900033. [Google Scholar] [CrossRef]
- Abdelaal, G.; Veuger, S. Reversing oncogenic transformation with iron chelation. Oncotarget 2021, 12, 106–124. [Google Scholar] [CrossRef]
- Fu, D.; Richardson, D.R. Iron chelation and regulation of the cell cycle: 2 mechanisms of posttranscriptional regulation of the universal cyclin-dependent kinase inhibitor p21(CIP1/WAF1) by iron depletion. Blood 2007, 110, 752–761. [Google Scholar] [CrossRef]
- Nurtjahja-Tjendraputra, E.; Fu, D.; Phang, J.M.; Richardson, D.R. Iron chelation regulates cyclin D1 expression via the proteasome: A link to iron deficiency-mediated growth suppression. Blood 2007, 109, 4045–4054. [Google Scholar] [CrossRef]
- Kulp, K.S.; Green, S.L.; Vulliet, P.R. Iron deprivation inhibits cyclin-dependent kinase activity and decreases cyclin D CDK4 protein levels in asynchronous MDA-MB-453 human breast cancer cells. Exp. Cell Res. 1996, 229, 60–68. [Google Scholar] [CrossRef]
- Gao, J.; Richardson, D.R. The potential of iron chelators of the pyridoxal isonicotinoyl hydrazone class as effective antiproliferative agents, IV: The mechanisms involved in inhibiting cell-cycle progression. Blood 2001, 98, 842–850. [Google Scholar] [CrossRef]
- Debebe, Z.; Ammosova, T.; Jerebtsova, M.; Kurantsin-Mills, J.; Niu, X.M.; Charles, S.; Richardson, D.R.; Ray, P.E.; Gordeuk, V.R.; Nekhai, S. Iron chelators ICL670 and 311 inhibit HIV-1 transcription. Virology 2007, 367, 324–333. [Google Scholar] [CrossRef]
- Choi, J.H.; Kim, J.S.; Won, Y.W.; Uhm, J.; Park, B.B.; Lee, Y.Y. The potential of deferasirox as a novel therapeutic modality in gastric cancer. World J. Surg. Oncol. 2016, 14, 77. [Google Scholar] [CrossRef] [PubMed]
- Mettert, E.L.; Kiley, P.J. Fe-S proteins that regulate gene expression. Biochim. Biophys. Acta-Mol. Cell Res. 2015, 1853, 1284–1293. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.K.; Huang, X.; Wang, X.Y.; Yan, S.Q.; Guo, S.Y.; Abdalla, A.E.; Huang, C.W.; Xie, J.P. L-Serine potentiates fluoroquinolone activity against Escherichia coli by enhancing endogenous reactive oxygen species production. J. Antimicrob. Chemother. 2016, 71, 2192–2199. [Google Scholar] [CrossRef] [PubMed]
- Vernis, L.; El Banna, N.; Baille, D.; Hatem, E.; Heneman, A.; Huang, M.E. Fe-S Clusters Emerging as Targets of Therapeutic Drugs. Oxidative Med. Cell. Longev. 2017, 2017, 12. [Google Scholar] [CrossRef]
- Rouault, T.A. Biogenesis of iron-sulfur clusters in mammalian cells: New insights and relevance to human disease. Dis. Model. Mech. 2012, 5, 155–164. [Google Scholar] [CrossRef]
- Jang, S.J.; Imlay, J.A. Hydrogen peroxide inactivates the Escherichia coli Isc iron-sulphur assembly system, and OxyR induces the Suf system to compensate. Mol. Microbiol. 2010, 78, 1448–1467. [Google Scholar] [CrossRef]
- Chen, G.; Chen, Z.; Hu, Y.M.; Huang, P. Inhibition of Mitochondrial Respiration and Rapid Depletion of Mitochondrial Glutathione by beta-Phenethyl Isothiocyanate: Mechanisms for Anti-Leukemia Activity. Antioxid. Redox Signal. 2011, 15, 2911–2921. [Google Scholar] [CrossRef]
- Italia, K.; Colah, R.; Ghosh, K. Hydroxyurea Could Be a Good Clinically Relevant Iron Chelator. PLoS ONE 2013, 8, 5. [Google Scholar] [CrossRef]
- Huang, M.E.; Facca, C.; Fatmi, Z.; Baille, D.; Benakli, S.; Vernis, L. DNA replication inhibitor hydroxyurea alters Fe-S centers by producing reactive oxygen species in vivo. Sci Rep 2016, 6, 12. [Google Scholar] [CrossRef]
- Sandoval-Acuna, C.; Torrealba, N.; Tomkova, V.; Jadhav, S.B.; Blazkova, K.; Merta, L.; Lettlova, S.; Adamcova, M.K.; Rosel, D.; Brabek, J.; et al. Targeting Mitochondrial Iron Metabolism Suppresses Tumor Growth and Metastasis by Inducing Mitochondrial Dysfunction and Mitophagy. Cancer Res. 2021, 81, 2289–2303. [Google Scholar] [CrossRef]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Valko, M.; Rhodes, C.J.; Moncol, J.; Izakovic, M.; Mazur, M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 2006, 160, 1–40. [Google Scholar] [CrossRef] [PubMed]
- Schieber, M.; Chandel, N.S. ROS Function in Redox Signaling and Oxidative Stress. Curr. Biol. 2014, 24, R453–R462. [Google Scholar] [CrossRef] [PubMed]
- Benassi, B.; Fanciulli, M.; Fiorentino, F.; Porrello, A.; Chiorino, G.; Loda, M.; Zupi, G.; Biroccio, A. c-Myc phosphorylation is required for cellular response to oxidative stress. Mol. Cell 2006, 21, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Galaris, D.; Skiada, V.; Barbouti, A. Redox signaling and cancer: The role of “labile” iron. Cancer Lett. 2008, 266, 21–29. [Google Scholar] [CrossRef]
- Cooke, M.S.; Evans, M.D.; Dizdaroglu, M.; Lunec, J. Oxidative DNA damage: Mechanisms, mutation, and disease. Faseb J. 2003, 17, 1195–1214. [Google Scholar] [CrossRef]
- Nakamura, T.; Naguro, I.; Ichijo, H. Iron homeostasis and iron-regulated ROS in cell death, senescence and human diseases. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 1398–1409. [Google Scholar] [CrossRef]
- Juan, C.A.; de la Lastra, J.M.P.; Plou, F.J.; Perez-Lebena, E. The Chemistry of Reactive Oxygen Species (ROS) Revisited: Outlining Their Role in Biological Macromolecules (DNA, Lipids and Proteins) and Induced Pathologies. Int. J. Mol. Sci. 2021, 22, 4642. [Google Scholar] [CrossRef]
- Cadet, J.; Wagner, J.R. DNA Base Damage by Reactive Oxygen Species, Oxidizing Agents, and UV Radiation. Cold Spring Harb. Perspect. Biol. 2013, 5, a012559. [Google Scholar] [CrossRef]
- Pujari, S.S.; Tretyakova, N. Chemical Biology of N-5-Substituted Formamidopyrimidine DNA Adducts. Chem. Res. Toxicol. 2017, 30, 434–452. [Google Scholar] [CrossRef]
- Benhar, M.; Engelberg, D.; Levitzki, A. ROS, stress-activated kinases and stress signaling in cancer. Embo Rep. 2002, 3, 420–425. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.M.; Dean, A.E.; Horikoshi, N.; Heer, C.; Spitz, D.R.; Gius, D. Emerging evidence for targeting mitochondrial metabolic dysfunction in cancer therapy. J. Clin. Investig. 2018, 128, 3682–3691. [Google Scholar] [CrossRef] [PubMed]
- Cragg, L.; Hebbel, R.P.; Miller, W.; Solovey, A.; Selby, S.; Enright, H. The iron chelator L1 potentiates oxidative DNA damage in iron-loaded liver cells. Blood 1998, 92, 632–638. [Google Scholar] [CrossRef] [PubMed]
- Dai, Y.M.; Gold, B.; Vishwanatha, J.K.; Rhode, S.L. Mimosine inhibits viral-dna synthesis through ribonucleotide reductase. Virology 1994, 205, 210–216. [Google Scholar] [CrossRef]
- Hallak, M.; Dvilansky, A.; Shpilberg, O.; Levi, I.; Mazar, J.; Nathan, I. Mimosine induces apoptosis through metal ion chelation, mitochondriai activation and reactive oxygen species production in human leukemic cells. Blood 2004, 104, 203B. [Google Scholar] [CrossRef]
- Hallak, M.; Vazana, L.; Shpilberg, O.; Levy, I.; Mazar, J.; Nathan, I. A molecular mechanism for mimosine-induced apoptosis involving oxidative stress and mitochondrial activation. Apoptosis 2008, 13, 147–155. [Google Scholar] [CrossRef]
- Mikhailov, I.; Russev, G.; Anachkova, B. Treatment of mammalian cells with mimosine generates DNA breaks. Mutat. Res.-DNA Repair. 2000, 459, 299–306. [Google Scholar] [CrossRef]
- Qiao, S.L.; Murakami, K.; Zhao, Q.H.; Wang, B.L.; Seo, H.; Yamashita, H.; Li, X.T.; Iwamoto, T.; Ichihara, M.; Yoshino, M. Mimosine-Induced Apoptosis in C6 Glioma Cells Requires the Release of Mitochondria-Derived Reactive Oxygen Species and p38, JNK Activation. Neurochem. Res. 2012, 37, 417–427. [Google Scholar] [CrossRef]
- Wada, T.; Penninger, J.M. Mitogen-activated protein kinases in apoptosis regulation. Oncogene 2004, 23, 2838–2849. [Google Scholar] [CrossRef]
- Murakami, K.; Yoshino, M. Generation of reactive oxygen species by hydroxypyridone compound/iron complexes. Redox Rep. 2020, 25, 59–63. [Google Scholar] [CrossRef]
- Kyriakou, S.; Mitsiogianni, M.; Mantso, T.; Cheung, W.; Todryk, S.; Veuger, S.; Pappa, A.; Tetard, D.; Panayiotidis, M.I. Anticancer activity of a novel methylated analogue of L-mimosine against an in vitro model of human malignant melanoma. Investig. New Drugs 2019, 38, 621–633. [Google Scholar] [CrossRef] [PubMed]
- Kyriakou, S.; Cheung, W.; Mantso, T.; Mitsiogianni, M.; Anestopoulos, I.; Veuger, S.; Trafalis, D.T.; Franco, R.; Pappa, A.; Tetard, D.; et al. A novel methylated analogue of L-Mimosine exerts its therapeutic potency through ROS production and ceramide-induced apoptosis in malignant melanoma. Investig. New Drugs 2021, 39, 971–986. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, C.M.; Bernhardt, P.V.; Chin, P.; Richardson, D.R. Structural variations and formation constants of first-row transition metal complexes of biologically active aroylhydrazones. Eur. J. Inorg. Chem. 2003, 6, 1145–1156. [Google Scholar] [CrossRef]
- Becker, E.M.; Lovejoy, D.B.; Greer, J.M.; Watts, R.; Richardson, D.R. Identification of the di-pyridyl ketone isonicotinoyl hydrazone (PKIH) analogues as potent iron chelators and anti-tumour agents. Br. J. Pharmacol. 2003, 138, 819–830. [Google Scholar] [CrossRef]
- Chaston, T.B.; Watts, R.N.; Yuan, J.; Richardson, D.R. Potent antitumor activity of novel iron chelators derived from di-2-pyridylketone isonicotinoyl hydrazone involves fenton-derived free radical generation. Clin. Cancer Res. 2004, 10, 7365–7374. [Google Scholar] [CrossRef]
- Bernhardt, P.V.; Caldwell, L.M.; Chaston, T.B.; Chin, P.; Richardson, D.R. Cytotoxic iron chelators: Characterization of the structure, solution chemistry and redox activity of ligands and iron complexes of the di-2-pyridyl ketone isonicotinoyl hydrazone (HPKIH) analogues. J. Biol. Inorg. Chem. 2003, 8, 866–880. [Google Scholar] [CrossRef]
- Rao, V.A.; Klein, S.R.; Agama, K.K.; Toyoda, E.; Adachi, N.; Pommier, Y.; Shacter, E.B. The Iron Chelator Dp44mT Causes DNA Damage and Selective Inhibition of Topoisomerase II alpha in Breast Cancer Cells. Cancer Res. 2009, 69, 948–957. [Google Scholar] [CrossRef]
- Yuan, J.; Lovejoy, D.B.; Richardson, D.R. Novel di-2-pyridyl-derived iron chelators with marked and selective antitumor activity: In vitro and in vivo assessment. Blood 2004, 104, 1450–1458. [Google Scholar] [CrossRef]
- Gutierrez, E.; Richardson, D.R.; Jansson, P.J. The Anticancer Agent Di-2-pyridylketone 4,4-Dimethyl-3-thiosemicarbazone (Dp44mT) Overcomes Prosurvival Autophagy by Two Mechanisms Persistent Induction of Autophagosome Synthesis and Impairment Of Lysosomal Integrity. J. Biol. Chem. 2014, 289, 33568–33589. [Google Scholar] [CrossRef]
- Whitnall, M.; Howard, J.; Ponka, P.; Richardson, D.R. A class of iron chelators with a wide spectrum of potent antitumor activity that overcomes resistance to chemotherapeutics. Proc. Natl. Acad. Sci. USA 2006, 103, 14901–14906. [Google Scholar] [CrossRef]
- Noulsri, E.; Richardson, D.R.; Lerdwana, S.; Fucharoen, S.; Yamagishi, T.; Kalinowski, D.S.; Pattanapanyasat, K. Antitumor activity and mechanism of action of the iron chelator, Dp44mT, against leukemic cells. Am. J. Hematol. 2009, 84, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Li, P.C.; Zheng, X.; Shou, K.Q.; Niu, Y.H.; Jian, C.; Zhao, Y.; Yi, W.R.; Hu, X.; Yu, A.X. The iron chelator Dp44mT suppresses osteosarcoma’s proliferation, invasion and migration: In vitro and in vivo. Am. J. Transl. Res. 2016, 8, 5370–5385. [Google Scholar] [PubMed]
- Dixon, K.M.; Lui, G.Y.L.; Kovacevic, Z.; Zhang, D.; Yao, M.; Chen, Z.; Dong, Q.; Assinder, S.J.; Richardson, D.R. Dp44mT targets the AKT, TGF-beta and ERK pathways via the metastasis suppressor NDRG1 in normal prostate epithelial cells and prostate cancer cells. Br. J. Cancer 2013, 108, 409–419. [Google Scholar] [CrossRef]
- Krishan, S.; Richardson, D.R.; Sahni, S. The Anticancer Agent, Di-2-Pyridylketone 4,4-Dimethyl-3-Thiosemicarbazone (Dp44mT), Up-Regulates the AMPK-Dependent Energy Homeostasis Pathway in Cancer Cells. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 2916–2933. [Google Scholar] [CrossRef] [PubMed]
- Kovacevic, Z.; Chikhani, S.; Lui, G.Y.L.; Sivagurunathan, S.; Richardson, D.R. The Iron-Regulated Metastasis Suppressor NDRG1 Targets NEDD4L, PTEN, and SMAD4 and Inhibits the PI3K and Ras Signaling Pathways. Antioxid. Redox Signal. 2013, 18, 874–887. [Google Scholar] [CrossRef]
- Shang, C.W.; Zhou, H.Y.; Liu, W.; Shen, T.; Luo, Y.; Huang, S.L. Iron chelation inhibits mTORC1 signaling involving activation of AMPK and REDD1/Bnip3 pathways. Oncogene 2020, 39, 5201–5213. [Google Scholar] [CrossRef]
- Lovejoy, D.B.; Jansson, P.J.; Brunk, U.T.; Wong, J.; Ponka, P.; Richardson, D.R. Antitumor Activity of Metal-Chelating Compound Dp44mT Is Mediated by Formation of a Redox-Active Copper Complex That Accumulates in Lysosomes. Cancer Res. 2011, 71, 5871–5880. [Google Scholar] [CrossRef]
- Lovejoy, D.B.; Sharp, D.M.; Seebacher, N.; Obeidy, P.; Prichard, T.; Stefani, C.; Basha, M.T.; Sharpe, P.C.; Jansson, P.J.; Kalinowski, D.S.; et al. Novel Second-Generation Di-2-Pyridylketone Thiosemicarbazones Show Synergism with Standard Chemotherapeutics and Demonstrate Potent Activity against Lung Cancer Xenografts after Oral and Intravenous Administration in Vivo. J. Med. Chem. 2012, 55, 7230–7244. [Google Scholar] [CrossRef]
- Stacy, A.E.; Palanimuthu, D.; Bernhardt, P.V.; Kalinowski, D.S.; Jansson, P.J.; Richardson, D.R. Structure-Activity Relationships of Di-2-pyridylketone, 2-Benzoylpyridine, and 2-Acetylpyridine Thiosemicarbazones for Overcoming Pgp-Mediated Drug Resistance. J. Med. Chem. 2016, 59, 8601–8620. [Google Scholar] [CrossRef]
- Quach, P.; Gutierrez, E.; Basha, M.T.; Kalinowski, D.S.; Sharpe, P.C.; Lovejoy, D.B.; Bernhardt, P.V.; Jansson, P.J.; Richardson, D.R. Methemoglobin Formation by Triapine, Di-2-pyridylketone-4,4-dimethyl-3-thiosemicarbazone (Dp44mT), and Other Anticancer Thiosemicarbazones: Identification of Novel Thiosemicarbazones and Therapeutics That Prevent This Effect. Mol. Pharmacol. 2012, 82, 105–114. [Google Scholar] [CrossRef]
- Guo, Z.L.; Richardson, D.R.; Kalinowski, D.S.; Kovacevic, Z.; Tan-Un, K.C.; Chan, G.C.F. The novel thiosemicarbazone, di-2-pyridylketone 4-cyclohexyl-4-methyl-3-thiosemicarbazone (DpC), inhibits neuroblastoma growth in vitro and in vivo via multiple mechanisms. J. Hematol. Oncol. 2016, 9, 98. [Google Scholar] [CrossRef] [PubMed]
- Geleta, B.R.; Tout, F.; Lim, S.C.; Sahni, S.; Jansson, P.J.; Apte, M.V.; Richardson, D.; Kovacevic, Z. Targeting Wnt/tenascin C-mediated cross talk between pancreatic cancer cells and stellate cells via activation of the metastasis suppressor NDRG1. J. Biol. Chem. 2022, 298, 101608. [Google Scholar] [CrossRef]
- Wang, T.T.; Fu, Y.; Huang, T.F.; Liu, Y.X.; Wu, M.H.; Yuan, Y.B.; Li, S.S.; Li, C.Z. Copper Ion Attenuated the Antiproliferative Activity of Di-2-pyridylhydrazone Dithiocarbamate Derivative; However, There Was a Lack of Correlation between ROS Generation and Antiproliferative Activity. Molecules 2016, 21, 1088. [Google Scholar] [CrossRef] [PubMed]
- Li, C.P.; Liu, Y.X.; Fu, Y.; Huang, T.F.; Kang, L.X.; Li, C.Z. The antiproliferative activity of di-2-pyridylketone dithiocarbamate is partly attributed to catalase inhibition: Detailing the interaction by spectroscopic methods. Mol. Biosyst. 2017, 13, 1817–1826. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, Y.X.; Guo, R.; Fu, Y.; Zhang, Z.H.; Zhang, P.F.; Zhou, P.X.; Wang, T.T.; Huang, T.F.; Li, X.T.; et al. The novel dithiocarbamate, DpdtC suppresses HER2-overexpressed cancer cells by up-regulating NDRG1 via inactivation of HER2-ERK 1/2 signaling. Sci. Rep. 2018, 8, 3398. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Tian, Z.Y.; Zhao, X.H.; Li, Y.; Duan, S.Y. A novel antitumor dithiocarbamate compound inhibits the EGFR/AKT signaling pathway and induces apoptosis in esophageal cancer cells. Oncol. Lett. 2020, 20, 877–883. [Google Scholar] [CrossRef]
- Kim, J.; Park, A.; Hwang, J.; Zhao, X.; Kwak, J.; Kim, H.W.; Ku, M.; Yang, J.; Kim, T.I.; Jeong, K.-S.; et al. KS10076, a chelator for redox-active metal ions, induces ROS-mediated STAT3 degradation in autophagic cell death and eliminates ALDH1+ stem cells. Cell Rep. 2022, 40, 111077. [Google Scholar] [CrossRef]
- Chaston, T.B.; Lovejoy, D.B.; Watts, R.N.; Richardson, D.R. Examination of the antiproliferative activity of iron chelators: Multiple cellular targets and the different mechanism of action of triapine compared with desferrioxamine and the potent pyridoxal isonicotinoyl hydrazone analogue 311. Clin. Cancer Res. 2003, 9, 402–414. [Google Scholar]
- Knox, J.J.; Hotte, S.J.; Kollmannsberger, C.; Winquist, E.; Fisher, B.; Eisenhauer, E.A. Phase II study of triapinea (R) in patients with metastatic renal cell carcinoma: A trial of the national cancer institute of canada clinical trials group (NCIC IND.161). Investig. New Drugs 2007, 25, 471–477. [Google Scholar] [CrossRef]
- Timoshnikov, V.A.; Kobzeva, T.V.; Polyakov, N.E.; Kontoghiorghes, G.J. Inhibition of Fe2+- and Fe3+- induced hydroxyl radical production by the iron-chelating drug deferiprone. Free Radic. Biol. Med. 2015, 78, 118–122. [Google Scholar] [CrossRef]
- Fiorillo, M.; Toth, F.; Brindisi, M.; Sotgia, F.; Lisanti, M.P. Deferiprone (DFP) Targets Cancer Stem Cell (CSC) Propagation by Inhibiting Mitochondrial Metabolism and Inducing ROS Production. Cells 2020, 9, 1529. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carter, A.; Racey, S.; Veuger, S. The Role of Iron in DNA and Genomic Instability in Cancer, a Target for Iron Chelators That Can Induce ROS. Appl. Sci. 2022, 12, 10161. https://doi.org/10.3390/app121910161
Carter A, Racey S, Veuger S. The Role of Iron in DNA and Genomic Instability in Cancer, a Target for Iron Chelators That Can Induce ROS. Applied Sciences. 2022; 12(19):10161. https://doi.org/10.3390/app121910161
Chicago/Turabian StyleCarter, Andrew, Seth Racey, and Stephany Veuger. 2022. "The Role of Iron in DNA and Genomic Instability in Cancer, a Target for Iron Chelators That Can Induce ROS" Applied Sciences 12, no. 19: 10161. https://doi.org/10.3390/app121910161
APA StyleCarter, A., Racey, S., & Veuger, S. (2022). The Role of Iron in DNA and Genomic Instability in Cancer, a Target for Iron Chelators That Can Induce ROS. Applied Sciences, 12(19), 10161. https://doi.org/10.3390/app121910161




