Comparison and Selection of Conventional PCR Primer Sets for Studies Associated with Nitrogen Cycle Microorganisms in Surface Soil
Abstract
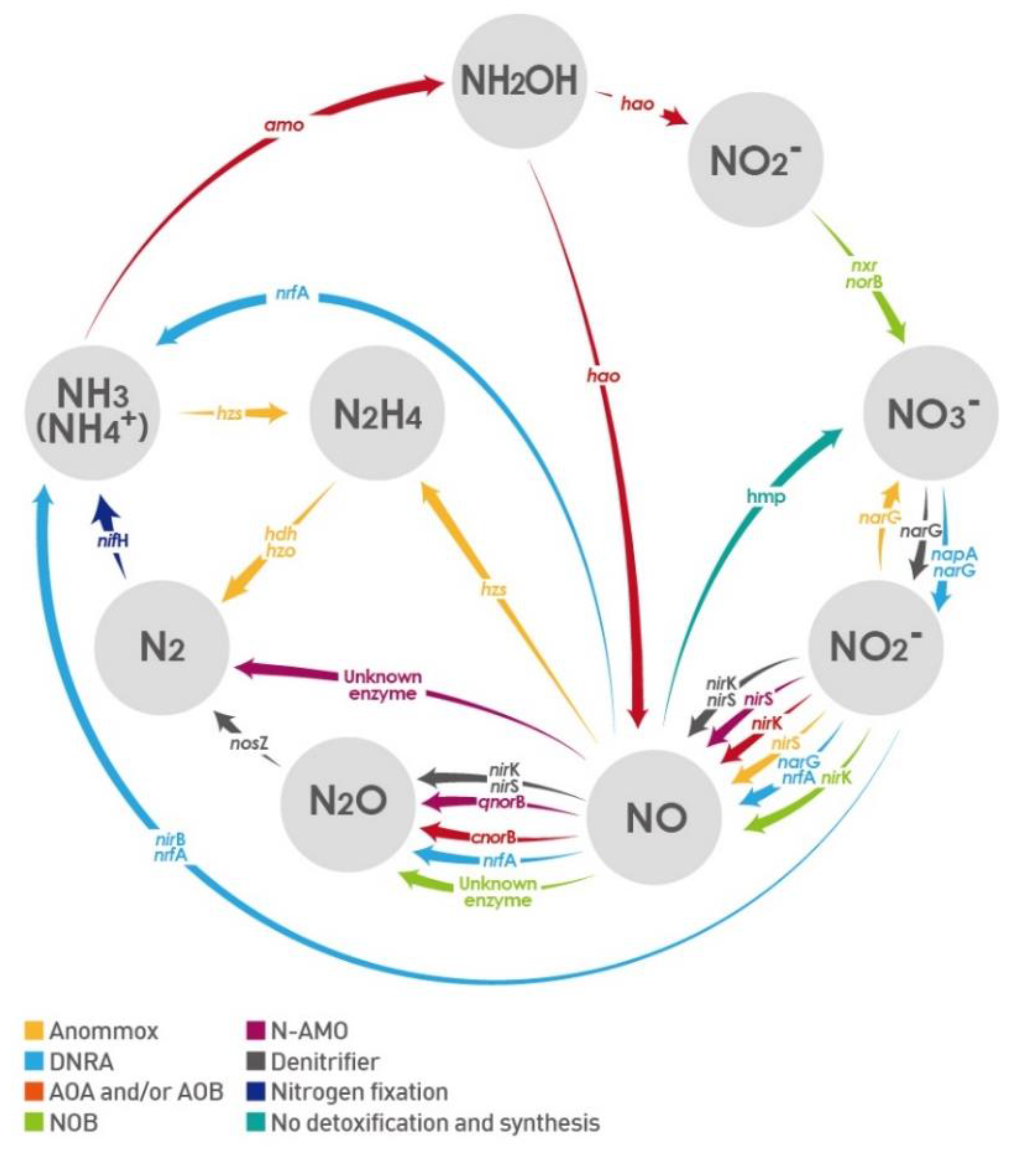
1. Introduction
2. Materials and Methods
2.1. Collection of Positive Controls
2.2. Comparison of PCR Methods for Nitrogen Cycle Microorganism Genes
3. Results
3.1. Securing the Positive Controls
3.2. PCR Method Selection for Each Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hirsch, P.R.; Mauchline, T.H. The importance of the microbial N cycle in soil for crop plant nutrition. Adv. Appl. 2015, 93, 45–71. [Google Scholar]
- Takai, K. The nitrogen cycle: A large, fast, and mystifying cycle. Microbes Environ. 2019, 34, 223–225. [Google Scholar] [CrossRef]
- Stein, L.Y.; Klotz, M.G. The nitrogen cycle. Curr. Biol. 2016, 26, 94–98. [Google Scholar] [CrossRef]
- Zhang, X.; Ward, B.B.; Sigman, D.M. Global nitrogen cycle: Critical enzymes, organisms, and processes for nitrogen budgets and dynamics. Chem. Rev. 2020, 120, 5308–5351. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, L.; Wang, S.; Ye, F.; Zhu, G. Global distribution of anaerobic ammonia oxidation (Anammox) bacteria: Field surveys in wetland, dryland, groundwater aquifer and snow. Front. Microbiol. 2019, 10, 2583. [Google Scholar] [CrossRef]
- Daims, H.; Lebedeva, E.V.; Pjevac, P.; Han, P.; Herbold, C.; Albertsen, M.; Jehmlich, N.; Palatinszky, M.; Vierheilig, J.; Bulaev, A.; et al. Complete nitrification by Nitrospira bacteria. Nature 2015, 528, 504–509. [Google Scholar] [CrossRef]
- Kits, K.D.; Sedlacek, C.J.; Lebedeva, E.V.; Han, P.; Bulaev, A.; Pjevac, P.; Daebeler, A.; Romano, S.; Albertsen, M.; Stein, L.Y.; et al. Kinetic analysis of a complete nitrifier reveals an oligotrophic lifestyle. Nature 2017, 549, 269–272. [Google Scholar] [CrossRef]
- Masuda, Y.; Itoh, H.; Shiratori, Y.; Isobe, K.; Otsuka, S.; Senoo, K. Predominant but previously-overlooked prokaryotic drivers of reductive nitrogen transformation in paddy soils, revealed by metatranscriptomics. Microbes Environ. 2017, 32, 180–183. [Google Scholar] [CrossRef]
- Luo, G.; Wang, T.; Li, K.; Li, L.; Zhang, J.; Guo, S.; Ling, N.; Shen, Q. Historical nitrogen deposition and straw addition facilitate the resistance of soil multifunctionality to drying—Wetting cycles. Appl. Environ. Microbiol. 2019, 85, e02251–18. [Google Scholar] [CrossRef]
- Konneke, M.; Bernhard, A.E.; de la Torre, J.R.; Walker, C.B.; Waterbury, J.B.; Stahl, D.A. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 2005, 437, 543–546. [Google Scholar] [CrossRef]
- Keeley, R.F.; Rodriguez-Gonzalez, L.; Class, U.S.F.G.; Briggs, G.E.; Frazier, V.E.; Mancera, P.A.; Manzer, H.S.; Ergas, S.J.; Scott, K.M. Degenerate PCR primers for assays to track steps of nitrogen metabolism by taxonomically diverse microorganisms in a variety of environments. J. Microbiol. Methods 2020, 175, 105990. [Google Scholar] [CrossRef] [PubMed]
- Kim, H. Comparison of PCR primers for analyzing denitrifying microorganisms in the hyporheic zone. Appl. Sci. 2020, 10, 4172. [Google Scholar] [CrossRef]
- Wang, Y.; Zhu, G.; Harhangi, H.R.; Zhu, B.; Jetten, M.S.; Yin, C.; Op den Camp, H.J. Co-occurrence and distribution of nitrite-dependent anaerobic ammonium and methane-oxidizing bacteria in a paddy soil. FEMS. Microbiol. Lett. 2012, 336, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kwon, J.; Kim, B.Y.; Kim, J.H. Development of an accurate and sensitive diagnostic system based on conventional PCR for detection of African swine fever virus in food waste. Indian J. Microbiol. 2022, 62, 293–306. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Kim, H.; Yang, J.E.; Ryu, H.S.; Moon, J.; Lee, J.Y.; Lee, H.J. Comparison of microbial gene diversity in grassland topsoil depending on soil quality. Appl. Sci. 2021, 11, 9569. [Google Scholar] [CrossRef]
- Bae, K.S.; Lee, S.; Lee, J.Y.; Kim, J.H.; Joo, Y.L.; Lee, S.H.; Chung, H.M.; You, K.A. Development of diagnostic systems for wide range and highly sensitive detection of two waterborne hepatitis viruses from groundwater using the conventional reverse-transcription nested PCR assay. J. Virol. Methods 2022, 299, 114344. [Google Scholar] [CrossRef]
- Ministry of the Environment. Soil Pollution Process Test Standard, and Sampling Distance Adjustment According to Field Conditions; No. 2017-22; National Institute of Environmental Research: Incheon, Korea, 2017.
- Ma, Y.; Zilles, J.L.; Kent, A.D. An evaluation of primers for detecting denitrifiers via their functional genes. Environ. Microbiol. 2019, 21, 1196–1210. [Google Scholar] [CrossRef] [PubMed]
- Alcantara-Hernandez, R.J.; Valenzuela-Encinas, C.; Marsch, R.; Dendooven, L. Respiratory and dissimilatory nitrate-reducing communities from an extreme saline alkaline soil of the former lake Texcoco (Mexico). Extremophiles 2009, 13, 169–178. [Google Scholar] [CrossRef]
- Xu, S.; Feng, S.; Sun, H.; Wu, S.; Zhuang, G.; Deng, Y.; Bai, Z.; Jing, C.; Zhuang, X. Linking N2O emissions from biofertilizer-amended soil of tea plantations to the abundance and structure of N2O-reducing microbial communities. Environ. Sci. Technol. 2018, 52, 11338–11345. [Google Scholar] [CrossRef]
- Achouak, W.; Abrouk, D.; Guyonnet, J.; Barakat, M.; Ortet, P.; Simon, L.; Lerondelle, C.; Heulin, T.; Haichar, F.E.Z. Plant hosts control microbial denitrification activity. FEMS Microbiol. 2019, 95, fiz021. [Google Scholar] [CrossRef]
- Song, B.; Lisa, J.A.; Tobias, C.R. Linking DNRA community structure and activity in a shallow lagoonal estuarine system. Front. Microbiol. 2014, 5, 460. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Ding, L.; Pan, Y.; Hu, H.; Ye, L.; Ren, H. Nitrogen loading effects on nitrification and denitrification with functional gene quantity/transcription analysis in biochar packed reactors at 5 °C. Sci. Rep. 2018, 8, 9844. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kwon, D.; Kim, C.; Lee, S. Analysis of bacterial diversity in water from the Han River water source protection area via a pyrosequencing assay. J. Environ. Health Sci. 2016, 42, 274–279. [Google Scholar] [CrossRef][Green Version]
- Shahi, S.K.; Zarei, K.; Guseva, N.V.; Mangalam, A.K. Microbiota analysis using two-step PCR and next-generation 16S rRNA gene sequencing. J. Vis. Exp. 2019, 152, 59980. [Google Scholar] [CrossRef]
- Blauda, A.; Zaan, B.V.D.; Menona, M.; Laircd, G.J.; Zhanga, D.; Huberc, P.; Schieferc, J.; Blumc, W.E.H.; Kitzlere, B.; Huanga, W.E.; et al. The abundance of nitrogen cycle genes and potential greenhouse gas fluxes depends on land use type and little on soil aggregate size. Appl. Soil Ecol. 2018, 125, 1–11. [Google Scholar] [CrossRef]
- Giraud, M.; Groh, J.; Gerke, H.H.; Bruggemann, N.; Vereecken, H.; Putz, T. Soil nitrogen dynamics in a managed temperate grassland under changed climatic conditions. Water 2021, 13, 931. [Google Scholar] [CrossRef]
- Wang, G.; Luo, Z.; Huang, Y.; Sun, W.; Wei, Y.; Xiao, L.; Deng, X.; Zhu, J.; Li, T.; Zhang, W. Simulating the spatiotemporal variations in aboveground biomass in Inner Mongolian grasslands under environmental changes. Atmos. Chem. Phys. 2021, 21, 3059–3071. [Google Scholar] [CrossRef]
- Gong, H.; Li, Y.; Li, S. Effects of the interaction between biochar and nutrients on soil organic carbon sequestration in soda saline–alkali grassland: A review. Glob. Ecol. 2021, 26, e01449. [Google Scholar] [CrossRef]
- Climate of Korea. Available online: https://www.weather.go.kr/w/obs-climate/climate/korea-climate/korea-char.do# (accessed on 29 September 2022).
- Jung, J.; Yeom, J.; Han, J.; Kim, J.; Park, W. Seasonal changes in nitrogen-cycle gene abundances and in bacterial communities in acidic forest soils. J. Microbiol. 2012, 50, 365–373. [Google Scholar] [CrossRef]
- Nevins, C.J.; Strauss, S.L.; Inglett, P. An overview of key soil nitrogen cycling transformations. EDIS 2020, 2020, 1. [Google Scholar] [CrossRef]
- Walworth, J. Nitrogen in soil and the environment. CALS 2013, 1, AZ1591. [Google Scholar]
- Zehr, J.P.; Kudela, R.M. Nitrogen cycle of the open ocean: From genes to ecosystems. Ann. Rev. Mar. Sci. 2011, 3, 197–225. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, K.; Makaju, S.; Ibrahim, R.; Missaoui, A. Current progress in nitrogen fixing plants and microbiome research. Plants 2020, 9, 97. [Google Scholar] [CrossRef] [PubMed]
- Ni, H.; Liu, C.; Sun, B.; Liang, Y. Response of global farmland soil organic carbon to nitrogen application over time depends on soil type. Geoderma 2022, 406, 115542. [Google Scholar] [CrossRef]
- Colloff, M.; Wakelin, S.; Gomez, D.; Rogers, S.L. Detection of nitrogen cycle genes in soils for measuring the effects of changes in land use and management. Soil Biol. Biochem. 2008, 40, 1637–1645. [Google Scholar] [CrossRef]
- Moring, A.; Hooda, S.; Raghuram, N.; Adhya, T.K.; Ahmad, A.; Bandyopadhyay, S.K.; Barsby, T.; Beig, G.; Bentley, A.R.; Bhatia, A.; et al. Nitrogen challenges and opportunities for agricultural and environmental science in India. Front. Sustain. Food Syst. 2021, 5, 505347. [Google Scholar] [CrossRef]
- Fowler, D.; Coyle, M.; Skiba, U.; Sutton, M.A.; Cape, J.N.; Reis, S.; Sheppard, L.J.; Jenkins, A.; Grizzetti, B.; Galloway, J.N.; et al. The global nitrogen cycle in the twenty-first century. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20130164. [Google Scholar] [CrossRef]
- Langenfeld, N.J.; Kusuma, P.; Wallentine, T.; Criddle, C.S.; Seefeldt, L.C.; Bugbee, B. Optimizing nitrogen fixation and recycling for food production in regenerative life support systems. Front. Astron. Space Sci. 2021, 8, 699688. [Google Scholar] [CrossRef]
| # | Land Use Types | Location (Latitude, Longitude) | Number of Samples | |
|---|---|---|---|---|
| 1 | Field (Fi) | Yeongjuk-ri, Angseong-myeon, Chungju-si, Chungcheongbuk-do, Korea (37°07’59.5” N, 127°47’17.7” E) | 5 samples/location | |
| 2 | Forest (Fo) | Goseongrisan, Cheongpyeong-myeon, Gapyeong-gun, Gyeonggi-do, Korea (37°42′40.3” N, 127°29′33.4” E) | ||
| 3 | Bare land (B) | Goseong-ri, Cheongpyeong-myeon, Gapyeong-gun, Gyeonggi-do, Korea (37°42’46.2” N, 127°30’40.0” E) | ||
| 4 | Grassland | (G1) | Hadam-ri, Geumga-myeon, Chungju-si, Chungcheongbuk-do, Korea (37°03’59.2” N, 127°53’45.0” E) | |
| (G2) | Munho-ri, Seojong-myeon, Yangpyeong-gun, Gyeonggi-do, Korea (37°37’36.9” N, 127°21’51.5” E) | |||
| Total | 5 locations (4 land use types) | 25 samples | ||
| # | Gene | Detailed Gene | Set # | Primer Set Information | Amplicon Size (Nt) | PCR Conditions (Temperature, Time) | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Sequence (5’–3’) | 1st Denaturation | Denaturation | Annealing | Extension | Final Extension | ||||||
| 1 | amo | amoA | amoA (6) | amoA-1F | GGGGTTTCTACTGGTGGT | 491 | 95 °C, 3 min | 95 °C, 45 s | 57 °C, 60 s | 72 °C, 60 s | 72 °C, 5 min | [9] |
| amoA-2R | CCCCTCKGSAAAGCCTTCTTC | (45) | ||||||||||
| amoB | amoB (2) | CrenAmo2.1F | CACGGTGTMCAAGCACA | 465 | 95 °C, 5 min | 95 °C, 45 s | 51 °C, 60 s | 72 °C, 60 s | 72 °C, 5 min | [10] | ||
| CrenAmo2.2R | RATTACYTGCCAVGGTC | (35) | ||||||||||
| amoC | Not selected | |||||||||||
| 2 | norB | cnorB | cnorB (2) | BF | AIGTGGTCGAGAAGTGGCTCTA | 177 | 95 °C, 5 min | 98 °C, 30 s | 57 °C, 30 s | 72 °C, 30 s | 72 °C, 5 min | [11] |
| BR | TCTGIACGGTGAAGATCACC | (40) | ||||||||||
| qnorB | qnorB (2) | 2F | GGNCAYCARGGNTAYGA | 638 | 95 °C, 5 min | 95 °C, 30 s | 1–10 cycles: 57 °C → 52.5 °C 11–40 cycles: 55 °C, 40 s | 72 °C, 30 s | 72 °C, 10 min | [12] | ||
| 7R | GGNGGRTTDATCADGAANCC | (40) | ||||||||||
| 3 | hao | Not selected | ||||||||||
| 4 | hzo | Not selected | ||||||||||
| 5 | hzs | hzs #5 | 396F | ARGGHTGGGGHAGYTGGAAG | 200–300 | 95 °C, 3 min | 95 °C, 30 s | 59 °C, 30 s | 72 °C, 30 s | 72 °C, 5 min | [13] | |
| 742R | GTYCCHACRTCATGVGTCTG | (40) | ||||||||||
| 6 | napA | napA #4 | V67m | AAYATGGCVGARATGCACCC | 484 | 94 °C, 3 min | 94 °C, 30 s | 1–8 cycles: 59 °C -55 °C, 9–38 cycles: 55 °C, 45 s | 72 °C, 45 s | 72 °C, 10 min | [18] | |
| V17m | GRTTRAARCCCATSGTCCA | (38) | ||||||||||
| 7 | narG | narG #1 | F1 | ACICAYGGIGTIAACTGYAC | 524 | 93 °C, 4 min | 93 °C, 60 s | 52 °C, 60 s | 72 °C, 60 s | 72 °C, 7 min | [19] | |
| R1 | TCGSMRTACCAGTCRTARAA | (25) | ||||||||||
| 8 | nifH | nifH #2 | F | AAAGGYGGWATCGGYAARTCCACCAC | 407 | 95 °C, 5 min | 98 °C, 30 s | 57 °C, 30 s | 72 °C, 30 s | 72 °C, 5 min | [11] | |
| Rb | TGSGCYTTGTCYTCRCGGATBGGCAT | (40) | ||||||||||
| 9 | nirB | Not selected | ||||||||||
| 10 | nirK | nirK #3 | F1aCu | ATCATGGTSCTGCCGCG | 473 | 94 °C, 3 min | 94 °C, 30 s | 57 °C, 60 s | 73 °C, 60 s | 75 °C, 10 min | [20] | |
| R3Cu | TTGGTGTTRGACTAGCTCCG | (35) | ||||||||||
| 11 | nirS | nirS #2 | nirS4QF | AACGYSAAGGARACSGG | 425 | 95 ℃, 10 min | 95 °C, 15 s | 1–6 cycles: 63 °C →58 °C, 7–41 cycles: 58 °C, 30 s | 72 °C, 30 s | 72 °C, 10 min | [21] | |
| nirS8QR | GASTTCGGRTGSGTCTTSAYGAA | (41) | ||||||||||
| 12 | nosZ | nosZ #1 | nosZ1840F | CGCRACGGCAASAAGGTSMSSGT | 267 | 95 °C, 10 min | 95 °C, 15 s | 1–6 cycles: 65 °C→60 °C, 7–41 cycles: 60 °C, 60 s | 72 °C, 30 s | 72 °C, 10 min | [12] | |
| nosZ2090R | CAKRTGCAKSGCRTGGCAGAA | (41) | ||||||||||
| 13 | nrfA | nrfA #5 | F2aw | CARTGYCAYGTBGARTA | 236 | 94 °C, 5 min | 94 °C, 30 s | 52 °C, 45 s | 72 °C, 20 s | 72 °C, 5 min | [22] | |
| R1 | TWNGGCATRTGRCARTC | (40) | ||||||||||
| 14 | nxr | nxrA | nxrA #1 | F | CAGACCGACGTGTGCGAAAG | 322 | 95 °C, 10 min | 95 °C, 15 s | 58 °C, 30 s | 72 °C, 30 s | 72 °C, 5 min | [23] |
| R | TCYACAAGGAACGGAAGGTC | (35) | ||||||||||
| nxrB | Not selected | |||||||||||
| # | Gene | Detail Gene | Field (Fi) | Forest (Fo) | Bareland (B) | Grassland | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grassland 1 (G1) | Grassland 2 (G2) | ||||||||||||||||||||||||||
| Sp | Su | R | A | W | Sp | Su | R | A | W | Sp | Su | R | A | W | Sp | Su | R | A | W | Sp | Su | R | A | W | |||
| 1 | amo | amoA | - | ● | - | - | - | - | - | - | ● | - | - | - | - | ● | - | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● |
| amoB | - | - | - | - | - | - | - | - | - | - | - | - | - | ● | - | ● | ● | ● | ● | ● | - | ● | - | - | ● | ||
| amoC | Not determind | ||||||||||||||||||||||||||
| 2 | norB | cnorB | - | ● | ● | - | - | - | - | ● | ● | - | - | - | ● | ● | - | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● |
| qnorB | - | - | - | - | - | - | - | - | - | - | - | - | - | ● | - | - | ● | ● | ● | - | ● | ● | ● | - | ● | ||
| 3 | hao | Not determind | |||||||||||||||||||||||||
| 4 | hzo | Not determind | |||||||||||||||||||||||||
| 5 | hzs | - | - | - | - | - | - | - | - | ● | - | - | - | - | ● | - | - | - | - | - | - | ● | ● | ● | ● | - | |
| 6 | napA | - | ● | - | - | - | - | - | - | ● | ● | - | - | - | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | |
| 7 | narG | - | - | - | - | - | - | - | - | ● | ● | - | - | - | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ○ | |
| 8 | nifH | - | - | ● | - | - | - | - | - | ● | - | - | - | - | ● | - | - | ● | ● | ● | - | ● | ● | ● | ● | - | |
| 9 | nirB | Not determind | |||||||||||||||||||||||||
| 10 | nirK | - | ● | ● | - | - | - | - | ● | - | - | - | ● | ● | - | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ||
| 11 | nirS | - | - | - | - | - | - | - | - | - | - | - | - | - | ● | - | ● | ● | ● | ● | - | ● | ● | ● | ● | - | |
| 12 | nosZ | - | ● | - | - | - | - | - | - | ● | - | - | - | - | - | ● | ● | ● | ● | ● | - | ● | ● | ● | ● | - | |
| 13 | nrfA | - | - | - | - | - | - | - | - | ● | - | - | - | - | ● | - | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | |
| 14 | nxr | nxrA | - | ● | ● | - | - | - | - | ● | ● | - | - | - | ● | ● | - | ● | - | - | - | - | - | ● | - | ● | ● |
| nxrB | Not determind | ||||||||||||||||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Jung, Y.-J.; Moon, J.; Lee, J.-Y.; Kim, H.; Yang, J.-E.; Lee, H.; Jung, J.; Kim, H.-R. Comparison and Selection of Conventional PCR Primer Sets for Studies Associated with Nitrogen Cycle Microorganisms in Surface Soil. Appl. Sci. 2022, 12, 10314. https://doi.org/10.3390/app122010314
Lee S, Jung Y-J, Moon J, Lee J-Y, Kim H, Yang J-E, Lee H, Jung J, Kim H-R. Comparison and Selection of Conventional PCR Primer Sets for Studies Associated with Nitrogen Cycle Microorganisms in Surface Soil. Applied Sciences. 2022; 12(20):10314. https://doi.org/10.3390/app122010314
Chicago/Turabian StyleLee, Siwon, Yong-Ju Jung, Jinah Moon, Jin-Young Lee, Heejung Kim, Jae-E Yang, Hyunji Lee, Jaewon Jung, and Ha-Rang Kim. 2022. "Comparison and Selection of Conventional PCR Primer Sets for Studies Associated with Nitrogen Cycle Microorganisms in Surface Soil" Applied Sciences 12, no. 20: 10314. https://doi.org/10.3390/app122010314
APA StyleLee, S., Jung, Y.-J., Moon, J., Lee, J.-Y., Kim, H., Yang, J.-E., Lee, H., Jung, J., & Kim, H.-R. (2022). Comparison and Selection of Conventional PCR Primer Sets for Studies Associated with Nitrogen Cycle Microorganisms in Surface Soil. Applied Sciences, 12(20), 10314. https://doi.org/10.3390/app122010314








