Region-of-Interest Optimization for Deep-Learning-Based Breast Cancer Detection in Mammograms
Abstract
:1. Introduction
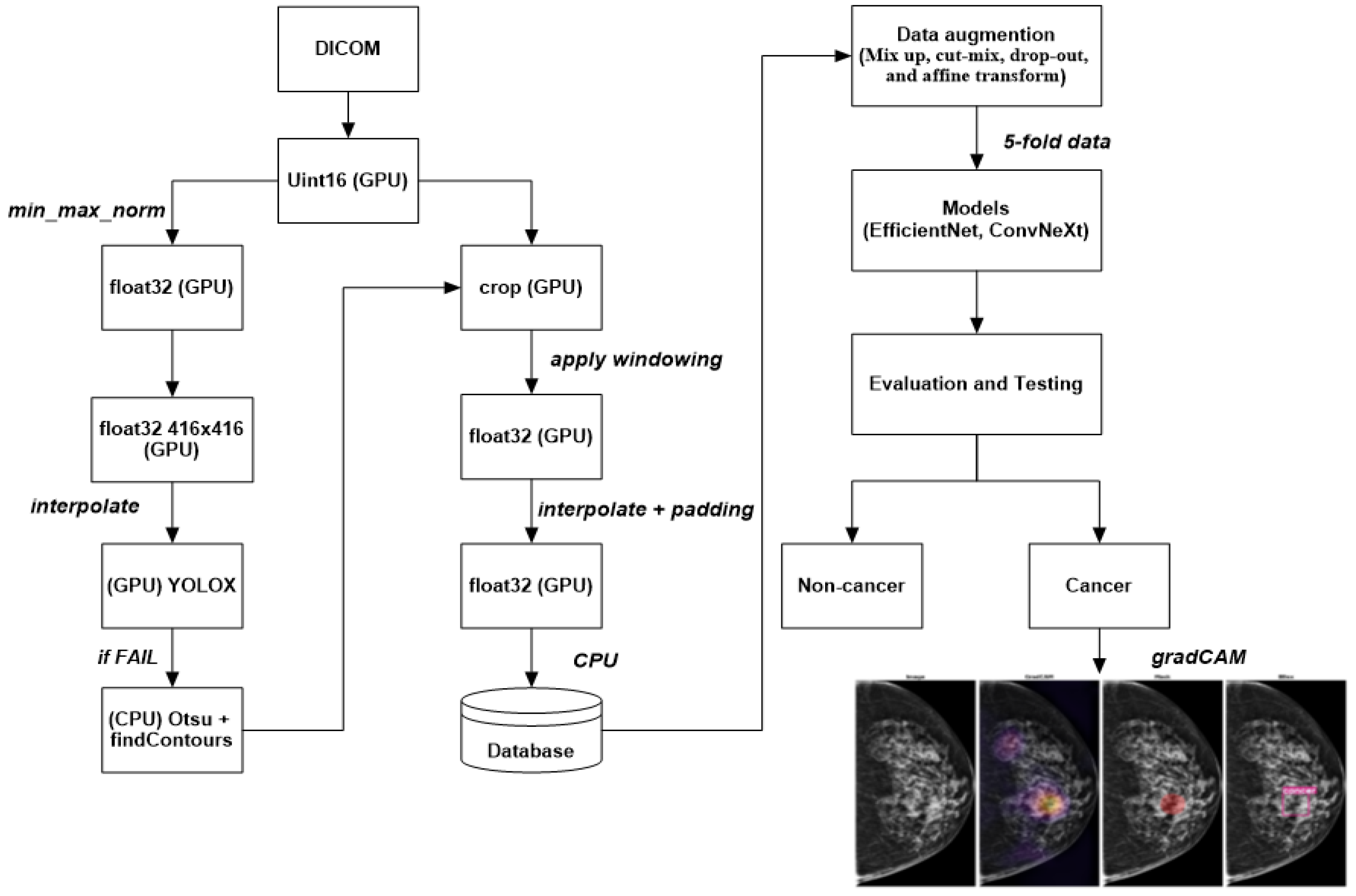
- A novel deep-learning approach for detecting breast cancer using mammography images is proposed in this paper. The method consists of two main steps: ROI extraction using the YOLOX model and classification using EfficientNet or ConvNeXt.
- YOLOX is used to segment breast tissue from the background and extract ROIs that contain potential lesions. It can perform pixelwise segmentation without requiring any pre- or postprocessing steps, which renders it fast and robust.
- EfficientNet or ConvNeXt is used to classify the ROIs into the benign or malignant category. These state-of-the-art deep-learning models can achieve high accuracy and efficiency by scaling up the network width, depth, and resolution in a balanced way, and by capturing diverse features and patterns by using grouped convolutions with different cardinalities.
- Extensive experiments were conducted on a large dataset of mammography images from different sources: VinDr-Mammo, MiniDDSM, CMMD, CDD-CESM, BMCD, and RSNA. The approach is compared with several baseline methods. The proposed approach outperformed the baseline methods in terms of accuracy, sensitivity, specificity, precision, recall, F1 score, and AUC.
- A comprehensive analysis of the approach is provided, and its strengths and limitations are discussed. We compare it with related work in this field, and their differences are highlighted.
2. Materials and Methods
2.1. Datasets
- VinDr-Mammo [12]: A large-scale benchmark dataset for computer-aided diagnosis in full-field digital mammography (FFDM) that consists of 5000 four-view exams with breast-level assessment and finding annotations following the Breast Imaging Report and Data System (BI-RADS). Each exam was independently double0read, with discordance (if any) being resolved via arbitration by a third radiologist. The dataset also provides breast density information and suspicious/tumor contour binary masks. The dataset was collected from VinDr Hospital in Vietnam.
- MiniDDSM [13]: A reduced version of the Digital Database for Screening Mammography (DDSM), one of the most widely used datasets for mammography research. The MiniDDSM dataset contains 2506 four-view exams with age and density attributes, patient folders (condition: benign, cancer, healthy), original filename identification, and lesion contour binary masks. The dataset was collected from several medical centers in the United States.
- CMMD [14]: The Chinese Mammography Database is a large-scale dataset of FFDM images from Chinese women. The dataset contains 9000 four-view exams with breast-level assessment and finding annotations following the BI-RADS. The dataset also provides age and density information. The dataset was collected from several hospitals in China.
- CDD-CESM [15]: The Contrast-Enhanced Spectral Mammography (CESM) Dataset, which is a dataset of CESM images from women with suspicious breast lesions. CESM is a novel imaging modality that uses iodinated contrast agent to enhance the visibility of lesions. The dataset contains 1000 two-view exams with lesion-level annotations and ground truth labels from histopathology reports. The dataset was collected from several hospitals in Spain.
- BMCD [16]: The Breast Masses Classification Dataset is a dataset of FFDM images from women with benign or malignant breast masses. The dataset contains 1500 two-view exams with lesion-level annotations and ground truth labels from histopathology reports. The dataset was collected from several hospitals in Turkey.
- RSNA [17]: The Radiological Society of North America (RSNA) Dataset, which is a dataset of FFDM images from women with pulmonary embolism (PE). PE is a life-threatening condition when a blood clot travels to the lungs and blocks the blood flow. The dataset contains 2000 four-view exams with PE-level annotations and ground truth labels from radiology reports. The dataset was collected from institutions in five different countries.
2.2. Models
2.3. Preprocessing Image Data
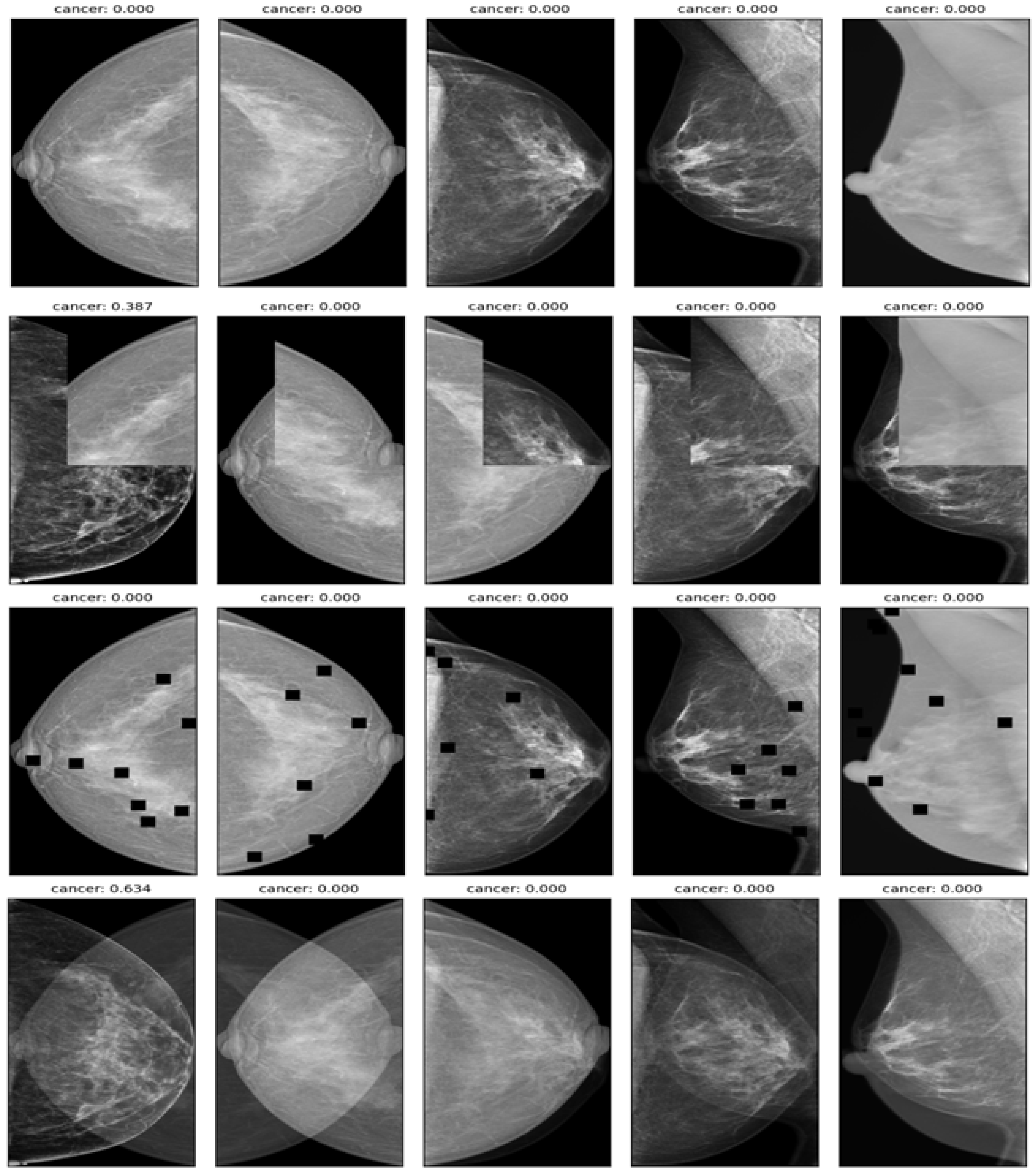
- Mix up: A technique that generates new training samples by linearly interpolating between two images and their labels. This technique can produce high-quality inter-class examples that prevent the model from memorizing the training distribution and improve its generalization ability.
- Cut-mix: A technique that generates new training samples by randomly cutting out patches from two images, pasting them together, and assigning the labels according to the area ratio of the patches. This technique can also produce interclass examples that enhance the model’s robustness to occlusion and localization errors.
- Drop-out: A technique randomly drops out units in a neural network layer during training to prevent overfitting. This technique can decrease the co-adaptation of features and increase the diversity of feature representations.
- Affine transform: A technique that applies geometric transformations such as scaling, rotation, translation, and shearing to the images. This technique can increase the invariance of the model to geometric variations and improve its performance on unseen images.
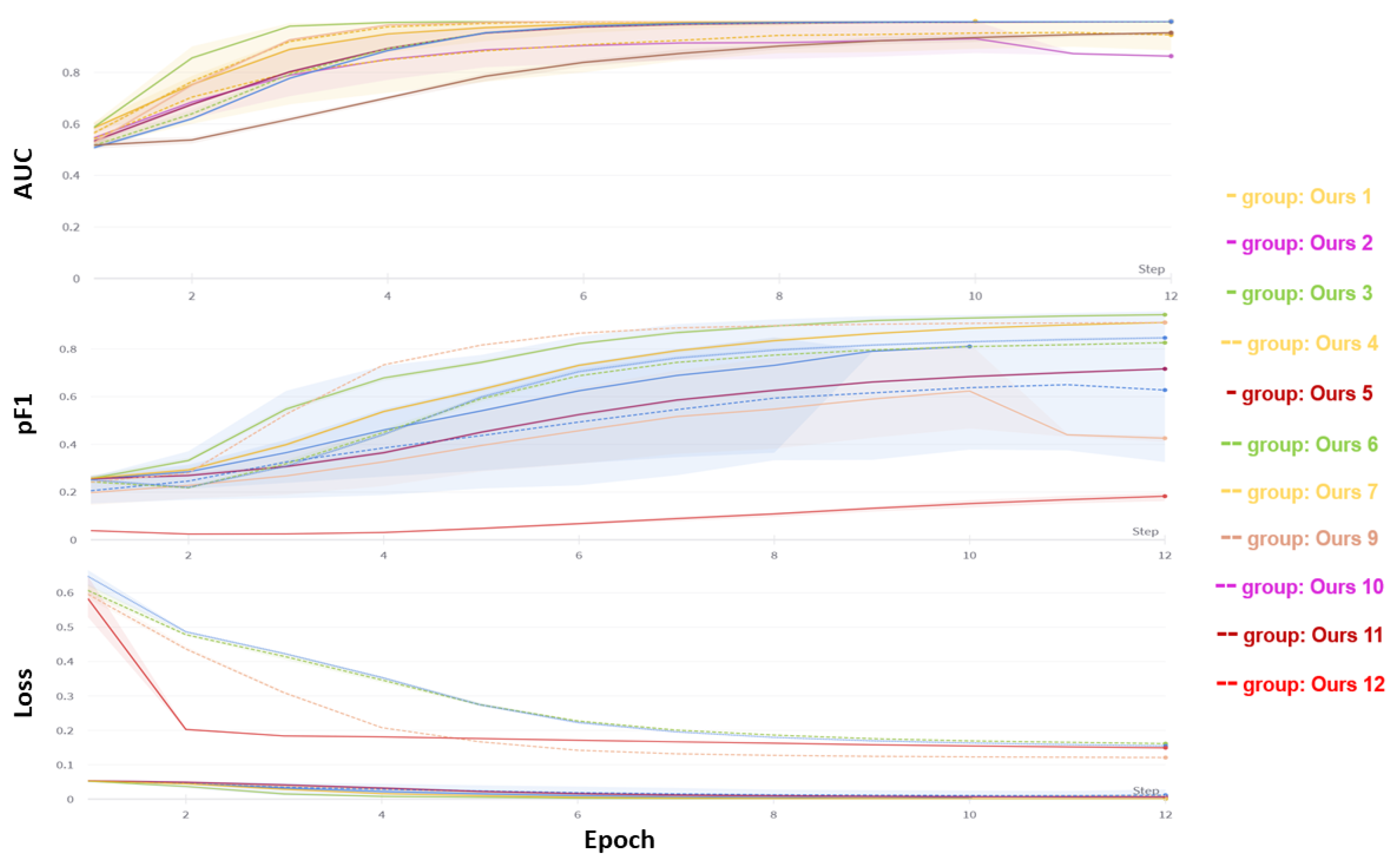
2.4. Metrics
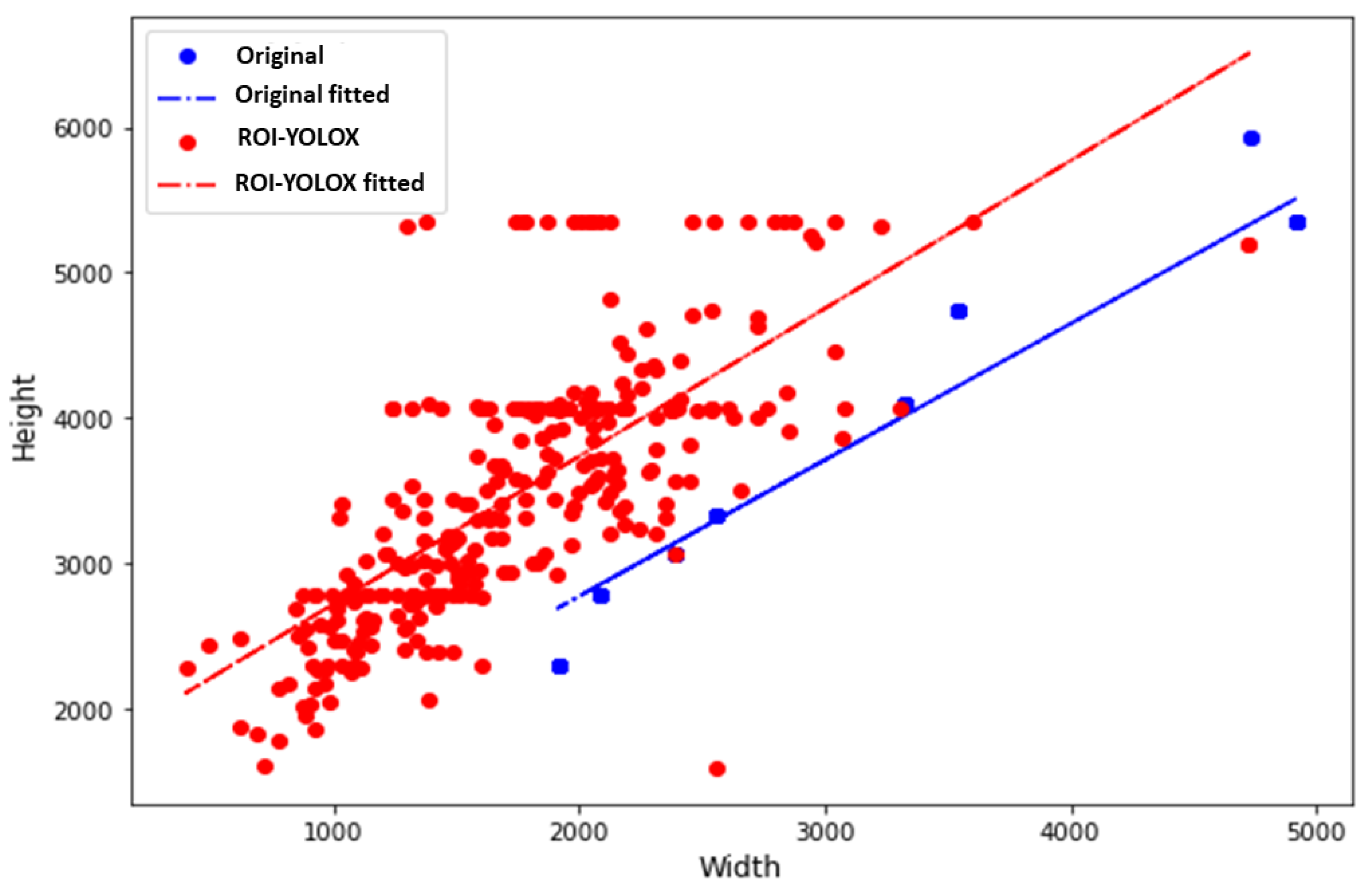
- Average precision (AP) is a performance metric that provides a summary of the precision-recall curve. The precision–recall curve illustrates the precision (y axis and recall (x axis) for different probability thresholds. Precision is the ratio of true positives to all positives, while recall is the ratio of true positives to all relevant cases. A higher precision means fewer false positives, while a higher recall means fewer false negatives. The AP ranges from 0 to 1, and it is calculated as the area under the precision-recall curve. A higher AP indicates better performance of the model. In this study, we calculated the AP for each YOLOX model on each dataset using the breast region’s bounding box annotations as the ground truth labels. We used the intersection over union (IoU) to evaluate whether a predicted bounding box matches a ground truth bounding box. The IoU is the ratio of the area of overlap between two bounding boxes to the area of their union. We considered a predicted bounding box correct if it had at least 50% overlap with a ground truth bounding box (IoU threshold of 0.5). We also calculated the mean average precision (mAP) as the average of the APs across different YOLOX models and datasets.
- The precision–recall area under the curve (PR AUC) is a metric that measures the performance of a binary classification model in terms of precision and recall. Precision is the ratio of true positives to the sum of true positives and false positives, while recall is the ratio of true positives to the sum of true positives and false negatives. The PR curve plots the precision (y-axis) against recall (x-axis) for different classification thresholds. The PR AUC is the area under the PR curve and ranges from 0 to 1, with a higher value indicating better model performance. This metric is particularly useful when dealing with imbalanced datasets, where positive cases are much fewer than negative cases, as it focuses on the ability of the model to identify true positives among all predicted positives.
- ROC AUC is the area under the receiver operating characteristic curve. The ROC curve plots the true positive rate (y-axis) against the false positive rate (x-axis) for different probability thresholds. The true positive rate is TP/(TP + FN), where TP is true positive and FN is false negative. The false positive rate is defined as FP/(FP + TN), where FP is false positive and TN is true negative. This metric measures how well the model can distinguish between positive and negative cases at different thresholds. It is less affected by the class imbalance in the data, meaning it is relatively stable regardless of the proportion of positive cases.
- Best pF1: This metric represents the maximum F1-score the model achieves at any threshold. The F1-score is the harmonic mean of precision and recall, defined as 2 ∗ (precision ∗ recall)/(precision + recall). The F1 score balances the two aspects of the classification task. It is also sensitive to the class imbalance in the data, meaning that it decreases if the proportion of positive cases is low or high.
- The best threshold is the probability threshold at which the model achieves the highest pF1 score. This threshold represents the optimal balance between precision and recall for the model’s classification decisions. Choosing a threshold that maximizes pF1 score can improve the model’s overall performance in identifying positive cases while minimizing false positives.
3. Experiment Results
3.1. ROI Method with YOLOX Model
3.2. Classification
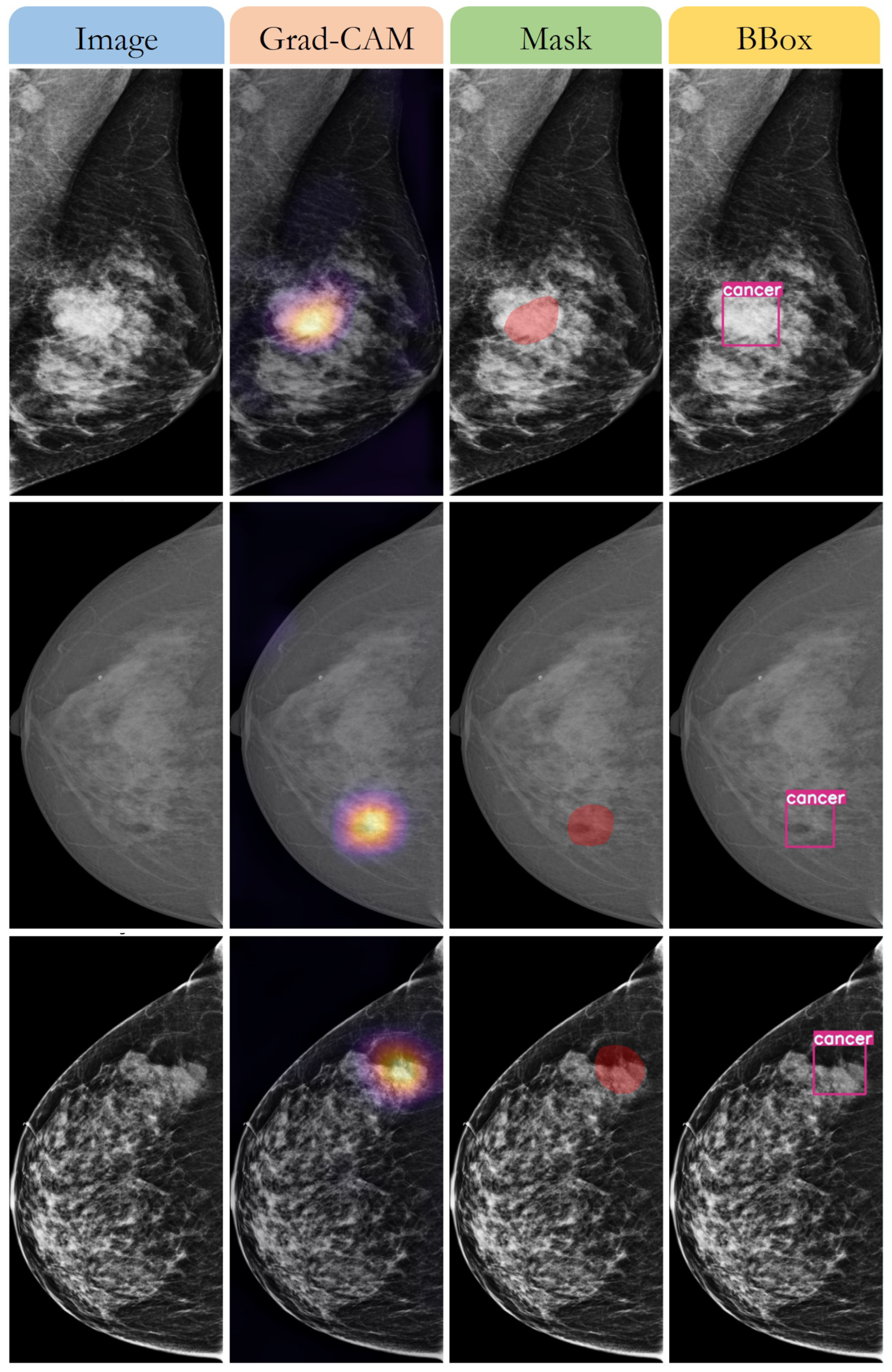
3.3. Detecting the Breast Cancer Area
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Breast Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/breast-cancer (accessed on 7 April 2023).
- Elmore, J.G.; Wells, C.K.; Lee, C.H.; Howard, D.H.; Feinstein, A.R. Variability in radiologists’ interpretations of mammograms. N. Engl. J. Med. 1994, 331, 1493–1499. [Google Scholar] [CrossRef] [PubMed]
- Welch, H.G.; Prorok, P.C.; O’Malley, A.J.; Kramer, B.S. Breast-cancer tumor size, overdiagnosis, and mammography screening effectiveness. N. Engl. J. Med. 2016, 375, 1438–1447. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef] [PubMed]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA; London, UK, 2016. [Google Scholar]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A survey on deep learning in medical image analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhungel, N.; Carneiro, G.; Bradley, A.P. Deep learning and structured prediction for the segmentation of mass in mammograms. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 605–612. [Google Scholar]
- Kooi, T.; Litjens, G.; van Ginneken, B.; Gubern-Mérida, A.; Sánchez, C.I.; Mann, R. Large scale deep learning for computer aided detection of mammographic lesions. Med. Image Anal. 2017, 35, 303–312. [Google Scholar] [CrossRef]
- Zhu, W.; Xie, X. Adversarial deep structural networks for mammographic mass segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; pp. 101–109. [Google Scholar]
- Ribli, D.; Horváth, A.; Unger, Z.; Pollner, P.; Csabai, I. Detecting and classifying lesions in mammograms with deep learning. Sci. Rep. 2018, 8, 4165. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Khosla, A.; Gargeya, R.; Irshad, H.; Beck, A.H. Deep learning for identifying metastatic breast cancer. arXiv 2016, arXiv:1606.05718. [Google Scholar]
- Nguyen, H.T.; Nguyen, H.Q.; Pham, H.H.; Lam, K.; Le, L.T.; Dao, M.; Vu, V. VinDr-Mammo: A large-scale benchmark dataset for computer-aided diagnosis in full-field digital mammography. Sci. Data 2023, 10, 277. [Google Scholar] [CrossRef]
- Looney, P.; Chen, J.; Giger, M.L. A mini-digital database for screening mammography: Mini-DDSM. J. Med. Imaging 2017, 4, 034501. [Google Scholar]
- Cui, C.; Li, L.; Cai, H.; Fan, Z.; Zhang, L.; Dan, T.; Li, J.; Wang, J. The Chinese Mammography Database (CMMD): An Online Mammography Database with Biopsy Confirmed Types for Machine Diagnosis of Breast; The Cancer Imaging Archive: Bethesda, MD, USA, 2021. [Google Scholar]
- Khaled, R.; Helal, M.; Alfarghaly, O.; Mokhtar, O.; Elkorany, A.; El Kassas, H.; Fahmy, A. Categorized contrast enhanced mammography dataset for diagnostic and artificial intelligence research. Sci. Data 2022, 9, 122. [Google Scholar] [CrossRef]
- Demir, Ö.; Güler, İ.N. Breast masses classification in mammograms using deep convolutional neural networks and transfer learning. Biomed. Signal Process. Control 2019, 53, 101567. [Google Scholar]
- Carr, C.; Kitamura, F.; Kalpathy-Cramer, J.; Mongan, J.; Andriole, K.; Vazirabad, M.; Riopel, M.; Ball, R.; Dane, S. RSNA Screening Mammography Breast Cancer Detection. 2022. Available online: https://kaggle.com/competitions/rsna-breast-cancer-detection (accessed on 27 February 2023).
- Tan, M.; Le, Q.V. EfficientNet: Rethinking model scaling for convolutional neural networks. arXiv 2019, arXiv:1905.11946. [Google Scholar]
- Liu, Z.; Mao, H.; Wu, C.-Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. arXiv 2022, arXiv:2201.03545. [Google Scholar]
- Zou, X.; Wu, Z.; Zhou, W.; Huang, J. YOLOX-PAI: An Improved YOLOX, Stronger and Faster than YOLOv6. arXiv 2022, arXiv:2208.13040. [Google Scholar]
- Wang, C.-Y.; Liao, H.-Y.M.; Yeh, I.-H.; Wu, Y.-H.; Chen, P.-Y.; Hsieh, J.-W. CSPNet: A New Backbone that can Enhance Learning Capability of CNN. arXiv 2020, arXiv:1911.11929. [Google Scholar]
- Li, J.; Wang, Y.; Liang, X.; Zhang, L. SFPN: Synthetic FPN for Object Detection. arXiv 2021, arXiv:2104.05746. [Google Scholar]
- Otsu, N. A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man Cybern. 1979, 9, 62–66. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Li, Y.; Sakaridis, C.; Dai, D.; Van Gool, L. ContourNet: Taking a further step toward accurate arbitrary-shaped scene text detection. In Proceedings of the 2019 IEEE/CVF International Conference on Computer Vision (ICCV), Seoul, Republic of Korea, 27 October–2 November 2019; pp. 9719–9728. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-CAM: Visual explanations from deep networks via gradient-based localization. Int. J. Comput. Vis. 2019, 128, 336–359. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Anguelov, D.; Erhan, D.; Szegedy, C.; Reed, S.; Fu, C.-Y.; Berg, A.C. SSD: Single Shot MultiBox Detector. arXiv 2016, arXiv:1512.02325. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. arXiv 2015, arXiv:1506.01497. [Google Scholar] [CrossRef] [Green Version]
- Dai, J.; Li, Y.; He, K.; Sun, J. R-FCN: Object Detection via Region-based Fully Convolutional Networks. arXiv 2016, arXiv:1605.06409. [Google Scholar]
- Singh, M.; Singh, T.; Soni, S. Pre-operative assessment of ablation margins for variable blood perfusion metrics in a magnetic resonance imaging-based complex breast tumor anatomy: Simulation paradigms in thermal therapies. Comput. Methods Programs Biomed. 2021, 198, 105781. [Google Scholar] [CrossRef] [PubMed]


| Data | Source | Country | Number of Exams | Number of Images | Number of Benign Cases | Number of Malignant Cases |
|---|---|---|---|---|---|---|
| VinDr-Mammo | VinDr Hospital | Vietnam | 5000 | 20,000 | 3500 | 1500 |
| MiniDDSM | DDSM | USA | 2506 | 10,024 | 1506 | 1000 |
| CMMD | Various hospitals | China | 9000 | 36,000 | 6000 | 3000 |
| CDD-CESM | Various hospitals | Spain | 1000 | 2000 | 500 | 500 |
| BMCD | Various hospitals | Turkey | 1500 | 3000 | 750 | 750 |
| RSNA | Various institutions | Multiple countries | 2000 | 8000 | - | - |
| Total | - | - | 21,006 | 79,024 | 12,256 (61.4%) | 6750 (33.9%) |
| Model Size | Image Size | Interpolation | AP New Validation (%) | AP Remake Validation (%) |
|---|---|---|---|---|
| Nano 1 | 416 | LINEAR | 96.26 | 94.21 |
| Nano 2 | 416 | AREA | 94.09 | 91.60 |
| Nano 3 | 640 | LINEAR | 95.85 | 88.40 |
| Nano 4 | 768 | LINEAR | 96.22 | 82.09 |
| Nano 5 | 1024 | LINEAR | 94.92 | 89.40 |
| Tiny 1 | 416 | LINEAR | 94.23 | 90.20 |
| Tiny 2 | 640 | LINEAR | 94.95 | 89.84 |
| Tiny 3 | 768 | AREA | 96.21 | 68.03 |
| Tiny 4 | 1024 | AREA | 93.69 | 73.70 |
| S 1 | 416 | LINEAR | 95.03 | 86.34 |
| S 2 | 640 | LINEAR | 96.10 | 70.80 |
| S 3 | 768 | LINEAR | 96.79 | 78.70 |
| Metric | Size | Precision | Recall | F1 Score |
|---|---|---|---|---|
| Negative | 12,256 | 0.92 | 0.93 | 0.97 |
| Positive | 6750 | 0.91 | 0.92 | 0.85 |
| Weighted Average | 17,514 | 0.92 | 0.92 | 0.97 |
| Method | Model | Dataset | Accuracy | Sensitivity | Specificity | F1-Score | ROC AUC | PR AUC |
|---|---|---|---|---|---|---|---|---|
| Original | EFN7 | VinDr-Mammo | 0.86 | 0.83 | 0.88 | 0.81 | 0.92 | 0.90 |
| Fs-ROI | EFN7 | VinDr-Mammo | 0.87 | 0.85 | 0.89 | 0.83 | 0.93 | 0.91 |
| Prediction | EFN7 | VinDr-Mammo | 0.90 | 0.88 | 0.92 | 0.86 | 0.96 | 0.94 |
| Original | CNX1 | VinDr-Mammo | 0.85 | 0.82 | 0.87 | 0.80 | 0.91 | 0.89 |
| Fs-ROI | CNX1 | VinDr Mammo | 0.87 | 0.84 | 0.89 | 0.82 | 0.93 | 0.90 |
| Prediction | CNX1 | VinDr-Mammo | 0.89 | 0.87 | 0.91 | 0.85 | 0.95 | 0.93 |
| Original | EFN7 | MiniDDSM | 0.84 | 0.81 | 0.86 | 0.80 | 0.90 | 0.88 |
| Fs-ROI | EFN7 | MiniDDSM | 0.85 | 0.83 | 0.87 | 0.81 | 0.91 | 0.89 |
| Prediction | EFN7 | MiniDDSM | 0.88 | 0.86 | 0.90 | 0.84 | 0.94 | 0.92 |
| Original | CNX1 | MiniDDSM | 0.83 | 0.80 | 0.85 | 0.79 | 0.89 | 0.87 |
| Fs-ROI | CNX1 | MiniDDSM | 0.84 | 0.82 | 0.86 | 0.80 | 0.90 | 0.88 |
| Prediction | CNX1 | MiniDDSM | 0.87 | 0.85 | 0.89 | 0.83 | 0.93 | 0.91 |
| Original | EFN7 | CMMD | 0.87 | 0.84 | 0.89 | 0.83 | 0.90 | 0.89 |
| Prediction | EFN7 | CMMD | 0.91 | 0.89 | 0.93 | 0.88 | 0.97 | 0.96 |
| Original | CNX1 | CMMD | 0.86 | 0.83 | 0.88 | 0.82 | 0.92 | 0.90 |
| Fs-ROI | CNX1 | CMMD | 0.87 | 0.85 | 0.89 | 0.83 | 0.93 | 0.91 |
| Prediction | CNX1 | CMMD | 0.92 | 0.90 | 0.94 | 0.89 | 0.98 | 0.97 |
| Original | EFN7 | CDD-CESM | 0.87 | 0.84 | 0.89 | 0.83 | 0.93 | 0.91 |
| Fs-ROI | EFN7 | CDD-CESM | 0.88 | 0.86 | 0.90 | 0.84 | 0.94 | 0.92 |
| Prediction | EFN7 | CDD-CESM | 0.92 | 0.90 | 0.94 | 0.89 | 0.98 | 0.97 |
| Original | CNX1 | CDD-CESM | 0.86 | 0.83 | 0.88 | 0.82 | 0.92 | 0.90 |
| Fs-ROI | CNX1 | CDD-CESM | 0.87 | 0.85 | 0.89 | 0.83 | 0.93 | 0.91 |
| Prediction | CNX1 | CDD-CESM | 0.92 | 0.90 | 0.94 | 0.89 | 0.98 | 0.97 |
| Original | EFN7 | BMCD | 0.87 | 0.84 | 0.89 | 0.83 | 0.93 | 0.91 |
| Fs-ROI | CNX1 | BMCD | 0.88 | 0.86 | 0.90 | 0.84 | 0.94 | 0.92 |
| Prediction | EFN7 | BMCD | 0.92 | 0.90 | 0.94 | 0.89 | 0.98 | 0.97 |
| Original | CNX1 | BMCD | 0.86 | 0.83 | 0.88 | 0.82 | 0.92 | 0.90 |
| Fs-ROI | CNX1 | BMCD | 0.87 | 0.85 | 0.89 | 0.83 | 0.93 | 0.91 |
| Prediction | CNX1 | BMCD | 0.92 | 0.90 | 0.94 | 0.89 | 0.98 | 0.97 |
| Original | EFN7 | RSNA | 0.86 | 0.83 | 0.88 | 0.82 | 0.91 | 0.89 |
| Fs-ROI | EFN7 | RSNA | 0.87 | 0.85 | 0.89 | 0.83 | 0.92 | 0.90 |
| Prediction | EFN7 | RSNA | 0.87 | 0.85 | 0.89 | 0.83 | 0.92 | 0.90 |
| Original | CNX1 | RSNA | 0.85 | 0.82 | 0.87 | 0.81 | 0.90 | 0.88 |
| Fs-ROI | CNX1 | RSNA | 0.86 | 0.84 | 0.88 | 0.82 | 0.91 | 0.89 |
| Prediction | CNX1 | RSNA | 0.86 | 0.84 | 0.88 | 0.82 | 0.91 | 0.89 |
| Method | AP (Benign) | AP (Malignant) | Best PF1 (Benign) | Best PF1 (Malignant) | Best Threshold (Benign) | Best Threshold (Malignant) |
|---|---|---|---|---|---|---|
| ROI-SSD [26] | 0.77 | 0.82 | 0.75 | 0.77 | 0.55 | 0.55 |
| ROI-RPN [27] | 0.75 | 0.80 | 0.73 | 0.75 | 0.54 | 0.54 |
| ROI-RFCN [28] | 0.73 | 0.78 | 0.71 | 0.73 | 0.52 | 0.52 |
| Ours | 0.81 | 0.86 | 0.79 | 0.81 | 0.56 | 0.56 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huynh, H.N.; Tran, A.T.; Tran, T.N. Region-of-Interest Optimization for Deep-Learning-Based Breast Cancer Detection in Mammograms. Appl. Sci. 2023, 13, 6894. https://doi.org/10.3390/app13126894
Huynh HN, Tran AT, Tran TN. Region-of-Interest Optimization for Deep-Learning-Based Breast Cancer Detection in Mammograms. Applied Sciences. 2023; 13(12):6894. https://doi.org/10.3390/app13126894
Chicago/Turabian StyleHuynh, Hoang Nhut, Anh Tu Tran, and Trung Nghia Tran. 2023. "Region-of-Interest Optimization for Deep-Learning-Based Breast Cancer Detection in Mammograms" Applied Sciences 13, no. 12: 6894. https://doi.org/10.3390/app13126894







